Abstract
OBJECTIVES: Brain metastases due to breast cancer are increasing, and the prognosis is poor. Lack of effective therapy is attributed to heterogeneity of breast cancers and their resulting metastases, as well as impermeability of the blood–brain barrier (BBB), which hinders delivery of therapeutics to the brain. This work investigates three experimental models of HER2 + breast cancer brain metastasis to better understand the inherent heterogeneity of the disease. We use magnetic resonance imaging (MRI) to quantify brain metastatic growth and explore its relationship with BBB permeability. DESIGN: Brain metastases due to breast cancer cells (SUM190-BR3, JIMT-1-BR3, or MDA-MB-231-BR-HER2) were imaged at 3 T using balanced steady-state free precession and contrast-enhanced T1-weighted spin echo sequences. The histology and immunohistochemistry corresponding to MRI were also analyzed. RESULTS: There were differences in metastatic tumor appearance by MRI, histology, and immunohistochemistry (Ki67, CD31, CD105) across the three models. The mean volume of an MDA-MB-231-BR-HER2 tumor was significantly larger compared to other models (F2,12 = 5.845, P < .05); interestingly, this model also had a significantly higher proportion of Gd-impermeable tumors (F2,12 = 22.18, P < .0001). Ki67 staining indicated that Gd-impermeable tumors had significantly more proliferative nuclei compared to Gd-permeable tumors (t[24] = 2.389, P < .05) in the MDA-MB-231-BR-HER2 model. CD31 and CD105 staining suggested no difference in new vasculature patterns between permeable and impermeable tumors in any model. CONCLUSION: Significant heterogeneity is present in these models of brain metastases from HER2 + breast cancer. Understanding this heterogeneity, especially as it relates to BBB permeability, is important for improvement in brain metastasis detection and treatment delivery.
Introduction
Breast cancer will affect approximately one in eight American women [1]. It is the most commonly diagnosed cancer in women and is the second leading cause of cancer death in American women [1]. The major challenge of this disease is its propensity to metastasize to distant sites such as bone, liver, lung, and brain [2]. The relative rate of breast cancer brain metastasis is extremely high, second only to lung cancer [3].
Breast cancer is a complex and heterogeneous disease and is classified into at least four molecular subtypes with different prognostic significance; further classification exists by histological type and grade [2,4,5]. No single subgroup is biologically homogeneous. The HER2 subtype—estrogen receptor negative (ER −), progesterone receptor negative (PR −), human epidermal growth factor receptor 2 positive (HER2 +) [6]—is of particular concern because it accounts for 10% to 15% of breast cancers and develops brain metastases at a significantly higher rate compared to other breast cancer subtypes [2,7].
Brain metastases that form from breast cancers are also complex and heterogeneous. These can manifest in three clinically distinct situations: multiple brain metastases (78%), solitary brain metastasis (14%), or leptomeningeal metastasis (8%) [8]. Each manifestation dictates a different course of treatment; however, responses to therapy vary among metastases. Some metastases will not respond to therapy, whereas others will appear to initially respond but develop resistance during treatment or will cause recurrence at a later time. This heterogeneity in tumor response complicates clinical management and contributes to the poor prognosis for this disease.
Clinical data suggest that HER2 overexpression is present in as many as 53% of breast cancer brain metastasis cases [7]. Median survival from diagnosis of HER2 + brain metastases to death is 4 to 6 months, and less than 20% of patients can expect to live 1 year [9]. Treatment for HER2 + metastatic breast cancer includes anti-HER2 therapy such as trastuzumab (Herceptin). Despite improvement in both disease-free and overall survival [10], central nervous system metastases develop in 31% to 48% of patients on trastuzumab [11–14]; and 33% to 50% of those are responding to therapy or have stable disease at other sites when diagnosed with central nervous system involvement [11,14]. This suggests that the brain offers a sanctuary site where metastases can persist, unaffected by therapy.
The poor prognosis for brain metastases has been largely attributed to obstacles created by the blood–brain barrier (BBB). Under normal conditions, the BBB acts to protect the brain by tightly regulating the diffusion of substrates from the blood into the brain parenchyma [15]; however, this also causes systemic therapies to be excluded from the brain. For example, chemo- and targeted therapies, such as doxorubicin and trastuzumab, which are highly successful at treating HER2 + metastases in other areas of the body, are ineffective for brain metastases [14,16] presumably because they are unable to cross the intact BBB.
Studying brain metastatic breast cancer is challenging because there are few experimental animal models [17–20]. The MDA-MB-231-BR cell line has been the most widely used to study brain metastasis from breast cancer [18,21–29]. Work performed in the Steeg laboratory has shown that this preclinical model shares many characteristics of human brain metastases [22]. The MDA-MB-231-BR cell line has also been transduced with HER2 cDNA (resulting in the MDA-MB-231-BR-HER2 cell line), and significant heterogeneity in the permeability of the BBB associated with these brain metastases was demonstrated by Lockman et al. [30] using dextran perfusion and ex vivo fluorescence microscopy.
Imaging is increasingly being used to study models of brain metastasis. Magnetic resonance imaging (MRI) allows for noninvasive, three-dimensional (3D), longitudinal studies at very high spatial resolution; and a wide range of contrast mechanisms are possible. Contrast-enhanced T1-weighted (T1w) MRI, which relies on tumor detection by diffusion of gadolinium-DTPA (Gd) across an impaired BBB in the local region of a tumor, is widely regarded as the most accurate method for clinical brain tumor detection. A previous imaging study performed in our laboratory showed that not all 231BR brain metastases were detectable after Gd-enhanced MRI and that the BBB permeability changed over time as brain metastases developed [24]. These findings suggest that brain metastases that are not permeable to Gd may go undetected by conventional MRI. In addition, heterogeneity in BBB permeability between metastases also means that therapeutic access to individual brain metastases varies [28,30,31].
In this study, we explore the relationship between the growth of brain metastases and their permeability status in three HER2 + human brain metastatic breast cancer cell lines, two of which are naturally HER2-expressing cell lines that have been recently developed. We present quantification of metastasis volume in the whole mouse brain using high-resolution 3D MRI as well as an assessment of the integrity of the BBB associated with each tumor (blood–tumor barrier, BTB). Correlative histology and immunohistochemistry provide image validation and further characterization of each cell line.
