Abstract
Objective
To evaluate the role of copy number abnormalities detectable by chromosomal microarray (CMA) testing in patients with epilepsy at a tertiary care center.
Methods
We identified patients with ICD-9 codes for epilepsy or seizures and clinical CMA testing performed between October 2006 and February 2011 at Boston Children’s Hospital. We reviewed medical records and included patients meeting criteria for epilepsy. We phenotypically characterized patients with epilepsy-associated abnormalities on CMA.
Results
Of 973 patients who had CMA and ICD-9 codes for epilepsy or seizures, 805 patients satisfied criteria for epilepsy. We observed 437 copy number variants (CNVs) in 323 patients (1–4 per patient), including 185 (42%) deletions and 252 (58%) duplications. Forty (9%) were confirmed de novo, 186 (43%) were inherited, and parental data were unavailable for 211 (48%). Excluding full chromosome trisomies, CNV size ranged from 18 kb to 142 Mb, and 34% were over 500 kb. In at least 40 cases (5%), the epilepsy phenotype was explained by a CNV, including 29 patients with epilepsy-associated syndromes and 11 with likely disease-associated CNVs involving epilepsy genes or “hotspots.” We observed numerous recurrent CNVs including 10 involving loss or gain of Xp22.31, a region described in patients with and without epilepsy.
Interpretation
Copy number abnormalities play an important role in patients with epilepsy. Given that the diagnostic yield of CMA for epilepsy patients is similar to the yield in autism spectrum disorders and in prenatal diagnosis, for which published guidelines recommend testing with CMA, we recommend the implementation of CMA in the evaluation of unexplained epilepsy.
Keywords: Epilepsy, array comparative genomic hybridization (aCGH), chromosomal microarray, copy number variants, deletions, duplications, chromosomal abnormalities
Introduction
Copy number variants (CNVs) play an increasingly recognized role in the genetics of epilepsy. Several large studies have identified recurrent genomic “hotspots” that predispose to idiopathic epilepsy, including 1q21.1, 15q11.2, 15q13.3, 15q11-q13, 16p11.2, and 16p13.11.1–4 CNVs occur in these regions due to non-allelic homologous recombination between flanking segmental duplications. The specific genes responsible for the susceptibility to epilepsy in these CNVs have not been clearly identified in many instances, though CNVs involving known epilepsy genes (e.g., SCN1A or KCNQ2) have also been found.5–11 Rare, non-recurrent CNVs have also been associated with epilepsy.3 “Large” CNVs, greater than 500 kb, are more often pathogenic than smaller CNVs.12 CNVs in patients with epilepsy, especially patients who also have intellectual disabilities (ID) and other neuropsychiatric symptoms, are often larger and more gene-rich than those in controls.13 One study of 102 patients with epilepsy, with or without additional neurodevelopmental abnormalities, identified a likely disease-causing CNV in 10 cases (almost 10%).14
Chromosomal microarrays (CMAs) have been increasingly included in the evaluation of epilepsy when a genetic etiology is suspected. Such suspicions may be raised by the presence of dysmorphic features, developmental delay (DD), autism spectrum disorder (ASD), or family history of epilepsy. Nonetheless, there are no formal guidelines yet that include CMA in the evaluation of epilepsy patients. We sought to describe our experience with diagnostic CMA in a cohort of 805 patients with epilepsy in order to demonstrate the yield of CMA in a series of patients with epilepsy ascertained clinically and to identify novel CNVs as possible candidate regions for future study.
Methods
Patient ascertainment
This study was approved by the Boston Children’s Hospital Institutional Review Board. We identified patients with an ICD-9 code for epilepsy or seizures who had a clinical CMA performed at Boston Children’s Hospital between October 2006 and February 2011. Two pediatric neurologists trained in epilepsy (HEO and JA) reviewed medical records for each patient to confirm the diagnosis of epilepsy, defined as recurrent unprovoked seizures or a single unprovoked seizure and abnormal EEG or MRI warranting treatment with anti-epileptic medication. For patients meeting inclusion criteria, we collected information about their CNVs (location, coordinates, size, genes encompassed, inheritance). For patients with CNVs consistent with known epilepsy syndromes, containing known epilepsy genes, in known epilepsy “hotspots,” or in areas with recurrent, overlapping CNVs, we analyzed phenotypic data and classified each patient with focal vs. generalized epilepsy, epilepsy syndrome, other diagnoses, imaging abnormalities, other genetic or metabolic testing results, risk factors for epilepsy, neuropsychiatric co-morbidities, and family history of epilepsy or febrile seizures. Pathogenicity of CNVs was determined based on CMA report, consistency with clinical phenotype, and updated information on known epilepsy syndromes, genes, and “hotspots.”
