Abstract
Previously, we reported that the transcription factor Nanog, which maintains the self-renewal of embryonic stem cells (ESCs), promotes the osteogenic differentiation of the mouse mesenchymal cell line C3H10T1/2 through a genome reprogramming process. In the present study, to clarify the mechanism underlying the multipotency of mesenchymal stem cells (MSCs) and to develop a novel approach to bone regenerative medicine, we attempted to identify the downstream effectors of Nanog in promoting osteogenic differentiation of mouse mesenchymal cells. We demonstrated that Runx1 and Runx3 are the downstream effectors of Nanog, especially in the early and intermediate osteogenic differentiation of the mouse mesenchymal cell line C3H10T1/2.
Introduction
The roles of transcription factors in the maintenance of stem cell pluripotency or multipotency are important, but the details of these roles have not yet been determined. Previously, we reported that the transcription factor Nanog promotes the osteogenic differentiation of the mouse mesenchymal cell line C3H10T1/2 cells through a genome reprogramming process (Ogasawara et al., 2013). Nanog is a homeodomain transcription factor that maintains the self-renewal of embryonic stem cells (ESCs) (Chambers et al., 2003; Mitsui et al., 2003). The forced expression of Nanog in mouse ESCs maintains the cells' pluripotency independently of leukemia inhibitory factor (LIF) signal transduction and activator of transcription 3 (Stat3) pathway (Chambers et al., 2003; Mitsui et al., 2003). Nanog is expressed in ESCs and the inner cell mass of human blastocysts (Hyslop et al., 2005) and is not expressed in somatic cells, except for some types of stem cells (Chambers et al., 2003; Mitsui et al., 2003; Riekstina et al., 2009). Thus, it is difficult to discuss the physiological role of Nanog in somatic cells. However, there have been several Nanog overexpression experiments reported in somatic cells (Go et al., 2008; Kochupurakkal et al., 2008; Ogasawara et al., 2013; Piestun et al., 2006; Zhang et al., 2005).
Regarding the cell source for tissue engineering, although it is expected that ESCs or induced pluripotent stem cells (iPSCs) will eventually be applied clinically, it is as yet difficult to do so except for very rare occasions because of their risk of malignant transformation or problems with ethics. In this situation, mesenchymal stem cells (MSCs) are regarded as one of the best cell sources for tissue engineering among the somatic stem cells because they also have multipotency; in addition, they present no problems with ethics and can be obtained easily in a minimally invasive manner. Therefore, MSCs have already been used for regenerative medicine in applications such as the treatment of patients with heart failure, bone or cartilage defects, and more (Adachi et al., 2005; Hare et al., 2009 Meijer et al., 2008).
When MSCs are used for regenerative medicine, it is important to obtain enough cells while also maintaining their multipotency. However, MSCs progressively lose their multipotency during long-term culture, resulting in the present limitations of tissue engineering, i.e., the small size and insufficient quality of the engineered tissues. To overcome this problem, the effects of several genes, including Nanog, were investigated (Go et al., 2008; Ogasawara et al., 2013; Simonsen et al., 2002; Terai et al., 2005). In the present study, to clarify the mechanisms underlying the multipotency of MSCs and to develop a novel approach to bone regenerative medicine, we sought to discover the downstream effectors of Nanog in promoting osteogenic differentiation of mesenchymal cells. Here we show that Runx1 and Runx3 are the downstream effectors of Nanog, especially in the early and intermediate osteogenic differentiation of the mouse mesenchymal cell line C3H10T1/2.
Materials and Methods
Growth factors and reagents
Dulbecco's Modified Eagle Medium (DMEM), fetal bovine serum (FBS), penicillin-streptomycin solution, and trypsin-EDTA solution were purchased from Sigma Chemical Co. (St. Louis, MO, USA). Isogen was purchased from Wako Pure Chemical Industries (Osaka, Japan). Puromycin was purchased from Life Technologies (Carlsbad, CA, USA). Recombinant human bone morphogenetic protein-2 (rhBMP-2) was generously provided by Astellas Pharma Inc. (Tokyo).