Methods
Cell Culture
The following three HER2 + human brain metastatic breast cancer cell lines were used: (1) MDA-MB-231-BR-HER2 [19], (2) JIMT-1-BR3 [32], and (3) SUM190-BR3. The MDA-MB-231-BR-HER2 cell line is derived from the breast carcinoma parental line, MDA-MB-231, and transfected with HER2 [19]; the other two cell lines were derived from their parental lines—JIMT-1 [20] and SUM190 [33], respectively—and are naturally HER2 +. All three cell lines were developed in the laboratory of Dr Patricia Steeg at the National Cancer Institute (Bethesda, MA). For MDA-MB-231-BR-HER2 and JIMT-1-BR3, cells were maintained in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum. MDA-MB-231-BR-HER2 media also contained penicillin and streptomycin; the JIMT-1-BR3 media contained 2 mM glutamine. SUM190-BR3 cells were grown in serum-free Ham F-12 medium supplemented with insulin and hydrocortisone. All cell lines were maintained at 37°C and 5% CO2. Cell viability was calculated by the trypan blue exclusion assay.
Cell Labeling
For a subset of in vivo experiments, cells were labeled with micron-sized iron oxide particles (0.9 μm MPIO, Bangs Laboratory, Fishers, IN) for cell tracking by MRI as previously described [21]. Labeling was achieved by coincubating 2 × 106 seeded cells with MPIO at a concentration of 25 μg Fe/mL for 24 hours in a T75-cm2 flask. The cells were harvested and thoroughly washed three times with Hanks balanced salt solution to remove unincorporated MPIO before cell injection. Perl Prussian Blue (PPB)–stained cytospin slides were analyzed to determine labeling efficiency; this was calculated in ImageJ software (Open-source, NIH, Bethesda, MD) as the ratio of PPB-positive cells to the total number of cells in five random fields of view at 40 × magnification using a Zeiss AXIO Imager A1 Microscope.
Iron-labeled cells create regions of signal void on MR images [21,23] and allow for the confirmation of successful cell injections and arrest of cancer cells in the brain; lack of signal voids results in exclusion from further study.
Animal Preparation
Female nude mice (nu/nu, aged 6-8 weeks, from Charles River Laboratories, Wilmington, MA) were housed in a pathogen-free barrier facility, and all experiments were approved by the Animal Use Subcommittee of the University Council on Animal Care at the University of Western Ontario. Cells suspended in 0.1 mL of Hanks balanced salt solution were delivered to female nude mice, anesthetized with 2% isoflurane in oxygen, by intracardiac injection to the beating left ventricle of the mouse heart.
The three brain metastatic breast cancer models were injected intracardially into mice. The first group (n = 5) was injected with 100,000 MDA-MB-231-BR-HER2 cells per mouse; another group of mice (n = 6) was injected with 175,000 JIMT-1-BR3 cells; the final group (n = 4) was injected with 500,000 SUM190-BR3 cells. The number of injected cells was selected based on in vivo tumorgenic potential determined from previous studies [28].
Experimental End Point
Mice were scanned using MRI at the experimental end point and then euthanized by an overdose injection of pentobarbital (Euthanyl). The end point was determined by the onset of neurological symptoms, unacceptable weight loss and body condition, or hind-limb paralysis. Different degrees of in vivo tumor growth and aggressiveness caused the end point to be reached at different times for each model; this is further discussed in our results. The end points were day 28 for JIMT1-BR3, day 36 for MDA-MB-231-BR-HER2, and day 64 for SUM190-BR3.
Magnetic Resonance Imaging
All images were acquired on a 3.0-T GE Excite MR750 clinical scanner (General Electric, Mississauga, Canada) using a custom-built insertable gradient coil (inner diameter = 17.5 cm, gradient strength = 500 mT/m, and peak slew rate = 3000 T/[m s]) and a custom solenoidal mouse head radiofrequency coil (inner diameter = 1.5 cm). Mice were anesthetized (1.5% isoflurane in oxygen), and temperature was maintained using warm saline bags during the scans. In vivo MRI was acquired using two pulse sequences: a 3D balanced steady-state free precession (bSSFP) scan and a two-dimensional post-gadolinium T1-weighted spin echo (T1wSE) scan.
The bSSFP sequence was used to detect signal voids post–cell injection on day 0 and to quantify metastases numbers and volumes in the whole mouse brain. The parameters for the bSSFP scans were as follows: resolution = 100 × 100 × 200 μm, repetition time = 8 or 10 milliseconds, echo time = 4 or 5 milliseconds, flip angle = 35°, signal averages = 2, radiofrequency phase cycles = 8, scan time = 28 or 36 minutes. The T1wSE was acquired approximately 40 minutes after the intraperitoneal administration of 0.2 mL of gadopentetate dimeglumine (Magnevist, Schering, US; 0.5 mmol/mL) and used to assess the BTB associated with individual tumors. The parameters for these scans were as follows: resolution = 156 × 156 × 400 μm, repetition time = 600 milliseconds, echo time = 20 milliseconds, signal averages = 8, scan time = 20 minutes. Zero filling interpolation was applied to all images.
Histology and Immunohistochemistry
At respective end points, all mice were sacrificed by pentobarbital overdose and perfused with saline and subsequently with 4% paraformaldehyde or 4% formalin. Brains were excised and placed in paraformaldehyde for at least another 24 hours. Fixed brains were processed, paraffin embedded, and serially cut into 5-μm sections. Tissue sections were deparaffinized, rehydrated, and either histologically stained with hematoxylin and eosin (H&E) or stained for CD31, CD105, or Ki67 by immunohistochemistry. Heat-mediated antigen retrieval with citrate buffer pH 6 was performed for sections that were to be stained for CD31 or Ki67. All tissue sections were blocked with Dako protein block, serum free (Dako Canada, Inc, Burlington, ON, Canada); and the following rabbit polyclonal antibodies were used: anti-CD31 (1:50, ab28364, Abcam Inc, Toronto, ON, Canada), anti-CD105 (1:1000, ab107595, Abcam Inc), and anti-Ki67 (1:100, ab833, Abcam Inc). Dako LSAB2 system-HRP (Dako Canada Inc) and Vector DAB peroxidase substrate kit (Vector Laboratories, Burlingame, CA) were used according to manufacturers’ instructions to detect positive staining. All stained sections were counterstained with hematoxylin before dehydration and mounting. All staining was imaged on an Axio Imager A1 microscope (Zeiss CANADA, Toronto, ON, Canada) with a Retiga EXi (QImaging Scientific Research Cameras, Surrey, BC, Canada) digital camera.