Array-CGH
Whole genome oligonucleotide array comparative genomic hybridization (aCGH) arrays (Agilent 244 K) were used to detect CNVs greater than 35–50 kb. CMA was performed when possible in the biological parents of individuals with CNVs of unknown or possible significance. Genomic coordinates provided refer to the human genome build hg18 except where specified.
Pathway analysis
Ingenuity Pathway Analysis (IPA) was used to evaluate gene content and identify potential candidate genes for epilepsy.
Results
Diagnostic yield of CMA in epilepsy patients
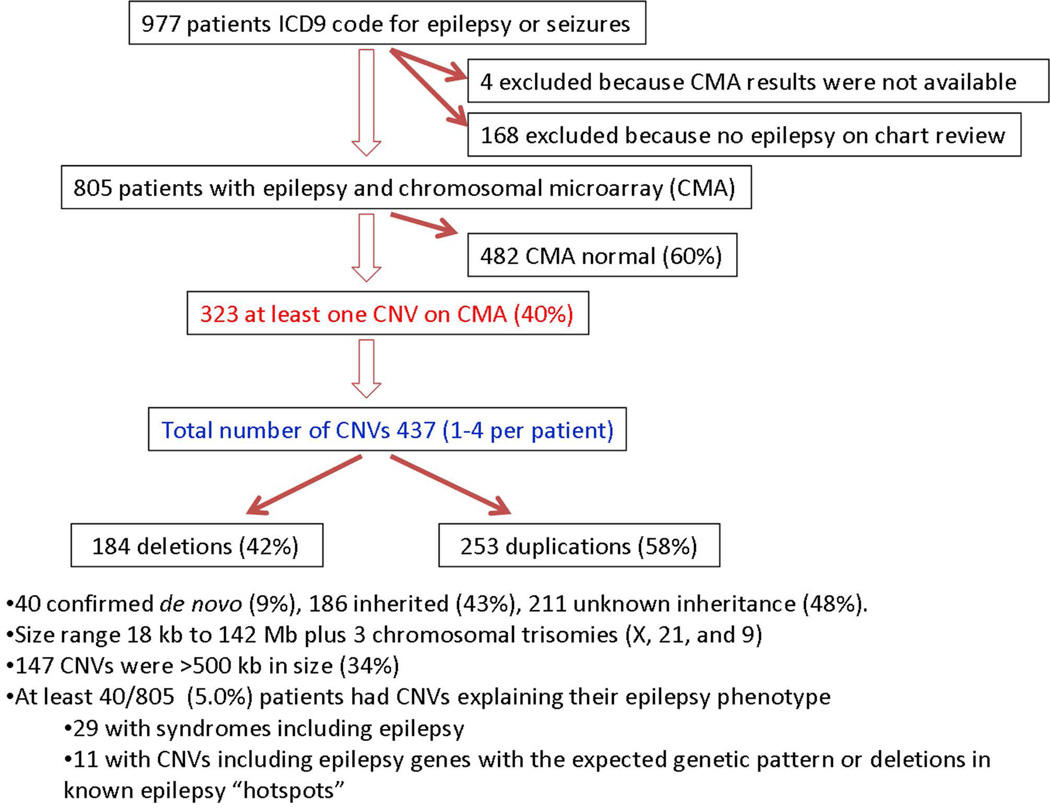
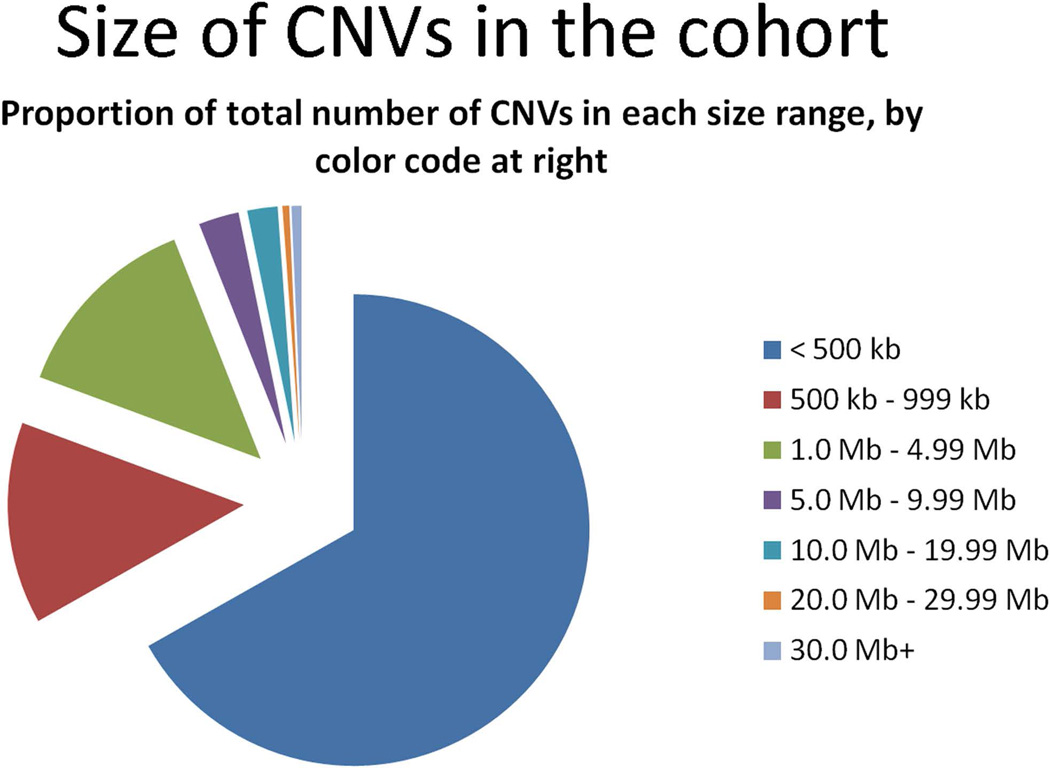
Of 6564 CMAs performed for all indications from October 2006 to February 2011, 973 patients had an ICD-9 code for epilepsy or seizures, and 168 of these were excluded because they did not have epilepsy based on record review (Figure 1). Of the 805 patients who met inclusion criteria, we identified 437 CNVs in 323 patients (40%), range 1–4 CNV per patient. This included 184 (42%) deletions and 253 (58%) duplications. Forty (9%) were confirmed de novo, 186 (43%) inherited, and 211 (48%) of unknown inheritance. Of the inherited CNVs, 101 were maternally inherited, 78 were paternally inherited, one was an inherited homozygous deletion (one copy from each parent), and six were reported inherited but the parent of origin was not specified. The size of the CNVs ranged from 18 kb to 142 Mb, not including three whole chromosomal trisomies (X, 21, and 9), and 147 (34%) were larger than 500 kb in size (Figure 2). CNVs provided an explanation for the epilepsy in at least 40 cases (5%), including 29 patients with known syndromes involving epilepsy and 11 patients with deletions in known epilepsy genes or hotspots (Tables 1–3).
Figure 1.
Flowchart of patient selection and chromosomal microarray genotype results
Figure 2.
Pie chart of CNV sizes in our cohort.
Table 1.
CNVs in our cohort in regions consistent with genetic syndromes associated with epilepsy.
| Known syndrome associated with epilepsy | Number of cases |
Coordinates |
|---|---|---|
| 22q11 duplication syndrome | 4 | Chr22: 17,086,001 – 17,390,449 Chr22: 17,274,135 – 19,835,414 Chr22: 17,274,835 – 19,835,417 Chr22: 17,270,271 – 19,891,514 |
| 1p36 deletion syndrome | 3 | Chr1: 4,010,525 – 15,294,730 Chr1: 836,543 – 2,623,228 Chr1: 1,239,023 – 2,033,572 |
| Mowat-Wilson syndrome (ZEB2 deletion, 2q22.3) | 3 | Chr2: 144,551,411 – 144898926 Chr2: 143,268,368 – 147,688,255 Chr2: 144,874,055 – 145,796,638 |
| Wolf-Hirschhorn syndrome (4p16.3 deletion) | 3 | Chr4: 0 – 8,759,904 Chr4: 41,413 – 2,762,699 Chr4: 62,447 – 13,267,439 |
| Dravet syndrome (SCN1A deletion, 2q24.3) | 2 | Chr2: 166,040,826 – 167,790,520 Chr2: 165,476,423 – 171,820,243 |
| Williams-Beuren region reciprocal syndrome (duplication 7q11.23) |
2 | Chr7: 72,039,022 – 73,780,204 Chr7: 72,338,350 – 74,993,703 |
| Kleefstra syndrome (9q34.3 deletion) | 2 | Chr9: 138,863,456 – 140,241,935 Chr9: 138,622,852 – 140,145,683 |
| Angelman syndrome (maternal 15q11-q13 deletion) | 2 | Chr15: 21,219,452 – 26,199,055 Chr15: 20,307,869 – 26,208,602 |
| Phelan-McDermid syndrome (22q13.3 deletion) | 2 | Chr22: 44,181,296 – 49,524,226 Chr22: 48,041,489 – 49,565,875 |
| MECP2 duplication syndrome (Xq28) | 2 | ChrX: 152,806,398 – 153,266,347 ChrX: 152,765,830 – 153,266,347 |
| 1q43-q44 deletion syndrome | 1 | Chr1: 235,963,770 – 247,190,718 |
| Terminal 6q deletion syndrome | 1 | Chr6: 161,333,762 – 170,748,662 |
| Benign familial neonatal convulsions (KCNQ2 deletion, 20q13.33) |
1 | Chr20: 61,040,936 – 62,379,059 |
| 22q11.2 deletion syndrome | 1 | Chr22: 17,403,824 – 19,835,417 |
|
Total # of patients with identified genetic syndromes involving epilepsy as a result of chromosomal microarray testing |
29/805 (3.6%) |
Table 3.
CNVs in cohort in regions of known epilepsy hotspots.