Cell culture and induction of osteogenic differentiation
The mouse mesenchymal cell line C3H10T1/2 was purchased from the Riken Cell Bank (Tsukuba, Japan). C3H10T1/2 cells constitutively expressing green fluorescent protein (GFP) or Nanog were established as described (Ogasawara et al., 2013). These cells were cultured in high-glucose DMEM containing 10% FBS and 1% penicillin/streptomycin. For the induction of osteogenic differentiation with BMP, we added rhBMP-2 to the medium at a concentration of 200 ng/mL. Cells constitutively expressing GFP were used as a control. In each experiment, we used multiple combinations of cells constitutively expressing GFP and cells constitutively expressing Nanog, all of which had been established from the same parental cells, and we present here representative results.
Total RNA extraction, real-time reverse-transcription PCA
Total RNA was isolated from the cells with Isogen following the manufacturer's protocol. Complementary DNA (cDNA) was synthesized from 1 μg of total RNA with a PrimeScript ® RT Reagent Kit with gDNA Eraser (Perfect Real Time, Takara Shuzo Co., Shiga, Japan) to generate single-stranded cDNA. For real-time PCR, we used the ABI Prism Sequence Detection System 7000 (Applied Biosystems, Foster City, CA, USA). Primers were designed based on the sequences obtained from the GenBank, and we selected amplicons of 50–250 bp with a melting temperature between 55°C and 60°C. Aliquots of the first-strand cDNA (1 μL) were amplified with the Fast SYBR® Green Master Mix (Applied Biosystems) under the following conditions: Initial denaturation for 10 min at 94°C followed by 40 cycles consisting of 15 sec at 94°C and 1 min at 60°C. The sequence of primers used in real-time PCR for mouse genes (Runx1, Runx2, Runx3, Col1, osteocalcin, and actin) are available upon request.
We confirmed that the expression levels of actin are steady under various experimental conditions in our experimental model, and we used actin as a housekeeping gene.
von Kossa staining
von Kossa staining was performed as described (Kugimiya et al., 2005). Briefly, C3H10T1/2 cells were inoculated at 1×105 cells per well in a 24-well plate and cultured in DMEM containing 10% FBS with or without rhBMP-2 (200 ng/mL). After being cultured for 21 days, the cells were rinsed with phosphate-buffered saline (PBS), fixed with 100% ethanol at room temperature for 15 min, stained with 5% silver nitrate solution (Wako) under ultraviolet light for 10 min, and incubated for 2 min with 1% sodium thiosulfate solution (Wako).
DNA microarray analysis
DNA microarray analysis was performed as described (Ogasawara et al., 2013). Briefly, a 250-ng aliquot of total RNA from Nanog-expressing cells or GFP-expressing cells cultured for 7 days with rhBMP-2 (200 ng/mL) was used for the first- and second-strand cDNA synthesis. RNA was isolated from the cells with a High Pure RNA Isolation Kit (Roche, Mannheim, Germany) following the manufacturer's protocol. The double-stranded cDNA was used for in vitro translation by a biotin-labeling reaction, using a GeneChip® 3′ IVT Express Kit (Affymetrix, Santa Clara, CA) according to the manufacturer's instructions. The resulting labeled cRNAs were hybridized in GeneChip® Mouse Genome 430 2.0 Arrays (Affymetrix) using a GeneChip® Hybridization, Wash, and Stain Kit (Affymetrix) and scanned according to the manufacturer's instructions.
RNA interference
Runx1siRNA(m) (sc-37678), Runx3siRNA(m) (sc-37680), siRNA TransfectionReagent (sc-29528), siRNA Transfection Medium (sc-36868), and Control siRNA-A (sc-37007) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). For RNA interference (RNAi), 2×105 cells were inoculated onto six-well plates and cultured in DMEM containing 10% FBS for 20–24 h, then transfected with 4 μL of siRNA duplex (10 μM) of Runx1siRNA or Runx3siRNA or Control siRNA-A, using siRNA Transfection Reagent and siRNA Transfection Medium according to the manufacturer's instructions. Twenty-four hours after transfection, the media were changed to fresh DMEM containing 10% FBS supplemented with or without rhBMP-2. The cells were then cultured for 48–72 h, and the total RNA was isolated.
Statistical analysis
The group means were compared by an analysis of variance (ANOVA), and the significance of differences was determined by post hoc testing using the Bonferroni method.