Data Analysis
Images were analyzed using open-source OsiriX image software, version 3.9.2. Qualitative analysis of signal voids on day 0 confirmed mice with successful iron-labeled cell injections; lack of signal voids excluded mice from further study. Brain metastases were counted manually. 3D tumor volumes were reconstructed using the OsiriX volume algorithm from the manual segmentation of a region of interest around each tumor boundary in every bSSFP image slice for each mouse. Comparison of corresponding bSSFP and T1wSE images was used to qualitatively assess BTB permeability to Gd—if metastases were detected on the T1wSE, these were classified as “Gd permeable”; in contrast, if they were detected on the bSSFP but not in the corresponding T1wSE, these were deemed “Gd impermeable.” If corresponding images were not available, permeability status was not decided. The “Gd-permeable fraction” was determined as the percentage of Gd-permeable metastases in relation to the total number of metastases in the whole brain at a specific time point. To evaluate the proliferative index of Gd-permeable and Gd-impermeable tumors, Ki67-stained tissue sections were matched to MRI data and imaged at 40 × magnification; all nuclei within a random field of view in the tumor were manually counted. The proliferative index was calculated as the percentage of Ki67-positively stained nuclei among all MDA-MB-231-BR-HER2 cancer cells. All values are presented graphically as mean ± SE. Statistical analysis was performed using unpaired t tests, or a one-way analysis of variance followed by Tukey honestly significant difference tests on GraphPad Prism version 6 software (GraphPad, San Diego, CA).
Results
Representative images of brain metastases from each group are shown in Figure 1. Contrast in bSSFP images is related to the ratio of spin–spin to spin–lattice relaxations (T2/T1); in general, metastases appear hyperintense compared to brain parenchyma because of relatively higher fluid content and therefore a longer T2 and T2/T1 ratio.
Figure 1.
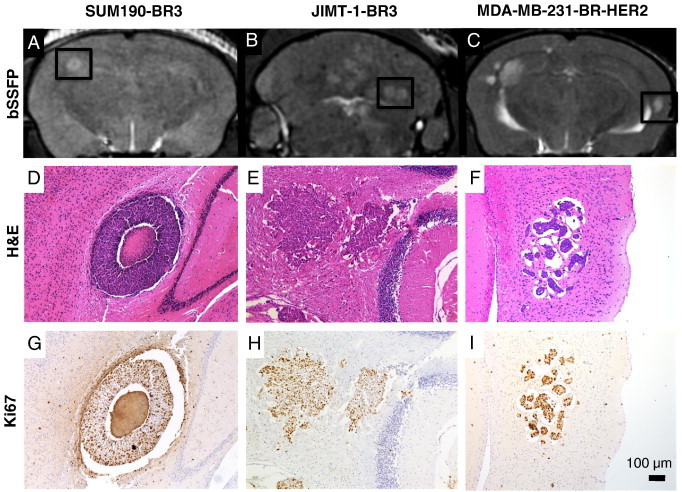
Representative images showing metastases resulting from SUM190-BR3, JIMT-1-BR3, or MDA-MB-231-BR-HER2 human brain metastatic breast cancer. (A-C) bSSFP images show metastasis burden (hyperintensities); (D-F) correlative H&E-stained sections of the area indicated by boxes show morphology of tumors in each model; (G-I) Ki67 staining indicates proliferative nuclei in brown. The region of signal void in the center of the SUM190-BR3 tumor (A) relates to the necrotic core seen in histology (D). JIMT-1-BR3 tumors are hyperintense in bSSFP (B) and grow as dense and proliferative nuclei clusters (E, H). 231-BR-HER2 tumors are the most hyperintense in bSSFP (C) and present as clusters of highly proliferative cell nuclei within pockets of edema in H&E (1, I).
The three models of HER2 + breast cancer brain metastasis each had a different appearance by MRI as well as in histology and immunohistochemistry. The SUM190-BR3 tumors were hyperintense with a hypointense core in bSSFP images (Figure 1A); this appearance was reflected in the corresponding H&E- and Ki67-stained tissue sections where a necrotic core was evident (Figure 1D) and proliferation was evident around the outer edge (Figure 1G). The JIMT-1-BR3 tumors also appeared hyperintense in bSSFP images (Figure 1B), and tissue staining showed tumors with dense clusters of proliferative nuclei (Figure 1E and H). The MDA-MB-231-BR-HER2 tumors appeared the brightest in bSSFP images (Figure 1C). In H&E-stained sections, they present as small, dense clusters of cell nuclei surrounded by substantial pockets of edema (Figure 1F); Ki67 staining indicated that these tumors were highly proliferative (Figure 1I).
The 3D high-resolution nature of MRI allowed us to quantify metastasis volume and permeability in the whole mouse brain for each animal in all three models. We evaluated 198 tumors at experimental end point across the three models. Tumor incidence and total tumor burden are shown in Table 1. The SUM190-BR3 model grew few tumors, and the total tumor volume per brain was relatively low despite having the most cells injected. The MDA-MB-231-BR-HER2 model was injected with the least cells, yet grew the most tumors and had the highest total tumor volume. The study end point was decided when mice displayed clinical symptoms of the disease and deteriorating health. The time to end point was different for each model: day 28 for JIMT-1-BR3, day 36 for MDA-MB-231-BR-HER2, and day 64 for SUM190-BR3. The mean volume of each tumor was calculated at these times for all mice in each of the three models (Figure 2). The average volume of a tumor was significantly different between models (F2,12 = 5.845, P < .05); but interestingly, a longer experiment did not mean larger tumors. Post hoc Tukey tests showed that MDA-MB-231-BR-HER2 tumors were significantly larger in size compared to JIMT-1-BR3 or SUM190-BR3 tumors despite the fact that the SUM190-BR3 model grew for longer. No significant difference in tumor volume was observed between JIMT-1-BR3 and SUM190-BR3 models, yet the SUM190-BR3 model grew for more than twice as long before the mice were noted to have neurological symptoms.
Table 1.