| Epilepsy “hotspot” |
# cases |
Coordinates (hg18) | Epilepsy Phenotype | Neuropsychiatric co- morbidities |
|---|---|---|---|---|
| 1q21.1 | 1 dup | chr1: 144,984,952–146,317,921 | Focal epilepsy with status epilepticus. |
DD, learning problems, ADHD |
| 15q11.2 | 1 dup, 1 del |
Duplication: chr15: 20,307,869–20,773,190 Deletion: chr15: 19,121,757–20,851,679 |
Duplication case - focal seizures Deletion case – neonatal seizures |
Duplication case - congenital hydrocephalus, DD, dysmorphic Deletion case – GDD |
| 15q13.3 | 1 dup, 1 del |
chr15: 29,803,656 – 30,298,096 chr15: 28,719,136 – 30,298,096 |
Duplication case - myoclonic astatic epilepsy Deletion case - primary generalized epilepsy |
Duplication case - speech delay Deletion case - GDD, ID, autistic features |
| 15q11-q13 duplication |
4 dup | 1. chr15: 18,362,555 – 30,232,544 2. chr15: 21,195,208 – 26,214,052 3. chr15: 21,208,377 – 26,208,602 4. chr15: 18,362,555 – 28,160,686 (4 copies) and 28,517,805 – 30686791 (3 copies) |
Generalized, focal or mixed epilepsy |
All with ID or autism |
| 16p11.2 | 5 dup, 4 del |
Duplications: 1. chr16: 29,572,030 −30,106,852 2. chr16: 29,258,288 – 30,106,808 3. chr16: 28,740,997 – 29,030,068 4. chr16: 21,476,688 – 29,238,851 5. chr16: 29,581,455 – 30,240,082 Deletions: 1. chr16: 2,958,202 – 30,106,252 2. chr16: 29,528,190 – 30,106,852 3. chr16: 28,394,123 – 29,238,851 4. chr16: 30308460 – 30,342,665 |
Duplication cases – focal, generalized or both with mixture of seizure types including spasms/drop attacks Deletion cases - focal or generalized seizures |
Duplication cases– one with polymicrogyria; all with DD/ID; one with autism Deletion cases – all with GDD and/or learning problems; one with macrocephaly |
| 16p13.11 | 2 dup, 3 del |
Duplications: 1. chr16: 15,034,210 – 1619,973 2. chr16: 14,687,636 – 16,199,736 Deletions: 1. chr16: 14,876,356 – 16,174,751 2. chr16: 14,876,356 – 16,199,682 3. chr16: 15,399,028 – 16,199,682 |
Duplication cases – one with neonatal seizures; one with focal seizures onset at 2 years Deletion cases – generalized or focal epilepsy |
Duplication cases – one with normal development; one with cortical dysplasia and DD/ID Deletion cases – all with DD/ID; one with autism spectrum disorder |
Because association with epilepsy is most established with deletions, only the deletions were considered in this report as pathogenic CNVs explaining the epilepsy syndromes. ADHD = attention deficit hyperactivity disorder, DD = developmental delay, GDD = global developmental delay, ID = intellectual disability.
CNVs associated with known epilepsy-associated syndromes
Twenty-nine patients (3.6%) had CNVs with well-established associations with syndromic epilepsy (Table 1). These included 22q11.2 duplication syndrome (n=4), 1p36 deletion syndrome (n=3), Mowat-Wilson syndrome (n=3), Wolf-Hirschhorn syndrome (n=3), Dravet syndrome (n=2), Williams-Beuren region reciprocal duplication syndrome (n=2), Kleefstra syndrome (2), Angelman syndrome (n=2), Phelan-McDermid syndrome (n=2), MECP2 duplication syndrome (n=2), 1q43-q44 deletion syndrome (n=1), terminal 6q deletion syndrome (n=1), benign familial neonatal convulsions (n=1), and 22q11.2 deletion syndrome (n=1). All of these patients’ signs and symptoms were consistent with the reported phenotypes associated with these CNVs, and the syndromes were considered responsible for their epilepsy. The duplication 17p11.2 was not considered the cause of the epilepsy in three patients with Potocki-Lupski syndrome because patients with this syndrome are described as having abnormal EEGs without clinical epilepsy.15
CNVs involving other known epilepsy genes
Twenty-two patients (2.7%) had CNVs containing known epilepsy genes not included in the known syndromes above, of which seven (0.9%) are likely disease-associated (Table 2). These included one homozygous deletion of PLCB116 and six heterozygous deletions involving CACNB4, CHRNA7, GABRA1, GABRG2, or PRRT2. Three of these seven (including CHRNA7 or PRRT2) overlap with the hotspots discussed below.
Table 2.
CNVs in cohort containing known epilepsy genes.
| Epilepsy gene | Pathogenic CNV(#) | Coordinates (hg18) | |
|---|---|---|---|
| PLCB1 (20p12.3) | 1 deletion, homozygous | chr20: 8,047,741 – 8,523,461 | Malignant migrating partial epilepsy of infancy (this case reported in Poduri et al. 2012) |
| CACNB4(2q23.3) | 2 deletions, heterozygous | chr2: 152,254,443 – 156,539,284 chr2: 152,062,510 – 15,285,180 |
Case 1 - GTCs and asymmetric tonic seizures, EEG generalized Case 2 - GTCs, normal EEG |
|
CHRNA7(15q13.3) *overlaps with hotspot |
1 deletion, heterozygous | chr15: 28,719,136 – 30,298,096 | Idiopathic generalized epilepsy (likely juvenile absence epilepsy) with moderate ID and hypotonia |
|
GABRA1 and GABRG2(5q34) |
1 deletion, heterozygous | chr5: 161,048,671 – 161,558,604 | Mixed focal and generalized epilepsy with GTCs and focal seizures, generalized and multifocal epileptiform activity |
|
PRRT2(16p11.2) *overlaps hotspot |
2 deletions, heterozygous | chr16: 29,582,020 – 30,106,252 chr16: 29,528,190 – 30,106,852 |
Case 1 - GTCs onset within 1st years of life and controlled in setting of GDD/ID Case 2 - GTCs in first year of life in setting of GDD, hypotonia, macrocephaly |
| Epilepsy gene | Possibly pathogenic CNV | Epilepsy phenotype | |
| NRXN1 (2p16.3) | 2 deletions, heterozygous | chr2: 51,079,674 – 51,236,317 chr2: 50,392,247 – 50,782,724 |
Case 1 - Slumping and vomiting then generalized convulsive seizures with normal EEGs, in setting of autism Case 2 - Myoclonic seizures, GTCs, and focal seizures with mixed generalized and multifocal epileptiform activity in the setting of autism |
| Epilepsy gene |
Not likely pathogenic, (duplication or heterozygous deletion in AR condition) |
Epilepsy phenotype | |
|
CNTNAP2 (7q35- q36.1) |
1 deletion, heterozygous | chr7: 145,919,587 – 146,373,456 | Focal seizures in the setting of GDD and tic disorder, right central spikes on EEG |
|
CLN3 (16p11.2) *overlaps with hotspot |
1 deletion, heterozygous | chr16: 28,394,123 – 29,238,851 | GTCs, focal onset in setting of GDD |
| GLDC(9p24.1) | 1 deletion, 1 duplication | chr9: 6,410,789 – 6,631,818 chr9: 6,516,152 – 7,003,793 |
Case 1 (deletion) – Refractory generalized epilepsy with tonic seizures onset at 2 years in setting of PVNH and autism Case 2 (duplication) – Infantile spasms then focal seizures with GDD, hypotonia |
|
KCNQ2 and CHRNA4(20q13.33) |
1 duplication | chr20: 59,721,605 – 61,720,694 | Landau-Kleffner variant |
|
CHRNA7(15q13.3) *overlaps with hotspot |
1 duplication | chr15: 29,803,656 – 30,298,096 | Myoclonic-astatic epilepsy |
| CLN5(13q22.3) | 1 duplication | chr13: 43,127,591 – 114,123,698 | Nocturnal frontal lobe epilepsy in the setting of hypomelanosis of Ito, hypotonia and DD |
| GABRA1(5q34) | 1 duplication | chr5: 161,130,609 – 161,317,552 | Focal seizures (eye deviation and limp) in the setting of fever, normal EEG, + microcephaly |
| GAMT(19p13.3) | 1 duplication | chr19: 848,315 – 1,942,330 | Benign rolandic epilepsy (+ learning disability and hypotonia). Bilateral central-temporal spikes |
| NRXN1(2p16.3) | 1 duplication | chr2: 50,412,268 – 50,729,054 | GTCs, easily controlled, in the setting of autism |
|
UBE3A(15q11.2) *overlaps with hotspot |
4 duplications | chr15: 21,195,208 – 26,214,052 chr15: 21,208,377 – 26,208,602 chr15: 18,362,555 – 28,160,686 (4 copies) and 28,517,805 – 30,686,791 (3 copies) chr15: 18,362,555 – 30,232,544 |
Case 1 - Focal seizures + autism Case 2 - Generalized seizures + autism Case 3 - Intractable mixed epilepsy (drop attacks, focal seizures, myoclonic seizures, GTC seizures) with ID Case 4 - Generalized seizures + autism |
Section 1 above are those that are likely causal of the epilepsy phenotype. Section 2 is a CNV possibly causal of the epilepsy phenotype. Those classified as possibly pathogenic are heterozygous deletions in NRXN1. Section 3 lists CNVs that are not likely to be pathogenic because they are 1) duplications in genes without known disease caused by duplications or 2) single deletions in genes associated with autosomal recessively inherited conditions. AR = autosomal recessive. DD = developmental delay, GDD = global developmental delay, GTC = generalized tonic clonic seizure, ID = intellectual disability, PVNH = periventricular nodular heterotopia.