Results
Calcification of C3H10T1/2 cells by forced Nanog expression
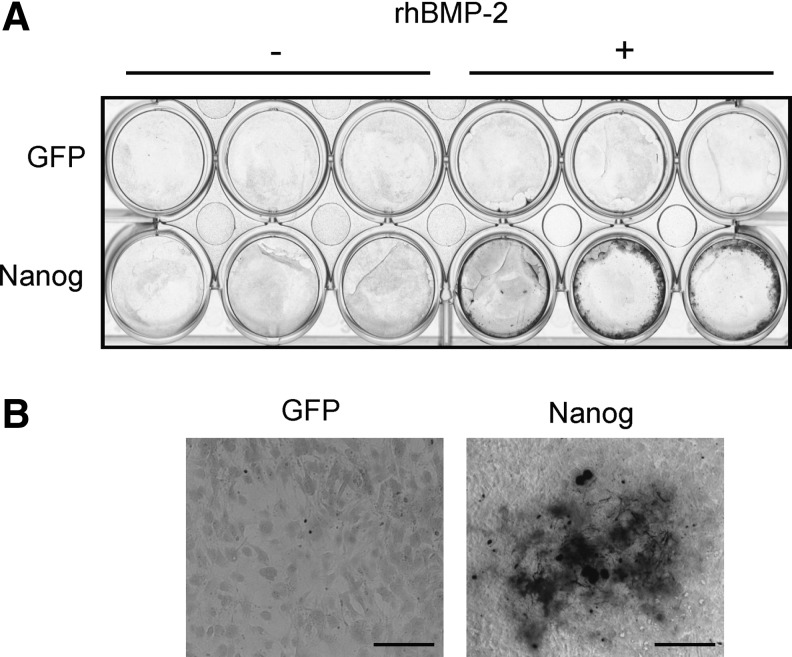
First, to reconfirm the promoted osteogenic differentiation by constitutively forced Nanog expression in mesenchymal cells (Ogasawara et al., 2013), we checked the calcification levels of C3H10T1/2 cells transduced with the Nanog or control GFP gene using a retrovirus vector. von Kossa staining showed that there was an increase of calcified cells among the cells showing constitutive Nanog expression compared to the cells constitutively expressing GFP, irrespective of the presence of rhBMP-2 (Fig. 1). Taking this finding and those of our previous study (Ogasawara et al., 2013) together, it was reconfirmed that the constitutive expression of Nanog promoted all steps of osteogenic differentiation.
FIG. 1.
Effects of constitutive Nanog expression on the calcification of C3H10T1/2 cells. (A) von Kossa staining of GFP- or Nanog-expressing cells cultured in the presence or absence of rhBMP-2 at 21 days. (B) High-magnification image of von Kossa staining of GFP- or Nanog-expressing cells cultured in the presence of rhBMP-2 at 21 days. Bar, 200 μm.
Runx1 and Runx3 as downstream effectors of Nanog
Following our previous report, we performed a DNA microarray analysis to identify downstream molecules involved in the promotion of osteogenic differentiation by forced Nanog expression. Between GFP-expressing and Nanog-expressing cells cultured with rhBMP-2, the expression of about 1000 genes showed a more than two-fold increase (log2Ratio >1) or decrease (log2Ratio <0.5). From the up-regulated genes, we selected several genes as candidates that could regulate osteogenic differentiation as downstream effectors of Nanog. Among these genes, we focused here on Runt-related transcription factor (Runx)1 and Runx3, in consideration of both the log2Ratio (Runx1, 3.45; Runx3, 1.61) and findings gathered from the literature (Kimura et al., 2010; Liakhovitskaia et al., 2010; Lian et al., 2003; Smith et al., 2005; Soung do et al., 2007; Wang et al., 2005; Yamashiro et al., 2004; Yano et al., 2013; Yoshida et al., 2004).
Because we performed the DNA microarray analysis with the culture period of 7 days, we monitored the change in the expression levels of Runx1/Runx3 in each cell line within a week. We also focused on the differences seen between Nanog-expressing cells and control GFP-expressing cells because the aim of the present study was to investigate the downstream effectors of Nanog in the promoted osteogenic differentiation of mesenchymal cells.