Tumor Incidence and Burden at End Point for Each Brain Metastasis Model
| SUM190-BR3 |
JIMT-1-BR3 |
231-BR-HER2 |
||||||
|---|---|---|---|---|---|---|---|---|
| 500,000 Cells Injected |
175,000 Cells Injected |
100,000 Cells Injected |
||||||
| Mouse ID | # of Tumors | Total Tumor Volume (mm3) | Mouse ID | # of Tumors | Total Tumor Volume (mm3) | Mouse ID | # of Tumors | Total Tumor Volume (mm3) |
| 1 | 1 | 0.0913 | 9 | 28 | 5.3570 | 1 | 9 | 1.6861 |
| 7 | 1 | 0.1697 | 10 | 26 | 7.5645 | 2 | 11 | 10.7690 |
| 8 | 2 | 0.6510 | 12 | 20 | 2.8284 | 3 | 54 | 30.9075 |
| 19 | 1 | 0.1665 | 13 | 5 | 1.4802 | 4 | 21 | 7.2567 |
| 15 | 8 | 1.0904 | 5 | 7 | 4.6846 | |||
| 17 | 4 | 0.6696 | ||||||
| Mean | 1.25 | 0.2696 | Mean | 15.17 | 3.1650 | Mean | 20.40 | 11.06 |
| SD | ± 0.50 | 0.2568 | SD | ± 10.82 | 2.7440 | SD | ± 19.54 | 11.59 |
Number of tumors and total tumor volume are shown for each mouse, as well as average values and standard deviations within each model. Overall, SUM190-BR3 mice have the smallest tumor burden, and MDA-MB-231-BR-HER2 mice have the largest, in terms of both number of tumors and total tumor volume.
Figure 2.
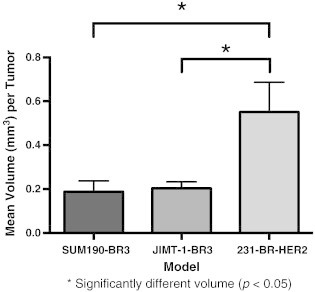
Quantification of mean volume per tumor (± SEM) at end point in SUM190-BR3, JIMT-1-BR3, and MDA-MB-231-BR-HER2 models. The average volume of a tumor was significantly different between groups (F2,12 = 5.845, P < .05), and MDA-MB-231-BR-HER2 tumors were significantly larger in size compared to JIMT-1-BR3 or SUM190-BR3 tumors. No other significant differences were observed.
The tumor permeability status for the three models is shown in Figure 3. A significant difference was observed in the percentage of Gd-permeable or Gd-impermeable tumors between models (F2,12 = 22.18, P < .0001). Post hoc Tukey tests showed the MDA-MB-231-BR-HER2 had a significantly different proportion of Gd-permeable and Gd-impermeable tumors compared to the JIMT-1-BR3 or SUM190-BR3 models. All of the SUM-190-BR3 tumors and most JIMT-1-BR3 tumors were Gd permeable, whereas only 63.6% of the MDA-MB-231-BR-HER2 tumors displayed BTB permeability to Gd. T1w SE post-Gd MR failed to detect 36.4% of MDA-MB-231-BR-HER2 metastases that were visible in bSSFP images. No other significant differences within or between groups were observed.
Figure 3.
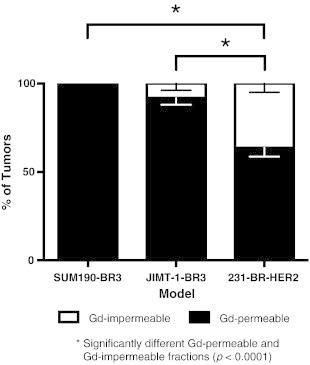
Quantification of the mean percentage of Gd-permeable (black) and Gd-permeable (white) tumors (± SEM). The percentage of Gd-permeable or Gd-impermeable tumors was significantly different between groups (F2,12 = 22.18, P < .0001). All SUM190-BR3 and most JIMT-1-BR3 tumors were Gd permeable, whereas only 63.6% of MDA-MB-231-BR-HER2 tumors display BTB permeability to Gd. Post hoc Tukey tests showed that the MDA-MB-231-BR-HER2 had a significantly different proportion of Gd-permeable and Gd-impermeable tumors compared to the JIMT-1-BR3 or SUM190-BR3 models. No other significant differences in permeability were observed.
Figure 4 shows representative bSSFP and T1w post-Gd images for a mouse with MDA-MB-231-BR-HER2 brain metastases. Three metastases were evident in the bSSFP image, yet only two of these were detectable in the T1wSE post-Gd. This suggested that the BTB associated with the third metastasis was intact.
Figure 4.
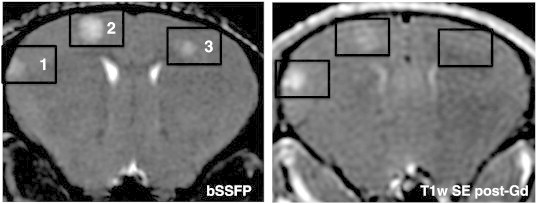
In vivo visualization of heterogeneous BTB permeability in the same animal. Axial MR images of the mouse brain; tumor burden is shown in the bSSFP image (left), and BTB permeability associated with the tumors is indicated in the T1w SE post-Gd (right). Three tumors are visualized in the bSSFP image, yet only two are detectable in the T1w SE post-Gd. This suggests that Gd cannot cross the BTB associated with tumor 3 and demonstrates the difference in tumor detection using contrast-enhanced MRI compared to the bSSFP sequence.