Suspected pathogenic CNVs included one patient with migrating partial epilepsy of infancy and a homozygous deletion of PLCB1,16 a deletion previously associated with early onset epilepsy.16, 17 Two deletions included CACNB4, associated with susceptibility to idiopathic generalized epilepsy,18 in one patient with generalized epilepsy and another with generalized tonic-clonic seizures (GTCs) and normal EEG. One patient with deletion of chromosome 15q13.3, including CHRNA7 and located within the epilepsy “hotspot” 15q13.3,3, 19 presented with generalized epilepsy, global developmental delay (GDD) and autistic features, all described in patients with the typical 15q13.3 deletion.19–23 One deletion of chromosome 5q34 involved the GABA-A receptor subunit-encoding genes GABRA1 and GABRG2. Both genes are associated with susceptibility for genetic generalized epilepsies (GGE), and GABRG2 has been identified in patients with febrile seizures, GEFS+ and Dravet syndrome.24, 25 The patient had focal and generalized seizures and generalized and multifocal epileptiform activity on EEG. Two deletions involved the gene PRRT2, included in the hotspot region 16p11.2 associated with ASD and DD.26 This gene has been associated with benign familial infantile epilepsy and paroxysmal kinesigenic dyskinesia;27 our two cases had GTCs within the first year of life and GDD, but no movement disorder at last follow-up (2 and 8 years).
Two heterozygous deletions involving NRXN1 (2p16.3) were considered possibly associated with epilepsy. Homozygous loss of function of NRXN1 is associated with Pitt Hopkins-like syndrome with epilepsy; heterozygous loss of function of NRXN1 has been associated with susceptibility to GGE.28–33 Both of our patients had GTCs, and one also had myoclonic and focal seizures; both had ASD, also previously associated with NRXN1.32, 34
Some CNVs involving epilepsy genes were not thought to be sufficient to cause disease, including three heterozygous deletions of genes known to cause autosomal recessive conditions (CNTNAP2, CLN3, GLDC) and ten duplications of genes associated with epilepsy only in the setting of loss of function (KCNQ2 and CHRNA4, GABRA1, GAMT, NRXN1, UBE3A, GLDC) (Table 2). Sequencing of genes associated with autosomal recessive conditions was not done.
Known epilepsy hotspots identified by CMA
Twenty-three patients (2.9%) had CNVs in the epilepsy hotspots 1q21.1, 15q11.2, 15q13.3, 15q11-q13, 16p11.2, and 16p13.11. The epilepsy phenotypes and neuropsychiatric comorbidities of these patients are summarized in Table 3. In some instances, the phenotypes are similar to those previously reported, especially in the patients with generalized epilepsy.1–3, 21, 35
Our series contained one duplication of 1q21.1 in a patient with focal epilepsy as well as DD and learning difficulties. Previous reports have described neurodevelopmental disabilities and epilepsy in association with deletions of 1q21.1, but the association with duplications is less established.1, 3, 36, 37
One patient with a deletion of 15q11.2, in the BP1-BP2 region, had neonatal seizures and GDD. One case with a duplication of 15q11.2 had congenital hydrocephalus, and the relative role of the CNV versus the hydrocephalus in this patient’s epilepsy can-not be determined. Duplications at this locus are seen both in patients with epilepsy and in the general population.3
The epilepsy candidate gene CHRNA7 lies on chromosome 15q13.3. One patient with a duplication in this region had myoclonic astatic epilepsy (Doose syndrome), not previously reported in association with this CNV. One patient with a deletion is described above;3, 19, 21 duplications have not previously been associated with epilepsy.3, 19, 21
Four duplications in the region 15q11-q13 were detected in patients with generalized, focal, or mixed epilepsy and ID or ASD.
In the 16p11.2 region, we identified five duplications and four deletions in our series (Table 3). The first two duplications and the first two deletions include the gene PRRT2, described above. Patients with duplications exhibited a mixture of seizures types notably including spasms and drop attacks (not previously reported). One patient had polymicrogyria. The patients with deletions had focal or generalized seizures. One patient had macrocephaly. All patients with CNVs in this region had DD or learning problems.
In the 16p13.11 region, we identified two patients with duplications and three cases with deletions. One patient with a duplication had neonatal seizures and normal development, and the other had focal epilepsy, ID, and a cortical dysplasia. The patients with deletions had a mixture of generalized and focal epilepsy and ID; one had ASD. Deletions of 16p13.11 are a known risk factor for both focal and generalized epilepsies; the role of duplications as a risk factor for epilepsy is not established.1, 2, 38
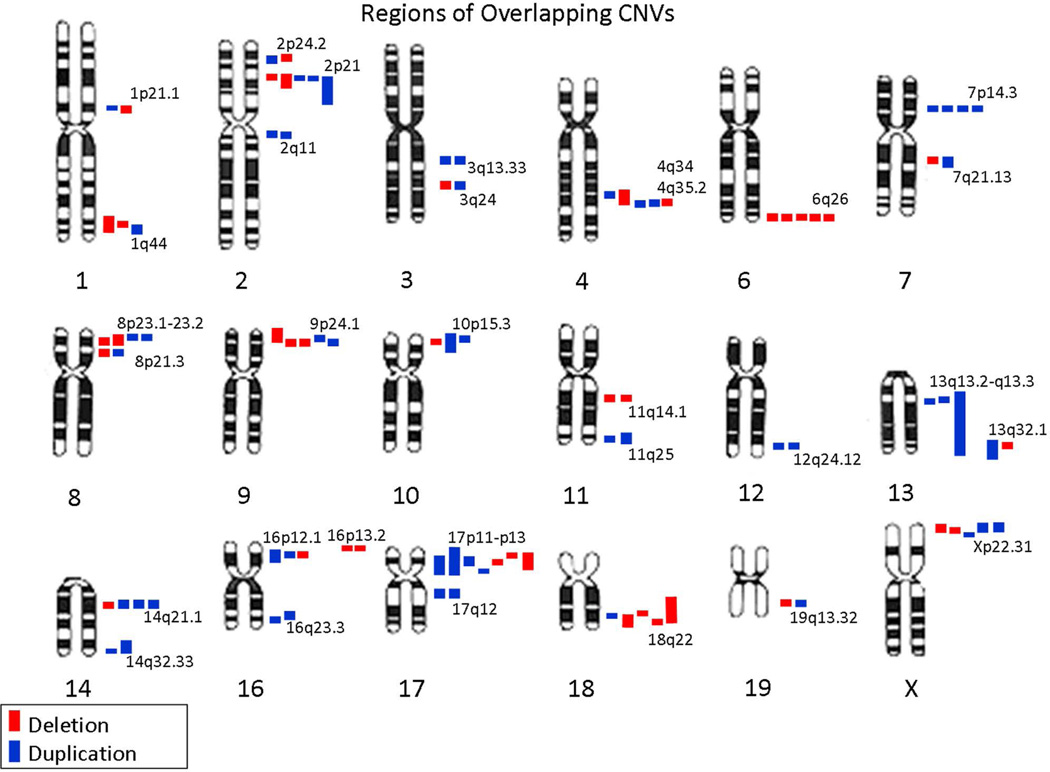
Additional regions with overlapping CNVs in our epilepsy series
Overlapping CNVs were found in numerous additional regions (Figure 3). One of these regions, Xp22.31, was identified as an area of interest because of multiple overlapping CNVs in our series and reports of patients with epilepsy. Extending the time frame from October 2006 to August 2012, we identified ten unrelated patients with epilepsy and overlapping CNVs in the Xp22.3 region (4 deletions, 6 duplications), ranging in size from 311 kb to 9.6 Mb (Table 4, Figure 4). Four had additional CNVs; the patients were a combination of males and females (Supplemental Tables 1 and 2). One of the deletions was de novo; the rest of the CNVs were inherited or of unknown inheritance. One of the duplication cases had another etiology for her epilepsy, a pathogenic missense mutation in the gene PCDH19. The other nine cases had no other identified etiology for epilepsy. The minimal region of overlap for the patients with deletions and epilepsy in this series was chrX: 7,061,905–7,920,059.