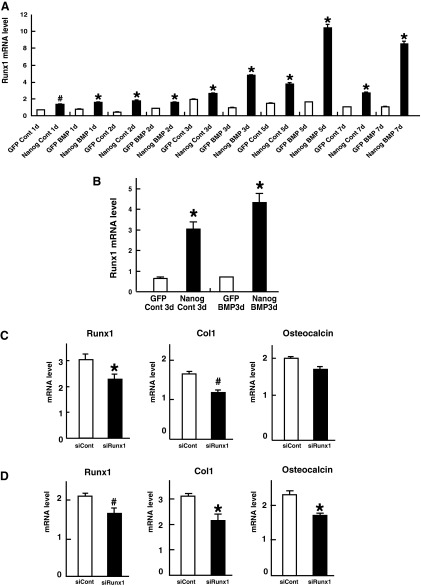
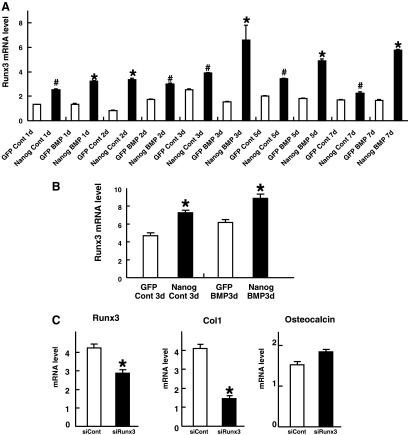
The real-time quantitative RT-PCR analyses confirmed that that there were significant up-regulations of both Runx1 (Fig. 2A) and Runx3 (Fig. 3A) in the Nanog-expressing cells compared to the level in the GFP-expressing cells. These were also true for the other groups of cells (Figs. 2B, 3B). Next, to investigate the functional relevance of Runx1 or Runx3 to the osteogenic differentiation promoted by Nanog, we knocked down Runx1 or Runx3 mRNA in Nanog-expressing cells through small RNA interference (siRNA). In the culture condition with rhBMP-2, type I collagen mRNA was suppressed by both siRunx1 (Fig. 2C) and siRunx3 (Fig. 3C), although osteocalcin mRNA was not affected (Fig. 2C and Fig. 3C). In the culture condition without rhBMP-2, siRunx1 suppressed both type I collagen and osteocalcin mRNA (Fig. 2D). As for siRunx3, we could not obtain enough reproducible data to make a conclusion about the function of Runx3 in the culture condition without rhBMP-2 (data not shown).
FIG. 2.
Runx1 as a downstream effector of Nanog. Results of the real-time RT-PCR conducted to measure the expression levels of Runx1 in GFP-expressing cells and Nanog-expressing cells cultured with or without rhBMP-2 for the indicated times (group A) (A) and those cultured with or without rhBMP-2 for 3 days (group B) (B). Quantities of mRNA are indicated as the levels in 1 μg of total RNA. Bars, means±standard deviation (SD) for three wells/group. (*) Significant stimulation at p<0.001 compared to GFP. (#) Significant stimulation at p<0.005 compared to GFP. We used multiple combinations of cells that had the same origin (parental cells) in each experiment. We measured the expression levels of Runx1, Col1, and osteocalcin in Nanog-expressing cells transfected with control siRNA or Runx1 siRNA and cultured with rhBMP-2 (C) or without rhBMP-2 (D) by quantitative real-time RT-PCR. Quantities of mRNA are indicated as the levels in 1 μg of total RNA. Bars, means±standard deviation (SD) for three wells/group. (*) Significant inhibition at p<0.01 compared to the control siRNA. (#) Significant inhibition at p<0.05 compared to the control siRNA. d, days.
FIG. 3.
Runx3 as a downstream effector of Nanog. We measured the expression levels of Runx3 in GFP-expressing cells and Nanog-expressing cells cultured with or without rhBMP-2 for the indicated times (group A) (A) and those cultured with or without rhBMP-2 for 3 days (group B) (B) by quantitative real-time RT-PCR. Quantities of mRNA are indicated as the levels in 1 μg of total RNA. Bars, means±standard deviation (SD) for three wells/group. (*) Significant stimulation at p<0.001 compared to GFP. (#) Significant stimulation at p<0.05 compared to GFP. We used multiple combinations of cells as described in the Fig. 2 legend. (C) We measured the expression levels of Runx3, Col1, and osteocalcin in Nanog-expressing cells transfected with control siRNA or Runx3 siRNA and cultured with rhBMP-2 by quantitative real-time RT-PCR. Quantities of mRNA are indicated as the levels in 1 μg of total RNA. Bars, means±standard deviation (SD) for three wells/group. (*) Significant inhibition at p<0.01 compared to the control siRNA. d, days.