Immunohistochemistry was performed and evaluated, including CD31, CD105, and Ki67 stains, in an effort to understand factors that might relate to differences between Gd-permeable and Gd-impermeable tumors. Individual tumors in whole brain sections were identified as either Gd permeable or Gd impermeable by comparing to MRI. For this analysis, Gd-impermeable tumors were assessed only from the MDA-MB-231-BR-HER2 model because of their low prevalence in the other models. Ki67 is a marker for proliferative nuclei and was used to calculate the mean proliferative index for Gd-permeable and Gd-impermeable brain metastases in the MDA-MB-231-BR-HER2 model. The proliferative index was determined as the percentage of positively stained nuclei among MDA-MB-231-BR-HER2 cells. The proliferative index for Gd-impermeable tumors was significantly higher than for Gd-permeable tumors (t[24] = 2.389, P < .05) (Figure 5). CD31 is a marker for endothelial cells, and CD105 marks proliferative endothelial cells. Together, these stains visualize vasculature patterns and can indicate where new vessels are being formed. This staining was only assessed qualitatively, and it is interesting to note the different staining patterns across the three models. In the SUM190-BR3 model, CD31 staining was strongly localized to the outer edge of the tumor rim, whereas CD105 staining was strongest on the inner edge of the tumor rim. This indicated the presence of vasculature around the tumor and new vasculature development inside, near the tumor core. The JIMT-1-BR3 and MDA-MB-231-BR-HER2 models had more similar patterns of CD31 and CD105 staining; the JIMT-1-BR3 model had existing and new vasculature dispersed throughout the tumor space, whereas in MDA-MB-231-BR-HER2 tumors, this appeared to be associated near areas of edema. Despite variance in vasculature patterns across the three models, no differences were observed between Gd-permeable and Gd-impermeable tumors (Figure 6).
Figure 5.
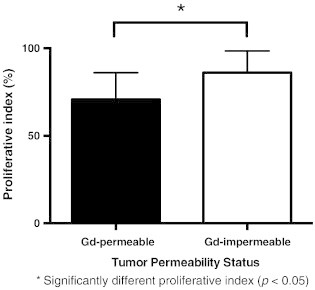
The mean proliferation index (± SD) for Gd-permeable and Gd-impermeable brain metastases in the MDA-MB-231-BR-HER2 model. This quantifies tumor proliferation by Ki67 staining and indicates that there is significantly more proliferative nuclei in Gd-impermeable compared to Gd-permeable brain metastases (t[24] = 2.389, P < .05).
Figure 6.
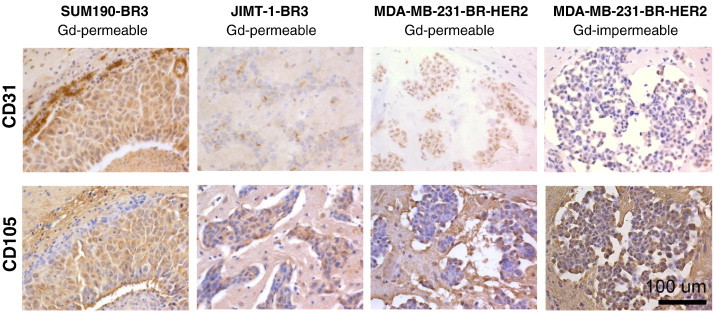
Representative images of CD31 (top) and CD105 (bottom) staining in SUM190-BR3, JIMT-1-BR3, and MDA-MB-231-BR-HER2 models. Staining patterns were qualitatively assessed for the distribution of endothelial cells and proliferative endothelial cells in tumor regions. Different staining patterns are evident between models; however, there is no obvious difference in the staining patterns between Gd-permeable and Gd-impermeable metastases within the MDA-MB-231-BR-HER2 model.
Discussion
In vivo MRI is a valuable tool for preclinical investigation of brain metastatic breast cancer and associated tumor permeability. In the current work, we used MRI to compare HER2 + brain metastases from the following three human breast cancer cell lines: (1) MDA-MB-231-BR-HER2 [19], (2) JIMT-1-BR3 [32], and (3) SUM190-BR3. We show that there are differences in MRI appearance and permeability to Gd in three different types of HER2 + brain metastases. Furthermore, these models exhibit the significant disease heterogeneity seen in the clinic; this is reflected by marked differences in tumor incidence, total tumor burden, and average size of a tumor in each of these models.
Our Gd-permeability analysis suggests that in vivo tumor permeability to Gd cannot be explained by lesion size, aggressiveness, or growth duration. Permeability to Gd indicates a disruption in the BBB, or BTB, when assessing brain tumors; this is important to understand because BBB disruption affects our ability to detect and treat brain metastases. Interestingly, models with smaller tumors—SUM190-BR3 and JIMT-1-BR3—had significantly higher proportions of Gd-permeable tumors than the MDA-MB-231-BR-HER2 model, which had many Gd-impermeable metastases despite forming much larger metastases more quickly than the other models. Moreover, qualitative analysis of staining for endothelial cells (CD31) and proliferative endothelial cells (CD105) shows different vasculature patterns across different models of HER2 + brain metastatic breast cancer but suggests no difference in development of new vasculature among Gd-permeable compared to Gd-impermeable tumors. This contradicts the dogma that large tumors have more neovasculature and are more “leaky” [24,34–36]. In addition to size, tumor aggressiveness does not explain permeability. Indeed, Gd-impermeable tumors in the MDA-MB-231-BR-HER2 model exhibited a significantly higher proliferative index than Gd-permeable tumors.
Our results align with preclinical studies by Lockman et al. [30] and Percy et al. [24] who have also demonstrated heterogeneity in the permeability status associated with brain metastases. Varying levels of passive permeability to Texas Red Dextran (3 kDa) or 14C-AIB (103 Da) were detected in MDA-MB-231-BR-HER2 metastases using ex vivo fluorescence microscopy or autoradiography, and it was found that tumor permeability was not correlated with tumor size; in addition, they suggest that it is also unrelated to morphology or vascular density [30]. Percy et al. [24] previously used the same in vivo MRI strategy used in our study and found that Gd-permeable tumors were significantly larger than Gd-impermeable tumors in the MDA-MB-231-BR model; however, size alone was not sufficient to predict permeability. Our work supplements these findings and further illustrates that tumor size and permeability are not directly related because models with smaller tumor size had higher proportions of Gd-permeable metastases. We demonstrate that tumor permeability is not the same in all models of HER2 + brain metastatic breast cancer.
The difference in permeability status across and within the models raises two important clinical implications. Firstly, systemic therapies may be ineffective because they rely on diffusion of a therapeutic across an impaired BBB for delivery to metastases. Trastuzumab is part of the standard treatment for HER2 + metastatic breast cancer; however, this molecule does not adequately cross the BBB [37] and is ineffective for brain metastatic breast cancer [11,12]. Interestingly, our study shows almost all SUM190-BR3 and JIMT-1-BR3 tumors are permeable to Gd. This molecule is much smaller (590 Da) than trastuzumab (145 kDa) and therefore may cross the BBB where size restrictions would exclude trastuzumab. Secondly, and perhaps more importantly, the population of potentially unresponsive metastases, due to limited drug uptake, are also not likely to be detected by current Gd-enhanced imaging strategies.