Figure 3.
Regions of overlapping CNVs in our series, not including known epilepsy genes, syndromes involving epilepsy, or epilepsy “hot spots.”
Table 4.
Epilepsy phenotypes in cases with CNVs at Xp22.31.
| Case | Xp22.31 deletions, loci and if de novo |
Other CNVs |
Gender | Age at sz onset |
Focal or generalized epilepsy |
Seizure types | EEG | Epilepsy syndrome |
|---|---|---|---|---|---|---|---|---|
| Deletion 1 | chrX:7,061,905- 7,920,059 (859 kb). De novo FISH confirmed |
Xq22.1 deletion (452 kb, de novo), 2p11.2 deletion (33kb, inherited) |
F | 2 wks |
Focal | Tonic, focal +/− 2ndary generalized |
1–2 month – Intermittent slowing right or left temporal, sharp waves right frontal and temporal, seizures right frontotemporal onset 9–10 months - normal |
None, neonatal/infantile seizures |
| Deletion 2 | chrX:6,477,006- 8,091,810 (1.6 Mb), unknown inheritance FISH confirmed |
None | M | 5 yrs | Unknown | 1 GTC and abnormal EEG |
“epileptiform activity” |
None |
| Deletion 3 | chrX:6,467,403- 8,091,751 (1.6 Mb), unknown inheritance |
None | M | 4 yrs | Focal | Focal tonic or clonic +/− 2ndary generalization |
Bilateral central- temporal spikes |
Rolandic epilepsy |
| Deletion 4 | Xp22.33-p22.2 deletion chrX:2,710,316- 12,342,188 (9.6 Mb), presumed de novo |
Xp22.2- q28 duplication (142 Mb) |
F | 11 mo |
Focal | focal, GTC | Left posterior quadrant slowing, generalized slowing, epileptiform activity left temporal and occipital or bi- posterior quadrants |
None |
|
Xp22.31 duplications, loci and if de novo |
||||||||
| Duplication 1 |
chrX: 8,413,301- 8,724,726 (311 kb, maternal inheritance) |
4p16.1 deletion (202 kb), de novo |
F | 21 mo |
Focal and Generalized |
GTC, Absence, Focal (hemi- clonic) |
Multifocal or generalized epileptiform activity |
Dravet-like onset and consistent with known mutation in PCDH19 |
| Duplication 2 |
chrX:7,788,165- 8,385,637 (597 kb), paternal inheritance |
None | F | 3 1/2 yrs |
Focal | GTC, focal | Intermittent bi- occipital slowing left more than right, slow PDR Normal at 6 yrs |
None |
| Duplication 3 |
chrX:6,455,262- 8,091,810 (1.6 Mb), unknown inheritance |
None | F | 2 mo | Generalized | Tonic, absence, myoclonic, drop attacks |
Multifocal and generalized epileptiform activity, poorly sustained PDR, lack of normal sleep features; Frequent electrographic seizures (staring and myoclonic jerks) with generalized spike and waves followed by decrements associated with jerks |
Not well defined, refractory myoclonic epilepsy |
| Duplication 4 |
chrX:6,463,313- 8,091,751 (1.6 Mb), paternal inheritance FISH confirmed |
Paternally inherited 4q32.3 duplication (609 kb) |
F | 1st week of life |
Focal | Clonic (facial twitching, never captured on EEG) |
Intermittent slowing left tempo-parietal at 8 yrs Normal as a neonate and at 9 yrs |
Neonatal seizures |
| Duplication 5 |
chrX:6,467,403- 8,091,751 (1.6 Mb), maternal inheritance |
None | F | 5 mo | Focal | Febrile SE, focal with 2ndary generalization, myoclonic |
Most normal, one with background slowing |
None |
| Duplication 6 |
chrX:6,461,822- 8,075,193 (1.7 Mb) Unknown inheritance |
None | M | 1 mo | Focal | Clonic | Normal | None |
GDD = global developmental delay, GTC = generalized tonic clonic seizure, mo = month, SE=status epilepticus, Yrs = years, Wks = weeks.
Figure 4.
Cases with epilepsy and deletions involving Xp22.31 in our October 2006 - August 2012 series, in the literature, and in the Decipher database.
In Table 4, we describe the epilepsy phenotypes: focal epilepsy in seven patients (3 deletions, 4 duplications), generalized epilepsy in one patient (duplication), focal plus generalized epilepsy in one patient (duplication), and not clearly defined as focal or generalized in one patient (deletion). The predominant seizure types were focal or GTCs. Age at seizure onset ranged from one week to five years. MRIs of the brain were normal except in two cases. One patient with a deletion of Xp22.3 had isodicentric X chromosome with duplication Xp22.2-q28 and deletion Xp22.33-p22.2. This patient had pons and cerebellar vermis hypoplasia, generalized volume loss, white matter T2 signal change, and malrotation of the hippocampi. One patient with a duplication had cerebellar hypoplasia. All of the patients had DD and ID; one had ASD. Other associated abnormalities included dysmorphic features (three deletions, one duplication) and hypotonia (three deletions, three duplications). There was no clear phenotypic distinction between patients with deletions and duplications or between males and females, and no strong evidence of epilepsy in relatives carrying the CNV (Supplemental Tables 1 and 2).
CNVs in this region were identified in 31 patients without epilepsy in the same time period. Of these 31 patients, seven had deletions (four males, three females). Fifty-eight percent (18/31) of the patients without epilepsy had DD and/or ASD. Phenotypic features of all patients with Xp22.31 deletions or duplications, with and without epilepsy, are reviewed (Supplemental Tables 1 and 2).
Alternate genetic etiologies identified in some patients with CNVs
In some patients, genetic etiologies other than the identified CNVs were identified for the epilepsy. For example, in one patient with a maternally inherited 17q12 duplication, a pathogenic mutation was identified in the gene CDKL5, and her phenotype was consistent with this gene mutation.11 One patient with four CNVs was found to have neuronal ceroid lipofuscinosis, identified by low PPT1 enzyme activity on skin biopsy. One patient with an inherited 17p13.3 deletion (not involving LIS1) was later found to have a likely pathogenic KCNQ2 mutation, and as above one patient with an Xp22.31 duplication had a pathogenic PCDH19 mutation. No alternate genetic etiology was identified in any of the patients whose CNVs were classified as likely pathogenic.
CNVs identify potential novel candidate genes for epilepsy
By evaluating the gene content of the CNVs in this series and performing pathway analysis using Ingenuity Pathway Analysis (IPA), we identified numerous potential candidate genes for epilepsy involved in neurodevelopment, synaptic transmission, ion channels, or pathways overlapping with known epilepsy genes. A summary of candidate genes identified in de novo CNVs is provided (Table 5). Additional candidate genes in CNVs of unknown inheritance >500 kb in size include EPHB1, KY, CTNNA, TDRDN, and NKAIN2 in duplications and HOMER2 in a deletion.
Table 5.