These data suggested that Runx1 and Runx3 might be downstream effectors of Nanog, especially in the early and intermediate osteogenic differentiation. We did not monitor alkaline phosphatase (ALP) mRNA because it was demonstrated that ALP is not suitable to evaluate the effects of Nanog, at least for the first 72 h (Ogasawara et al. 2013). Concerning Runx2, which is the master regulator of osteoblast differentiation and bone formation, the DNA microarray analysis did not show a more than two-fold increase (log2Ratio=−0.76) of Runx2. In agreement with the results of our DNA microarray analysis, the real-time quantitative RT-PCR analyses also demonstrated that the expression levels of Runx2 were not so dramatically upregulated by forced Nanog expression, compared with Runx1 or Runx3. In addition, the patterns of Runx2 upregulation varied depending on the groups of clones (data not shown).
Discussion
Transcription factors are important in the mechanisms underlying the maintenance of stem cell pluripotency. For instance, it has been shown that Oct3/4, Sox2, and Nanog function as core transcription factors in maintaining the pluripotency of ESCs (Boyer et al., 2005; Loh et al., 2006). In addition, iPSCs were initially established from somatic cells by introducing four transcription factors—Oct3/4, Sox2, c-Myc, and Klf4 (Takahashi and Yamanaka, 2006; Takahashi et al., 2007). As for the maintenance of the multipotency of MSCs, the involvement of Nanog and Sox2 was reported in the promotion of the osteogenic differentiation of MSCs (Go et al., 2008; Ogasawara et al., 2013).
From the clinical point of view, it is important to identify the downstream effectors of the critical transcription factors regulating the pluripotency or multipotency of stem cells because the targets of downstream effectors are thought to be more specific than those of upstream molecules. In other words, the clinical application of downstream effectors might circumvent or decrease side effects compared with the application of their upstream molecules.
We reported that the transcription factor Nanog promotes the osteogenic differentiation of mouse mesenchymal C3H10T1/2 cells through a genome reprogramming process, and we also identified nuclear factor of activated T cells c1 (NFATc1) as one of the downstream effectors (Ogasawara et al., 2013). In the present study, to clarify the mechanism underlying the pluripotency or multipotency of stem cells for the advancement of stem cell biology and bone regenerative medicine, we sought to find downstream effectors of Nanog other than NFATc1.
First, we reconfirmed that Nanog promotes osteogenic differentiation of C3H10T1/2 cells, including calcification. Next, we discovered that Runx1 and Runx3 were upregulated by forced Nanog expression in C3H10T1/2 cells. We also demonstrated that knockdown of Runx1 or Runx3 mRNA in Nanog-expressing cells through siRNA inhibits the expression levels of osteogenic differentiation markers' mRNA. It is noteworthy that the promoted osteogenic differentiation of C3H10T1/2 cells by Nanog was accompanied by the significant upregulation of both Runx1 and Runx3, but not by that of Runx2, which is a master regulator of osteoblast differentiation (Komori et al., 1997; Otto et al., 1997).
The Runx family has been studied extensively. Runx1 is an essential transcription factor for definitive hematopoiesis (Okuda et al., 1996), Runx2 is a master transcription factor in osteoblast differentiation and bone formation (Komori et al., 1997; Otto et al., 1997), and Runx3 plays a role in the development of certain neurons (Inoue et al., 2003; Levanon et al., 2002) in addition to its antioncogenic role in the gastric mucosa (Li et al., 2002).
Because all three Runx proteins bind to the same consensus sequence, it is possible that Runx1, Runx2, and Runx3 can mutually compensate for each function in most developing skeletal structures. In fact, there is evidence that not only Runx2 but also Runx1 and Runx3 are involved in the regulation of skeletal systems both in vivo and in vitro. In the mesenchymal condensations during early skeletal development, the co-expression of Runx1 and Runx2 was detected (Smith et al., 2005; Wang et al., 2005; Yamashiro et al., 2004). Runx1 plays an essential role in the development of the sternum (Kimura et al., 2010; Liakhovitskaia et al., 2010) and Runx1 functions in concert with Runx2 during sternal morphogenesis (Kimura et al., 2010).
Moreover, Runx2 and Runx3 have been shown to be functionally redundant in chondrocyte maturation on the basis of the finding that chondrocyte maturation is completely absent in Runx2 and Runx3 double-deficient mice, but not in Runx2-deficient mice (Yoshida et al., 2004).
In vitro studies revealed that Runx1 and Runx2 are co-expressed during the early differentiation of skeletal mesenchymal cells, but Runx1 expression decreases during the maturation of chondrocytes and osteoblasts (Lian et al., 2003; Wang et al., 2005). The forced expression of Runx1 in C3H10T1/2 cells promotes chondrogenic differentiation (Wang et al., 2005; Yano et al., 2013). In addition, the knockdown of Runx1 in both RCJ3.1C5.18 chondrogenic cells and C3H10T1/2 cells stimulated them to differentiate into chondrocytes, leading to the inhibition of chondrocyte differentiation (Wang et al., 2005; Yano et al., 2013).