Importantly, it is possible for brain metastases to develop while maintaining an intact BBB. This may happen when tumors co-opt the pre-existing rich vasculature of the brain and develop alongside pre-existing microvessels [41–43]. In this situation, a large brain metastasis can develop with an intact BTB because the tumor grows around a preformed and properly constructed blood vessel. These tumors are radiographically invisible by Gd-enhanced MRI [41] because Gd does not extravasate from the intact blood vessel; however, they are easily detectable in bSSFP images. The bSSFP sequence is advantageous to detect “Gd-impermeable” tumors because it offers high spatial resolution and contrast is related to the ratio of spin–spin to spin–lattice relaxations (T2/T1)—rather than extravasation of a contrast agent. This allows for the visualization of all brain metastases—regardless of the degree of BBB permeability. The bSSFP pulse sequence is available on clinical MRI systems but has been predominantly used for cardiac applications [38]. Further investigation of this pulse sequence for brain metastatic evaluation is warranted to determine its clinical utility.
High-resolution MRI also allowed us to quantify the number and volume of metastases in the whole brain for every mouse in each model. We found marked differences in tumor incidence, total tumor burden, and appearance of metastases across the three models despite the fact they are all meant to represent HER2 + brain metastatic breast cancer. The mean volume of MDA-MB-231-BR-HER2 metastases at end point was significantly larger than metastases in the JIMT-1-BR3 and SUM190-BR3 models. It is important to also consider the end point for each model to keep this result in context. The MDA-MB-231-BR-HER2 model is the most aggressive; it grows many large tumors, despite the least number of cells injected, and has a relatively short timeline with end point typically required by day 36. In comparison, the JIMT-1-BR3 model has a lesser tumor burden and grows significantly smaller metastases; yet end point is reached earlier at day 27/28. The SUM190-BR3 model grows tumors similar in size to the JIMT-1-BR3 model; but the total tumor burden is small, and these animals can survive more than twice as long to day 64.
In summary, the differences in appearance, number, volume, and permeability across the three models highlight significant heterogeneity in HER2 + brain metastatic breast cancer. Our findings have implications for both preclinical and clinical understanding of this disease. It is important to study new diagnostics and therapeutics in animal models that accurately reflect human disease, and the heterogeneity presented here suggests that a panel of different models may be necessary to adequately accomplish this. From a clinical standpoint, understanding the heterogeneity presented in this work will be important for improvements in detection strategies and delivery of therapeutic agents. This work emphasizes the need for personalized medicine; for example, if a patient has brain metastases similar to the 231-BR-HER2 model, it could be assumed that MRI is “blind” to approximately one third of the total brain metastatic burden present in that patient and that systemic therapies may not be effective against a large proportion of their disease; other treatment avenues, such as radiotherapy, might be pursued in this case.
3D MRI is a powerful tool to provide an in vivo comprehensive analysis of tumor incidence, tumor burden, and permeability status in preclinical models of HER2 + brain metastatic breast cancer. This information is challenging to obtain solely with ex vivo methods such as histology and immunohistochemistry, but it is important for understanding the natural heterogeneity present in experimental brain metastasis models. Understanding the disease heterogeneity presented here will be important for advancements in cancer diagnostics and therapy and for improving patient management and survival.
Acknowledgements
The authors thank Yuhua Chen for her assistance with cell culture and immunohistochemistry. We also gratefully acknowledge Drs Patricia Steeg and Brunilde Gril of the Center for Cancer Research, National Cancer Institute, for providing the cell lines used in this study and for their advice for cell culture.
Supported in part by the US Department of Defense Breast Cancer Research Program (grant W81XWH-06-2-0033), the Brain Tumour Foundation of Canada, and the Canadian Institutes of Health Research.
D. H. M. is supported by a fellowship funded by the Canadian Breast Cancer Foundation—Ontario Region, a Translational Breast Cancer Traineeship sponsored in part by the Breast Cancer Society of Canada, and a CIHR Strategic Training Program in Cancer Research and Technology Transfer award. A. F. C. is a Canada Research Chair in Oncology supported by the Canada Research Chairs Program.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that they have no competing interests.
References
- 1.Howlader N, Noone AM, Krapcho M, Garshell J, Neyman N, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, et al. SEER Cancer Statistics Review 1975-2010. Natl Cancer Inst [Internet]. Bethesda, MD; Available from: http://seer.cancer.gov/csr/1975_2010/, based on November 2012 SEER data submission, posted to the SEER website, April 2013.