Additional candidate genes for epilepsy in confirmedde novo CNVs.
| Deletions | ||||
|---|---|---|---|---|
| AGRN | GALR1 | MAPK8IP2 | PARK2 | RTN4R |
| AKT3 | GRIN1 | MBD5 | PI4KA | SNAP29 |
| ARID1B | JAKMIP1 | MBP | PI4KAP1 | THBS2 |
| CACNA1B | KCNJ3 | NIPA1 | PMP22 | WNT7B |
| DVL1 | KIF5C | PAFAH1B1 | PRKCZ | |
| Duplications | ||||
| APBA3 | EFNB3 | MYH10 | PCDH gene cluster | |
| ARHGEF7 | EGR3 | NLGN2 | PPP3CC | |
| COL4A1 | FGF14 | NSTR1 | PURA | |
| DLG4 | GABARAP | P2RX1 | SOX6 | |
| DOC2A | MAPK3 | P2RX5 | SH3GL1 | |
A) deletions and B) duplications. These include genes coding for ion channels, proteins involved in structure or organization of the synapse or in synaptic transmission of neurotransmitters, gene regulatory proteins, and proteins involved in neurodevelopment. The PCDH gene cluster on chromosome 5q31.2-q22 includes the following genes: PCDHA1, PCDHA2, PCDHA3, PCDHA4, PCDHA7, PCDHA8, PCDHA9, PCDHA10, PCDHA11, PCDHA12, PCDHA13, PCDHAC1, PCDHAC2, PCDHB1, PCDHB2, PCDHB3, PCDHB4, PCDHB5, PCDHB6, PCDHB7, PCDHB8, PCDHB9, PCDHB10, PCDHB11, PCDHB12, PCDHB13, PCDHB14, PCDHB15, PCDHB16, PCDHB17, PCDHB18, PCDHB19P, PCDHGA1, PCDHGA2, PCDHGA3, PCDHGA4, PCDHGA5, PCDHGA6, PCDHGA7, PCDHGA8, PCDHGA9, PCDHGA10, PCDHGA11, PCDHGA12, PCDHGB1, PCDHGB2, PCDHGB3, PCDHGB4, PCDHGB5, PCDHGB6, PCDHGB7, PCDHB8P, PCDHGC3, PCDHGC4, PCDHGC5, PCDH1.
Discussion
At least 5% of pediatric epilepsy patients in our series harbor pathogenic CNVs, and additional patients harbor possibly pathogenic CNVs. While more directed testing for specific epilepsy genes or genetic syndromes is indicated for some patients based on clinical phenotype, CMA is an excellent diagnostic tool for unexplained epilepsy as it can identify structural genomic rearrangements that contribute to epileptogenesis. CMA may identify genetic causes or risk factors for epilepsy among patients for whom the phenotype does not suggest a more focused diagnostic evaluation or patients for whom the full phenotype has not yet manifested at the time when the child presents with epilepsy. We advocate for caution in attributing the etiology of epilepsy to novel CNVs. It is possible that some CNVs serve as risk factors or modifiers of disease in combination with other factors, genetic and non-genetic. More evidence is needed to determine the pathogenicity of more rare CNVs with epilepsy.
We identified patients with well-defined deletion/duplication syndromes with epilepsy included in the phenotypes. Our patients with CNVs involving known epilepsy genes had epilepsy phenotypes consistent with the literature. Two patients with heterozygous deletions of the gene NRXN1 provide further support for a gene dose effect with increased risk for epilepsy even with single copy disruptions.28–33 Further study of other CNVs identified in patients with epilepsy compared to control populations may help to further elucidate the role of chromosomal regions and individual genes in epileptogenesis and to clarify the range of associated epilepsy phenotypes.
Our data show that the epilepsy “hotspots” 1q21.1, 15q11.2, 15q13.3, 15q11-q13, 16p11.2, and 16p13.11 have relevance in a clinical setting. Prior studies identifying these hotspots focused on GGE or idiopathic focal epilepsies, but some of our patients harboring these CNVs had other epilepsy phenotypes, expanding the clinical spectrum of these CNVs. One patient with a 15q13.3 duplication involving the gene CHRNA7 had myoclonic astatic epilepsy, a novel association that needs to be replicated in additional patients to be considered robust. Two patients had neonatal seizures, one with a 15q11.2 deletion and one with a 16p13.11 duplication. It is likely, in fact, that there is considerable phenotypic variability associated with these and other CNVs, as is proving to be the case for mutations in many of the known epilepsy genes. These data thus provide support for the use of CMA as a diagnostic test in a wide range of epilepsy phenotypes if the etiology is unknown.
We reviewed the Xp22.31 region in depth. Contiguous gene syndromes caused by deletions in the region of Xp22.3 are well established.39, 40 While DD and ID are commonly reported,39–42 epilepsy is reported in only a small number of patients with CNVs involving this region.42–51 In our series, we observed epilepsy in 24% of the patients with CNVs in this region, and the CNVs overlap with those of patients with epilepsy reported in the literature and in the DECIPHER database. The critical overlapping region in patients with seizures in our series (chrX: 7,061,905–7,920,059, overlapping with most but not all cases in the literature) is not an identified hotspot for deletions in the general population, based on the Database of Genomic Variants (DGV, http://dgv.tcag.ca/dgv/app/home, accessed 9/30/13). This study provides additional evidence that CNVs in the Xp22.31 region may predispose to epilepsy, albeit with variable penetrance. Further evidence from additional cases and controls is required to determine the strength of this relationship.
Because this is an observational study, there may have been a bias toward patients with epilepsy who also had multiple congenital anomalies and/or neurodevelopmental disorders in addition to epilepsy, increasing their likelihood of having pathogenic CNVs.13 Thus, the yield of CMA in epilepsy overall may be overestimated by this series. However, the group that we studied is representative of the patient population seen in referral centers for pediatric neurology, where much of the genetic testing for epilepsy occurs.
The diagnostic yield we observed is comparable to the yield in other clinical indications for CMA, including ID, ASD, and prenatal diagnosis.4, 38, 52–55 Based on available evidence, a consensus statement in 2010 estimated that the diagnostic yield of CMA for patients with unexplained DD/ID, ASD, or multiple congenital anomalies is about 15–20%.52 Prospective studies of patients with GGE, idiopathic focal epilepsies or epileptic encephalopathies have provided similar yields, and there is evidence that up to 10% of patients with epilepsy and ID have disease associated CNVs.1–4, 56
The advantages to the patient and clinician of having an identified CNV include the following: 1) ending the often long diagnostic process, 2) providing families with prognostic information and additional features of the syndrome that may be medically relevant, and 3) in some cases providing families access to syndrome-specific research or support resources and organizations. It less often directly impacts epilepsy treatment, but as chromosomal disorders are better defined, additional prospective data on efficacy of anti-epileptic medications in each syndrome can be collected.
In conclusion, CNVs play a causative role in some cases of pediatric epilepsy. Coupled, when appropriate, with evaluation of one or more epilepsy genes, or in some cases whole exome sequencing, CMA should be included in the diagnostic evaluation of patients with unexplained epilepsy. Specifically, we recommend CMA as part of the initial evaluation of unexplained epilepsy, particularly among patients with associated dysmorphic features, DD/ID, ASD, malformations, or a history of parental consanguinity. Furthermore, identifying regions of overlapping CNVs between patients with epilepsy is a starting point toward identifying candidate genes and additional genetic causes of epilepsy.
Supplementary Material
Acknowledgments
AP receives support from the NINDS (1K23NS069784)
HEO receives support from the NINDS (5K12NS079414-02)
DM receives support from NHGRI
The authors thank all of the clinicians who were involved in the care of the patients in this report, including Mark Gorman, M.D, Audrey Woerner, M.D., and others. We also thank Ann Seman, C.G.C., from the Boston Children’s Hospital DNA Diagnostic Laboratory (now Claritas Genomics).
Footnotes
- DM is a consultant for Claritas Genomics, Inc., a majority owned subsidiary of Boston Children’s Hospital (but does not own equity).
- YS is a consultant for Claritas Genomics, Inc., a majority owned subsidiary of Boston Children’s Hospital (but does not own equity).
- BW is a consultant for Claritas Genomics, Inc., a majority owned subsidiary of Boston Children’s Hospital.