In contrast, the role of Runx3 in the control of mesenchymal cell differentiation is thought to be more complex and diverse. For example, the gain or loss of the function of Runx3 in the MLB13MYC clone 17 (a prechondroblastic cell line) promotes or inhibits representative chondrogenic differentiation markers time dependently (Soung do et al., 2007). Concerning molecules interacting with Runx1, Yano and co-workers reported that a small thienoindazole derivative compound promoted the chondrogenic differentiation of C3H10T1/2 cells through the induction of Runx1, Sox5, and Sox6 (Yano et al., 2013). In addition, Runx1 has been shown to regulate both sternal morphogenesis and the commitment of mesenchymal cells to differentiate to chondrocytes via the induction of Sox5 and Sox6 (Kimura et al., 2010).
Taking these previous studies and our present work into consideration, it is tempting to propose that Runx1 and Runx3 are definitive regulators that maintain MSC multipotency and that the roles of Runx1 and Runx3 in the promotion of osteogenic or chondrogenic differentiation could be different, depending on the differentiation stage and their interaction with the other molecules involved. We consider these points as one of the causes of several complexities seen in the results of our siRunx1/Runx3 experiments. In the culture condition without rhBMP-2, osteocalcin expression was suppressed by siRunx1. The effects of siRunx3 varied, and we could not make any conclusion about the role of Runx3 induced by Nanog in the present study.
Further studies are required for a deeper understanding of the roles of Runx1 and Runx3 in the maintenance of the multipotency of MSCs. In particular, we speculate that the identification of the molecules interacting with Runx1/Runx3 might provide clues that could help answer this question. We also consider it important to determine the downstream factors of Nanog that control late osteogenic differentiation, because we found that NFATc1, Runx1, and Runx3 mainly regulated Col1, which is an early to intermediate osteogenic differentiation marker. To this end, we have been analyzing other molecules with a focus on transcription factors, on the basis of the results obtained from the DNA microarray analysis.
In conclusion, the findings obtained in this study provide several lines of evidence that Runx1 and Runx3 are downstream effectors of Nanog in the promoted osteogenic differentiation of the mouse mesenchymal cell line C3H10T1/2, which function especially in early to intermediate osteogenic differentiation.
Acknowledgments
We thank Astellas Pharma Inc. for kindly providing materials. We also thank Drs. T. Akagawa and K. Hoshi for their helpful discussion and support. This work was supported by JSPS KAKENHI (grants nos. 22659365, 23592912, 24390450, and 25670846) from the Japan Society for the Promotion of Science (JSPS), and grants from the Uemura Fund, Nihon University School of Dentistry.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Adachi N., Ochi M., Deie M., and Ito Y. (2005). Transplant of mesenchymal stem cells and hydroxyapatite ceramics to treat severe osteochondral damage after septic arthritis of the knee. J. Rheumatol. 32, 1615–1618 [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Gifford D.K., Melton D.A., Jaenisch R., and Young R.A. (2005). Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., and Smith A. (2003). Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, 643–655 [DOI] [PubMed] [Google Scholar]
- Go M.J., Takenaka C., and Ohgushi H. (2008). Forced expression of Sox2 or Nanog in human bone marrow derived mesenchymal stem cells maintains their expansion and differentiation capabilities. Exp. Cell Res. 314, 1147–1154 [DOI] [PubMed] [Google Scholar]