- 2.Kennecke H, Yerushalmi R, Woods R, Cheang MCU, Voduc D, Speers CH, Nielsen TO, Gelmon K. Metastatic behavior of breast cancer subtypes. J Clin Oncol. 2010;28(20):3271–3277. doi: 10.1200/JCO.2009.25.9820. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/20498394] [DOI] [PubMed] [Google Scholar]
- 3.Ohno S, Ishida M, Kataoka A, Murakami S. Brain metastasis of breast cancer. Breast Cancer. 2004;11(1):27–29. doi: 10.1007/BF02967997. [DOI] [PubMed] [Google Scholar]
- 4.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360(8):790–800. doi: 10.1056/NEJMra0801289. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/19228622] [DOI] [PubMed] [Google Scholar]
- 5.Vanden Bempt I, Drijkoningen M, De Wolf-Peeters C. The complexity of genotypic alterations underlying HER2-positive breast cancer: an explanation for its clinical heterogeneity. Curr Opin Oncol. 2007;19(6):552–557. doi: 10.1097/CCO.0b013e3282f0ad8e. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/17906451] [DOI] [PubMed] [Google Scholar]
- 6.Holliday DL, Speirs V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011;13(4):215–222. doi: 10.1186/bcr2889. [[Internet] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3236329&tool=pmcentrez&rendertype=abstract] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niwińska A, Murawska M, Pogoda K. Breast cancer brain metastases: differences in survival depending on biological subtype, RPA RTOG prognostic class and systemic treatment after whole-brain radiotherapy (WBRT) Ann Oncol. 2010;21(5):942–948. doi: 10.1093/annonc/mdp407. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/19840953] [DOI] [PubMed] [Google Scholar]
- 8.Pestalozzi BC. Brain metastases and subtypes of breast cancer. Ann Oncol. 2009;20(5):803–805. doi: 10.1093/annonc/mdp246. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/19403934] [DOI] [PubMed] [Google Scholar]
- 9.Clayton AJ, Danson S, Jolly S, Ryder WDJ, Burt PA, Stewart AL, Wilkinson PM, Welch RS, Magee B, Wilson G. Incidence of cerebral metastases in patients treated with trastuzumab for metastatic breast cancer. Br J Cancer. 2004;91(4):639–643. doi: 10.1038/sj.bjc.6601970. [[Internet] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2364775&tool=pmcentrez&rendertype=abstract] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slamon D, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344(11):783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 11.Bendell JC, Domchek SM, Burstein HJ, Harris L, Younger J, Kuter I, Bunnell C, Rue M, Gelman R, Winer E. Central nervous system metastases in women who receive trastuzumab-based therapy for metastatic breast carcinoma. Cancer. 2003;97(12):2972–2977. doi: 10.1002/cncr.11436. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/12784331] [DOI] [PubMed] [Google Scholar]
- 12.Lai R, Dang CT, Malkin MG, Abrey LE. The risk of central nervous system metastases after trastuzumab therapy in patients with breast carcinoma. Cancer. 2004;101(4):810–816. doi: 10.1002/cncr.20418. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/15305414] [DOI] [PubMed] [Google Scholar]
- 13.Shmueli E, Wigler N, Inbar M. Central nervous system progression among patients with metastatic breast cancer responding to trastuzumab treatment. Eur J Cancer. 2004;40(3):379–382. doi: 10.1016/j.ejca.2003.09.018. [[Internet] Available from: http://linkinghub.elsevier.com/retrieve/pii/S095980490300813X] [DOI] [PubMed] [Google Scholar]
- 14.Stemmler HJ, Kahlert S, Siekiera W, Untch M, Heinrich B, Heinemann V. Characteristics of patients with brain metastases receiving trastuzumab for HER2 overexpressing metastatic breast cancer. Breast. 2006;15(2):219–225. doi: 10.1016/j.breast.2005.04.017. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/16026983] [DOI] [PubMed] [Google Scholar]
- 15.Deeken JF, Löscher W. The blood–brain barrier and cancer: transporters, treatment, and Trojan horses. Clin Cancer Res. 2007;13(6):1663–1674. doi: 10.1158/1078-0432.CCR-06-2854. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/17363519] [DOI] [PubMed] [Google Scholar]
- 16.Connell JJ, Chatain G, Cornelissen B, Vallis KA, Hamilton A, Seymour L, Anthony DC, Sibson NR. Selective permeabilization of the blood–brain barrier at sites of metastasis. J Natl Cancer Inst. 2013;105(21):1634–1643. doi: 10.1093/jnci/djt276. [[Internet] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3818170&tool=pmcentrez&rendertype=abstract] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rye PD, Norum L, Olsen D-R, Garman-Vik S, Kaul S, Fodstad O. Brain metastasis model in athymic nude mice using a novel MUC1-secreting human breast-cancer cell line, MA11. Int J Cancer. 1996;68:682–687. doi: 10.1002/(SICI)1097-0215(19961127)68:5<682::AID-IJC20>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 18.Yoneda T, Williams PJ, Hiraga T, Niewolna M, Nishimura R. A bone-seeking clone exhibits different biological properties from the MDA-MB-231 parental human breast cancer cells and a brain-seeking clone in vivo and in vitro. J Bone Miner Res. 2001;16(8):1486–1495. doi: 10.1359/jbmr.2001.16.8.1486. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/11499871] [DOI] [PubMed] [Google Scholar]
- 19.Palmieri D, Bronder JL, Herring JM, Yoneda T, Weil RJ, Stark AM, Kurek R, Vega-Walle E, Feigenbaum L, Halverson D. Her-2 overexpression increases the metastatic outgrowth of breast cancer cells in the brain. Cancer Res. 2007;67(9):4190–4198. doi: 10.1158/0008-5472.CAN-06-3316. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/17483330] [DOI] [PubMed] [Google Scholar]
- 20.Tanner M, Kapanen AI, Junttila T, Raheem O, Grenman S, Elo J, Elenius K, Isola J. 2004. Characterization of a novel cell line established from a patient with Herceptin-resistant breast cancer; pp. 1585–1592. [PubMed] [Google Scholar]
- 21.Heyn C, Ronald JA, Ramadan SS, Snir JA, Barry AM, MacKenzie LT, Mikulis DJ, Palmieri D, Bronder JL, Steeg PS. In vivo MRI of cancer cell fate at the single-cell level in a mouse model of breast cancer metastasis to the brain. Magn Reson Med. 2006;56(5):1001–1010. doi: 10.1002/mrm.21029. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/17029229] [DOI] [PubMed] [Google Scholar]