Supplemental Table 1. Xp22.31 duplications with and without epilepsy, clinical phenotypes
Supplemental Table 2. Xp22.31 deletions with and without epilepsy, clinical phenotypes
Author contributions:
H.E.O. conceived of the study and participated in study design, acquisition of data, analysis and interpretation of data, and drafting/revising the manuscript. Y.S. participated in study design, acquisition of data, analysis and interpretation of data, and critical review of the manuscript. J.A. participated in acquisition of data, analysis and interpretation of data, and critical review of the manuscript. B.R.S. participated in the analysis and interpretation of data and provided critical review of the manuscript. R.P. participated in interpretation of the data and provided critical review of the manuscript. A.M.R, G.B., F.H.D., Y.E., D.J.H., F.M.H., E.H., S.K., O.K., J.L., T.L., J.M., K.M., J.T.M., E.N.., P.C.R., M.R., A.R., E.R., C.R., M.S., D.S., A.S., S.E.S., J.S., J.M.S., M.T., W-H.T., A.R.T., P.T., D.K.U., L.W., and R.W. participated in the analysis and interpretation of clinical data and provided critical review of the manuscript. B-L.W. participated in study design and provided critical review of the manuscript. D.T.M. participated in the analysis and interpretation of data and provided critical review of the manuscript. A.P. participated in study design, analysis and interpretation of data, and drafting/revising the manuscript and provided supervision for the project. All authors approved the final version of the manuscript.
References
- 1.de Kovel CG, Trucks H, Helbig I, et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain : a journal of neurology. 2010 Jan;133(Pt 1):23–32. doi: 10.1093/brain/awp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heinzen EL, Radtke RA, Urban TJ, et al. Rare deletions at 16p13.11 predispose to a diverse spectrum of sporadic epilepsy syndromes. American journal of human genetics. 2010 May 14;86(5):707–718. doi: 10.1016/j.ajhg.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mefford HC, Muhle H, Ostertag P, et al. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS genetics. 2010 May;6(5):e1000962. doi: 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mullen SA, Carvill GL, Bellows S, et al. Copy number variants are frequent in genetic generalized epilepsy with intellectual disability. Neurology. 2013 Oct 22;81(17):1507–1514. doi: 10.1212/WNL.0b013e3182a95829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heron SE, Cox K, Grinton BE, et al. Deletions or duplications in KCNQ2 can cause benign familial neonatal seizures. Journal of medical genetics. 2007 Dec;44(12):791–796. doi: 10.1136/jmg.2007.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurahashi H, Wang JW, Ishii A, et al. Deletions involving both KCNQ2 and CHRNA4 present with benign familial neonatal seizures. Neurology. 2009 Oct 13;73(15):1214–1217. doi: 10.1212/WNL.0b013e3181bc0158. [DOI] [PubMed] [Google Scholar]
- 7.Liang JS, Shimojima K, Takayama R, et al. CDKL5 alterations lead to early epileptic encephalopathy in both genders. Epilepsia. 2011 Oct;52(10):1835–1842. doi: 10.1111/j.1528-1167.2011.03174.x. [DOI] [PubMed] [Google Scholar]
- 8.Mulley JC, Nelson P, Guerrero S, et al. A new molecular mechanism for severe myoclonic epilepsy of infancy: exonic deletions in SCN1A. Neurology. 2006 Sep 26;67(6):1094–1095. doi: 10.1212/01.wnl.0000237322.04338.2b. [DOI] [PubMed] [Google Scholar]
- 9.Striano P, Paravidino R, Sicca F, et al. West syndrome associated with 14q12 duplications harboring FOXG1. Neurology. 2011 May 3;76(18):1600–1602. doi: 10.1212/WNL.0b013e3182194bbf. [DOI] [PubMed] [Google Scholar]
- 10.Wang JW, Kurahashi H, Ishii A, et al. Microchromosomal deletions involving SCN1A and adjacent genes in severe myoclonic epilepsy in infancy. Epilepsia. 2008 Sep;49(9):1528–1534. doi: 10.1111/j.1528-1167.2008.01609.x. [DOI] [PubMed] [Google Scholar]
- 11.Olson H, Poduri A. CDKL5 mutations in early onset epilepsy: Case report and review of the literature. Journal of Pediatric Epilepsy. 2012;1:151–159. [Google Scholar]
- 12.Itsara A, Cooper GM, Baker C, et al. Population analysis of large copy number variants and hotspots of human genetic disease. American journal of human genetics. 2009 Feb;84(2):148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Striano P, Coppola A, Paravidino R, et al. Clinical significance of rare copy number variations in epilepsy: a case-control survey using microarray-based comparative genomic hybridization. Archives of neurology. 2012 Mar;69(3):322–330. doi: 10.1001/archneurol.2011.1999. [DOI] [PubMed] [Google Scholar]
- 14.Bartnik M, Szczepanik E, Derwinska K, et al. Application of array comparative genomic hybridization in 102 patients with epilepsy and additional neurodevelopmental disorders. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2012 Oct;159B(7):760–771. doi: 10.1002/ajmg.b.32081. [DOI] [PubMed] [Google Scholar]
- 15.Potocki L, Bi W, Treadwell-Deering D, et al. Characterization of Potocki-Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. American journal of human genetics. 2007 Apr;80(4):633–649. doi: 10.1086/512864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poduri A, Chopra SS, Neilan EG, et al. Homozygous PLCB1 deletion associated with malignant migrating partial seizures in infancy. Epilepsia. 2012 Aug;53(8):e146–e150. doi: 10.1111/j.1528-1167.2012.03538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurian MA, Meyer E, Vassallo G, et al. Phospholipase C beta 1 deficiency is associated with early-onset epileptic encephalopathy. Brain : a journal of neurology. 2010 Oct;133(10):2964–2970. doi: 10.1093/brain/awq238. [DOI] [PubMed] [Google Scholar]
- 18.Escayg A, De Waard M, Lee DD, et al. Coding and noncoding variation of the human calcium-channel beta4-subunit gene CACNB4 in patients with idiopathic generalized epilepsy and episodic ataxia. American journal of human genetics. 2000 May;66(5):1531–1539. doi: 10.1086/302909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masurel-Paulet A, Andrieux J, Callier P, et al. Delineation of 15q13.3 microdeletions. Clinical genetics. 2010 Aug;78(2):149–161. doi: 10.1111/j.1399-0004.2010.01374.x. [DOI] [PubMed] [Google Scholar]
- 20.Dibbens LM, Mullen S, Helbig I, et al. Familial and sporadic 15q13.3 microdeletions in idiopathic generalized epilepsy: precedent for disorders with complex inheritance. Human molecular genetics. 2009 Oct 1;18(19):3626–3631. doi: 10.1093/hmg/ddp311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helbig I, Mefford HC, Sharp AJ, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nature genetics. 2009 Feb;41(2):160–162. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller DT, Shen Y, Weiss LA, et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. Journal of medical genetics. 2009 Apr;46(4):242–248. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muhle H, Mefford HC, Obermeier T, et al. Absence seizures with intellectual disability as a phenotype of the 15q13.3 microdeletion syndrome. Epilepsia. 2011 Dec;52(12):e194–e198. doi: 10.1111/j.1528-1167.2011.03301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cossette P, Liu L, Brisebois K, et al. Mutation of GABRA1 in an autosomal dominant form of juvenile myoclonic epilepsy. Nature genetics. 2002 Jun;31(2):184–189. doi: 10.1038/ng885. [DOI] [PubMed] [Google Scholar]
- 25.Macdonald RL, Kang JQ, Gallagher MJ. GABAA Receptor Subunit Mutations and Genetic Epilepsies. In: Noebels JL, Avoli M, Rogawski MA, Olsen RW, Delgado-Escueta AV, editors. Jasper’s Basic Mechanisms of the Epilepsies. 4th. Bethesda (MD); 2012. [PubMed] [Google Scholar]
- 26.Weiss LA, Shen Y, Korn JM, et al. Association between microdeletion and microduplication at 16p11.2 and autism. The New England journal of medicine. 2008 Feb 14;358(7):667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 27.Becker F, Schubert J, Striano P, et al. PRRT2-related disorders: further PKD and ICCA cases and review of the literature. Journal of neurology. 2013 May;260(5):1234–1244. doi: 10.1007/s00415-012-6777-y. [DOI] [PubMed] [Google Scholar]