- Hare J.M., Traverse J.H., Henry T.D., Dib N., Strumpf R.K., Schulman S.P., Gerstenblith G., DeMaria A.N., Denktas A.E., Gammon R.S., Hermiller J.B., Reisman M.A., Schaer G.L., and Sherman W. (2009). A randomized, double-blind, placebo-controlled, dose-escalation study of intravenous adult human mesenchymal stem cells (prochymal) after acute myocardial infarction. J. Am. Coll. Cardiol. 54, 2277–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyslop L., Stojkovic M., Armstrong L., Walter T., Stojkovic P., Przyborski S., Herbert M., Murdoch A., Strachan T., and Lako M. (2005). Downregulation of NANOG induces differentiation of human embryonic stem cells to extraembryonic lineages. Stem Cells 23, 1035–1043 [DOI] [PubMed] [Google Scholar]
- Inoue K., Ozaki S., Ito K., Iseda T., Kawaguchi S., Ogawa M., Bae S.C., Yamashita N., Itohara S., Kudo N., and Ito Y. (2003). Runx3 is essential for the target-specific axon pathfinding of trkc-expressing dorsal root ganglion neurons. Blood Cells Mol. Dis. 30, 157–160 [DOI] [PubMed] [Google Scholar]
- Kimura A., Inose H., Yano F., Fujita K., Ikeda T., Sato S., Iwasaki M., Jinno T., Ae K., Fukumoto S., Takeuchi Y., Itoh H., Imamura T., Kawaguchi H., Chung U.I., Martin J.F., Iseki S., Shinomiya K., and Takeda S. (2010). Runx1 and Runx2 cooperate during sternal morphogenesis. Development 137, 1159–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochupurakkal B.S., Sarig R., Fuchs O., Piestun D., Rechavi G., and Givol D. (2008). Nanog inhibits the switch of myogenic cells towards the osteogenic lineage. Biochem. Biophys. Res. Commun. 365, 846–850 [DOI] [PubMed] [Google Scholar]
- Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R.T., Gao Y.H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., and Kishimoto T. (1997). Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89, 755–764 [DOI] [PubMed] [Google Scholar]
- Kugimiya F., Kawaguchi H., Kamekura S., Chikuda H., Ohba S., Yano F., Ogata N., Katagiri T., Harada Y., Azuma Y., Nakamura K., and Chung U.I. (2005). Involvement of endogenous bone morphogenetic protein (BMP) 2 and BMP6 in bone formation. J. Biol.Chem. 280, 35704–35712 [DOI] [PubMed] [Google Scholar]
- Levanon D., Bettoun D., Harris-Cerruti C., Woolf E., Negreanu V., Eilam R., Bernstein Y., Goldenberg D., Xiao C., Fliegauf M., Kremer E., Otto F., Brenner O., Lev-Tov A., and Groner Y. (2002). The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J. 21, 3454–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.L., Ito K., Sakakura C., Fukamachi H., Inoue K., Chi X.Z., Lee K.Y., Nomura S., Lee C.W., Han S.B., Kim H.M., Kim W.J., Yamamoto H., Yamashita N., Yano T., Ikeda T., Itohara S., Inazawa J., Abe T., Hagiwara A., Yamagishi H., Ooe A., Kaneda A., Sugimura T., Ushijima T., Bae S.C., and Ito Y. (2002). Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 109, 113–124 [DOI] [PubMed] [Google Scholar]
- Liakhovitskaia A., Lana-Elola E., Stamateris E., Rice D.P., van 't Hof R.J., and Medvinsky A. (2010). The essential requirement for Runx1 in the development of the sternum. Dev. Biol. 340, 539–546 [DOI] [PubMed] [Google Scholar]
- Lian J.B., Balint E., Javed A., Drissi H., Vitti R., Quinlan E.J., Zhang L., Van Wijnen A.J., Stein J.L., Speck N., and Stein G.S. (2003). Runx1/AML1 hematopoietic transcription factor contributes to skeletal development in vivo. J. Cell. Physiol. 196, 301–311 [DOI] [PubMed] [Google Scholar]
- Loh Y.H., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Wong K.Y., Sung K.W., Lee C.W., Zhao X.D., Chiu K.P., Lipovich L., Kuznetsov V.A., Robson P., Stanton L.W., Wei C.L., Ruan Y., Lim B., and Ng H.H. (2006). The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38, 431–440 [DOI] [PubMed] [Google Scholar]
- Meijer G.J., de Bruijn J.D., Koole R., and van Blitterswijk C.A. (2008). Cell based bone tissue engineering in jaw defects. Biomaterials 29, 3053–3061 [DOI] [PubMed] [Google Scholar]
- Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., and Yamanaka S. (2003). The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642 [DOI] [PubMed] [Google Scholar]