- 22.Fitzgerald DP, Palmieri D, Hua E, Hargrave E, Herring JM, Qian Y, Vega-Valle E, Weil RJ, Stark AM, Vortmeyer AO. Reactive glia are recruited by highly proliferative brain metastases of breast cancer and promote tumor cell colonization. Clin Exp Metastasis. 2008;25(7):799–810. doi: 10.1007/s10585-008-9193-z. [[Internet] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2679391&tool=pmcentrez&rendertype=abstract] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ribot EJ, Martinez-Santiesteban FM, Simedrea C, Steeg PS, Chambers AF, Rutt BK, Foster PJ. In vivo single scan detection of both iron-labeled cells and breast cancer metastases in the mouse brain using balanced steady-state free precession imaging at 1.5 T. J Magn Reson Imaging. 2011;34(1):231–238. doi: 10.1002/jmri.22593. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/21698713] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Percy DB, Ribot EJ, Chen Y, McFadden C, Simedrea C, Steeg PS, Chambers AF, Foster PJ. In vivo characterization of changing blood-tumor barrier permeability in a mouse model of breast cancer metastasis: a complementary magnetic resonance imaging approach. Invest Radiol. 2011;46(11):718–725. doi: 10.1097/RLI.0b013e318226c427. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/21788908] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perera M, Ribot E, Percy DB, McFadden C, Simedrea C, Palmieri D, Chambers AF, Foster PJ. In vivo magnetic resonance imaging for investigating the development and distribution of experimental brain metastases due to breast cancer. Transl Oncol. 2012;5(3):217–225. doi: 10.1593/tlo.12109. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3384276/] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kil WJ, Cerna D, Burgan WE, Beam K, Carter D, Steeg PS, Tofilon PJ, Camphausen K. In vitro and in vivo radiosensitization induced by the DNA methylating agent temozolomide. Clin Cancer Res. 2008;14(3):931–938. doi: 10.1158/1078-0432.CCR-07-1856. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/18245557] [DOI] [PubMed] [Google Scholar]
- 27.McGowan PM, Simedrea C, Ribot EJ, Foster PJ, Palmieri D, Steeg PS, Allan AL, Chambers AF. Notch1 inhibition alters the CD44hi/CD24lo population and reduces the formation of brain metastases from breast cancer. Mol Cancer Res. 2011;9(7):834–844. doi: 10.1158/1541-7786.MCR-10-0457. [[Internet] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3140630&tool=pmcentrez&rendertype=abstract] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taskar KS, Rudraraju V, Mittapalli RK, Samala R, Thorsheim HR, Lockman J, Gril B, Hua E, Palmieri D, Polli JW. Lapatinib distribution in HER2 overexpressing experimental brain metastases of breast cancer. Pharm Res. 2012;29(3):770–781. doi: 10.1007/s11095-011-0601-8. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/22011930] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gril B, Palmieri D, Bronder JL, Herring JM, Vega-Valle E, Feigenbaum L, Liewehr DJ, Steinberg SM, Merino MJ, Rubin SD. Effect of lapatinib on the outgrowth of metastatic breast cancer cells to the brain. J Natl Cancer Inst. 2008;100(15):1092–1103. doi: 10.1093/jnci/djn216. [[Internet] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2575427&tool=pmcentrez&rendertype=abstract] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lockman PR, Mittapalli RK, Taskar KS, Rudraraju V, Gril B, Bohn K, Adkins CE, Roberts A, Thorsheim HR, Gaasch JA. Heterogeneous blood–tumor barrier permeability determines drug efficacy in experimental brain metastases of breast cancer. Clin Cancer Res. 2010;16(23):5664–5678. doi: 10.1158/1078-0432.CCR-10-1564. [[Internet] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2999649&tool=pmcentrez&rendertype=abstract] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morikawa A, Peereboom DM, Thorsheim HR, Samala R, Balyan R, Murphy CG, Lockman PR, Simmons A, Weil RJ, Tabar V. Capecitabine and lapatinib uptake in surgically resected brain metastases from metastatic breast cancer patients: a prospective study. Neuro Oncol. 2014:1–7. doi: 10.1093/neuonc/nou141. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/25015089] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmieri D, Duchnowska R, Woditschka S, Hua E, Qian Y, Biernat W, Sosinsa-Mielcarek K, Gril B, Stark AM, Hewitt SM. Profound prevention of experimental brain metastases of breast cancer by temozolomide in an MGMT-dependent manner. Clin Cancer Res. 2014;20(10):2727–2739. doi: 10.1158/1078-0432.CCR-13-2588. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/24634373] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forozan F, Veldman R, Ammerman CA, Parsa NZ, Kallioniemi A, Kallioniemi O, Ethier SP. Molecular cytogenetic analysis of 11 new breast cancer cell lines. Br J Cancer. 1999;81(8):1328–1334. doi: 10.1038/sj.bjc.6695007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang R, Price JE, Fujimaki T, Bucana CD, Fidler IJ. Differential permeability of the blood–brain barrier in experimental brain metastases produced by human neoplasms implanted into nude mice. Am J Pathol. 1992;141(5):1115–1124. [PMC free article] [PubMed] [Google Scholar]
- 35.Strugar J, Rothbart D, Harrington W, Criscuolo G. Vascular permeability factor in brain metastases: correlation with vasogenic brain edema and tumor angiogenesis. J Neurosurg. 1994;81:560–566. doi: 10.3171/jns.1994.81.4.0560. [DOI] [PubMed] [Google Scholar]
- 36.Thorsen F, Fite B, Mahakian LM, Seo JW, Qin S, Harrison V, Johnson S, Ingham E, Caskey C, Sundstrøm T. Multimodal imaging enables early detection and characterization of changes in tumor permeability of brain metastases. J Control Release. 2013;172(3):812–822. doi: 10.1016/j.jconrel.2013.10.019. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/24161382] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stemmler H-J, Schmitt M, Willems A, Bernhard H, Harbeck N, Heinemann V. Ratio of trastuzumab levels in serum and cerebrospinal fluid is altered in HER2-positive breast cancer patients with brain metastases and impairment of blood–brain barrier. Anticancer Drugs. 2007;18(1):23–28. doi: 10.1097/01.cad.0000236313.50833.ee. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/17159499] [DOI] [PubMed] [Google Scholar]
- 38.Leenders WPJ, Küsters B, Verrijp K, Maass C, Wesseling P, Heerschap A, Ruiter D, Ryan A, de Waal R. Antiangiogenic therapy of cerebral melanoma metastases results in sustained tumor progression via vessel co-option. Clin Cancer Res. 2004;10:6222–6230. doi: 10.1158/1078-0432.CCR-04-0823. [DOI] [PubMed] [Google Scholar]
- 39.Döme B, Hendrix MJC, Paku S, Tóvári J, Tímár J. Alternative vascularization mechanisms in cancer: pathology and therapeutic implications. Am J Pathol. 2007;170(1):1–15. doi: 10.2353/ajpath.2007.060302. [[Internet] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1762709&tool=pmcentrez&rendertype=abstract] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donnem T, Hu J, Ferguson M, Adighibe O, Snell C, Harris AL, Gatter KC, Pezzella F. Vessel co-option in primary human tumors and metastases: an obstacle to effective anti-angiogenic treatment? Cancer Med. 2013;2(4):427–436. doi: 10.1002/cam4.105. [[Internet] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3799277&tool=pmcentrez&rendertype=abstract] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sung K, Lee H-L, Hu HH, Nayak KS. Prediction of myocardial signal during CINE balanced SSFP imaging. Magn Reson Mater Phys. 2010;23(2):85–91. doi: 10.1007/s10334-010-0202-2. [[Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/20229086] [DOI] [PMC free article] [PubMed] [Google Scholar]