- 28.Duong L, Klitten LL, Moller RS, et al. Mutations in NRXN1 in a family multiply affected with brain disorders: NRXN1 mutations and brain disorders. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2012 Apr;159B(3):354–358. doi: 10.1002/ajmg.b.32036. [DOI] [PubMed] [Google Scholar]
- 29.Gregor A, Albrecht B, Bader I, et al. Expanding the clinical spectrum associated with defects in CNTNAP2 and NRXN1. BMC medical genetics. 2011;12:106. doi: 10.1186/1471-2350-12-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrison V, Connell L, Hayesmoore J, McParland J, Pike MG, Blair E. Compound heterozygous deletion of NRXN1 causing severe developmental delay with early onset epilepsy in two sisters. American journal of medical genetics Part A. 2011 Nov;155A(11):2826–2831. doi: 10.1002/ajmg.a.34255. [DOI] [PubMed] [Google Scholar]
- 31.Moller RS, Weber YG, Klitten LL, et al. Exon-disrupting deletions of NRXN1 in idiopathic generalized epilepsy. Epilepsia. 2013 Feb;54(2):256–264. doi: 10.1111/epi.12078. [DOI] [PubMed] [Google Scholar]
- 32.Schaaf CP, Boone PM, Sampath S, et al. Phenotypic spectrum and genotype-phenotype correlations of NRXN1 exon deletions. European journal of human genetics : EJHG. 2012 Dec;20(12):1240–1247. doi: 10.1038/ejhg.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dabell MP, Rosenfeld JA, Bader P, et al. Investigation of NRXN1 deletions: clinical and molecular characterization. American journal of medical genetics Part A. 2013 Apr;161A(4):717–731. doi: 10.1002/ajmg.a.35780. [DOI] [PubMed] [Google Scholar]
- 34.Ching MS, Shen Y, Tan WH, et al. Deletions of NRXN1 (neurexin-1) predispose to a wide spectrum of developmental disorders. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2010 Jun 5;153B(4):937–947. doi: 10.1002/ajmg.b.31063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shinawi M, Liu P, Kang SH, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. Journal of medical genetics. 2010 May;47(5):332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Basel-Vanagaite L, Goldberg-Stern H, Mimouni-Bloch A, Shkalim V, Bohm D, Kohlhase J. An emerging 1q21.1 deletion-associated neurodevelopmental phenotype. Journal of child neurology. 2011 Jan;26(1):113–116. doi: 10.1177/0883073810377658. [DOI] [PubMed] [Google Scholar]
- 37.Brunetti-Pierri N, Berg JS, Scaglia F, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nature genetics. 2008 Dec;40(12):1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hannes FD, Sharp AJ, Mefford HC, et al. Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. Journal of medical genetics. 2009 Apr;46(4):223–232. doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ballabio A, Carrozzo R, Gil A, et al. Molecular characterization of human X/Y translocations suggests their aetiology through aberrant exchange between homologous sequences on Xp and Yq. Annals of human genetics. 1989 Jan;53(Pt 1):9–14. doi: 10.1111/j.1469-1809.1989.tb01117.x. [DOI] [PubMed] [Google Scholar]
- 40.Shinawi M, Patel A, Panichkul P, Zascavage R, Peters SU, Scaglia F. The Xp contiguous deletion syndrome and autism. American journal of medical genetics Part A. 2009 Jun;149A(6):1138–1148. doi: 10.1002/ajmg.a.32833. [DOI] [PubMed] [Google Scholar]
- 41.Faletra F, D’Adamo AP, Santa Rocca M, et al. Does the 1.5 Mb microduplication in chromosome band Xp22.31 have a pathogenetic role? New contribution and a review of the literature. American journal of medical genetics Part A. 2012 Feb;158A(2):461–464. doi: 10.1002/ajmg.a.34398. [DOI] [PubMed] [Google Scholar]
- 42.Li F, Shen Y, Kohler U, et al. Interstitial microduplication of Xp22.31: Causative of intellectual disability or benign copy number variant? European journal of medical genetics. 2010 Mar-Apr;53(2):93–99. doi: 10.1016/j.ejmg.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 43.Melichar VO, Guth S, Hellebrand H, et al. A male infant with a 9.6 Mb terminal Xp deletion including the OA1 locus: Limit of viability of Xp deletions in males. American journal of medical genetics Part A. 2007 Jan 15;143(2):135–141. doi: 10.1002/ajmg.a.31451. [DOI] [PubMed] [Google Scholar]
- 44.Meindl A, Hosenfeld D, Bruckl W, et al. Analysis of a terminal Xp22.3 deletion in a patient with six monogenic disorders: implications for the mapping of X linked ocular albinism. Journal of medical genetics. 1993 Oct;30(10):838–842. doi: 10.1136/jmg.30.10.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doherty MJ, Glass IA, Bennett CL, et al. An Xp; Yq translocation causing a novel contiguous gene syndrome in brothers with generalized epilepsy, ichthyosis, and attention deficits. Epilepsia. 2003 Dec;44(12):1529–1535. doi: 10.1111/j.0013-9580.2003.61702.x. [DOI] [PubMed] [Google Scholar]
- 46.Gohlke BC, Haug K, Fukami M, et al. Interstitial deletion in Xp22.3 is associated with X linked ichthyosis, mental retardation, and epilepsy. Journal of medical genetics. 2000 Aug;37(8):600–602. doi: 10.1136/jmg.37.8.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puusepp H, Zordania R, Paal M, Bartsch O, Ounap K. Girl with partial Turner syndrome and absence epilepsy. Pediatric neurology. 2008 Apr;38(4):289–292. doi: 10.1016/j.pediatrneurol.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 48.Spranger S, Schiller S, Jauch A, et al. Leri-Weill syndrome as part of a contiguous gene syndrome at Xp22.3. American journal of medical genetics. 1999 Apr 23;83(5):367–371. doi: 10.1002/(sici)1096-8628(19990423)83:5<367::aid-ajmg5>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 49.Tobias ES, Bryce G, Farmer G, et al. Absence of learning difficulties in a hyperactive boy with a terminal Xp deletion encompassing the MRX49 locus. Journal of medical genetics. 2001 Jul;38(7):466–470. doi: 10.1136/jmg.38.7.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Steensel MA, Vreeburg M, Engelen J, et al. Contiguous gene syndrome due to a maternally inherited 8.41 Mb distal deletion of chromosome band Xp22.3 in a boy with short stature, ichthyosis, epilepsy, mental retardation, cerebral cortical heterotopias and Dandy-Walker malformation. American journal of medical genetics Part A. 2008 Nov 15;146A(22):2944–2949. doi: 10.1002/ajmg.a.32473. [DOI] [PubMed] [Google Scholar]
- 51.Milunsky J, Huang XL, Wyandt HE, Milunsky A. Schizophrenia susceptibility gene locus at Xp22.3. Clinical genetics. 1999 Jun;55(6):455–460. doi: 10.1034/j.1399-0004.1999.550610.x. [DOI] [PubMed] [Google Scholar]
- 52.Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. American journal of human genetics. 2010 May 14;86(5):749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen Y, Dies KA, Holm IA, et al. Clinical genetic testing for patients with autism spectrum disorders. Pediatrics. 2010 Apr;125(4):e727–e735. doi: 10.1542/peds.2009-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wapner RJ, Martin CL, Levy B, et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. The New England journal of medicine. 2012 Dec 6;367(23):2175–2184. doi: 10.1056/NEJMoa1203382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Battaglia A, Doccini V, Bernardini L, et al. Confirmation of chromosomal microarray as a first-tier clinical diagnostic test for individuals with developmental delay, intellectual disability, autism spectrum disorders and dysmorphic features. European journal of paediatric neurology : EJPN : official journal of the European Paediatric Neurology Society. 2013 May;:24. doi: 10.1016/j.ejpn.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 56.Mefford HC, Yendle SC, Hsu C, et al. Rare copy number variants are an important cause of epileptic encephalopathies. Annals of neurology. 2011 Dec;70(6):974–985. doi: 10.1002/ana.22645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.