- Ogasawara T., Ohba S., Yano F., Kawaguchi H., Chung U.I., Saito T., Yonehara Y., Nakatsuka T., Mori Y., Takato T., and Hoshi K. (2013). Nanog promotes osteogenic differentiation of the mouse mesenchymal cell line C3H10T1/2 by modulating bone morphogenetic protein (BMP) signaling. J. Cell Physiol. 228, 163–171 [DOI] [PubMed] [Google Scholar]
- Okuda T., van Deursen J., Hiebert S.W., Grosveld G., and Downing J.R. (1996). AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84, 321–330 [DOI] [PubMed] [Google Scholar]
- Otto F., Thornell A.P., Crompton T., Denzel A., Gilmour K.C., Rosewell I.R., Stamp G.W., Beddington R.S., Mundlos S., Olsen B.R., Selby P.B., and Owen M.J. (1997). Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89, 765–771 [DOI] [PubMed] [Google Scholar]
- Piestun D., Kochupurakkal B.S., Jacob-Hirsch J., Zeligson S., Koudritsky M., Domany E., Amariglio N., Rechavi G., and Givol D. (2006). Nanog transforms NIH3T3 cells and targets cell-type restricted genes. Biochem. Biophys. Res. Commun. 343, 279–285 [DOI] [PubMed] [Google Scholar]
- Riekstina U., Cakstina I., Parfejevs V., Hoogduijn M., Jankovskis G., Muiznieks I., Muceniece R., and Ancans J. (2009). Embryonic stem cell marker expression pattern in human mesenchymal stem cells derived from bone marrow, adipose tissue, heart and dermis. Stem Cell Rev. 5, 378–386 [DOI] [PubMed] [Google Scholar]
- Simonsen J.L., Rosada C., Serakinci N., Justesen J., Stenderup K., Rattan S.I., Jensen T.G., and Kassem M. (2002). Telomerase expression extends the proliferative life-span and maintains the osteogenic potential of human bone marrow stromal cells. Nat. Biotechnol. 20, 592–596 [DOI] [PubMed] [Google Scholar]
- Smith N., Dong Y., Lian J.B., Pratap J., Kingsley P.D., van Wijnen A.J., Stein J.L., Schwarz E.M., O'Keefe R.J., Stein G.S., Schwarz E.M., O'Keefe R.J., Stein G.S., and Drissi M.H. (2005). Overlapping expression of Runx1(Cbfa2) and Runx2(Cbfa1) transcription factors supports cooperative induction of skeletal development. J. Cell Physiol. 203, 133–143 [DOI] [PubMed] [Google Scholar]
- Soung do Y., Dong Y., Wang Y., Zuscik M.J., Schwarz E.M., O'Keefe R.J., and Drissi H. (2007). Runx3/AML2/Cbfa3 regulates early and late chondrocyte differentiation. J. Bone Miner. Res. 22, 1260–1270 [DOI] [PubMed] [Google Scholar]
- Takahashi K., and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., and Yamanaka S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- Terai M., Uyama T., Sugiki T., Li X.K., Umezawa A., and Kiyono T. (2005). Immortalization of human fetal cells: The life span of umbilical cord blood-derived cells can be prolonged without manipulating p16INK4a/RB braking pathway. Mol. Biol. Cell 16, 1491–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Belflower R.M., Dong Y.F., Schwarz E.M., O'Keefe R.J., and Drissi H. (2005). Runx1/AML1/Cbfa2 mediates onset of mesenchymal cell differentiation toward chondrogenesis. J. Bone Miner. Res. 20, 1624–1636 [DOI] [PubMed] [Google Scholar]
- Yamashiro T., Wang X.P., Li Z., Oya S., Aberg T., Fukunaga T., Kamioka H., Speck N.A., Takano-Yamamoto T., and Thesleff I. (2004). Possible roles of Runx1 and Sox9 in incipient intramembranous ossification. J. Bone Miner. Res. 19, 1671–1677 [DOI] [PubMed] [Google Scholar]
- Yano F., Hojo H., Ohba S., Fukai A., Hosaka Y., Ikeda T., Saito T., Hirata M., Chikuda H., Takato T., Kawaguchi H., and Chung U.I. (2013). A novel disease-modifying osteoarthritis drug candidate targeting Runx1. Ann. Rheum. Dis. 72, 748–753 [DOI] [PubMed] [Google Scholar]
- Yoshida C.A., Yamamoto H., Fujita T., Furuichi T., Ito K., Inoue K., Yamana K., Zanma A., Takada K., Ito Y., and Komori T. (2004). Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev 18, 952–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wang X., Chen B., Suo G., Zhao Y., Duan Z., and Dai J. (2005). Expression of Nanog gene promotes NIH3T3 cell proliferation. Biochem. Biophys. Res. Commun. 338, 1098–1102 [DOI] [PubMed] [Google Scholar]