Abstract
N6-methyladenosine (m6A) is the most abundant modified base in eukaryotic mRNA and has been linked to diverse effects on mRNA fate. Current m6A mapping approaches localize m6A residues to 100–200 nt-long regions of transcripts. The precise position of m6A in mRNAs cannot be identified on a transcriptome-wide level because there are no chemical methods to distinguish between m6A and adenosine. Here we show that anti-m6A antibodies can induce specific mutational signatures at m6A residues after ultraviolet light-induced antibody-RNA crosslinking and reverse transcription. We find these antibodies similarly induce mutational signatures at N6,2′-O-dimethyladenosine (m6Am), a nucleotide found at the first encoded position of certain mRNAs. Using these mutational signatures, we map m6A and m6Am at single-nucleotide resolution in human and mouse mRNA and identify snoRNAs as a novel class of m6A-containing ncRNAs.
N6-methyladenosine (m6A) is the most prevalent modified base in mRNA1–4. Although m6A was detected in poly(A)+ RNA in the 1970s3,4, the presence of m6A in mRNA was not widely accepted until recent transcriptome-wide m6A mapping studies revealed that m6A is found in several thousand transcripts, typically near the stop codon, but also in the coding sequence, 3′UTR, and 5′UTR of mRNAs1,2.
The current m6A mapping approach, methyl-RNA immunoprecipitation and sequencing (MeRIP-Seq, also called m6A-Seq)1,2, involves immunoprecipitation of ~100 nt-long RNA fragments using m6A-specific antibodies. Because immunoprecipitated RNA fragments can contain m6A anywhere along their length, multiple different m6A-containing fragments generate overlapping reads. These reads produce a peak whose summit reflects an underlying m6A residue1. Thus, MeRIP-Seq generates m6A peaks, but does not identify specific m6A residues.
Identifying m6A residues is challenging. Adenosine methylation appears to be restricted to adenosines in a R-A*-C context (R=G or A; A*=methylatable A). Early studies linked m6A to an extended consensus motif of N1-R-A*-C-N2 where N1 is A or G in 90% of the cases and N2 is very rarely a G5. Recent transcriptome-wide m6A mapping approaches suggest this broader consensus is best reflected by the motif DRACH (D=A, G or U; H=A, C or U)1,2,6. Although DRACH motifs are prevalent, only some are methylated in vivo1,2. Exact positions of m6A residues can be bioinformatically predicted from MeRIP-Seq peaks by searching for the presence of a subset of DRACH motifs near the point of highest read coverage6. However, this approach is complicated because m6A often appears in clusters which can result in large peaks spanning several m6A residues1. Additionally, multiple DRACH motifs can be present underneath a peak, making it difficult to predict the specific methylated adenosine.
There is no chemical method that results in selective modification and detection of m6A residues. This contrasts with other base modifications, such as 5-methylcytosine (5mC), in which chemical conversion of cytosine is used to map 5mC at single-nucleotide resolution in RNA7. The nearly identical chemical properties of A and m6A have prevented the development of a chemical method to distinguish these nucleotides. Additionally, unlike other base modifications, m6A does not introduce errors during reverse transcription that would allow direct mapping of its position8. Thus, a major goal is to develop a method that provides a specific chemical signature that indicates the precise location of m6A residues in the transcriptome.
We reasoned anti-m6A antibodies could be used to create such a highly selective mark. In this approach, the antibody would be crosslinked to RNA using UV light to create antibody-RNA crosslinks. Reverse transcription of crosslinked RNA would then result in mutations or truncations in the cDNA that indicate the presence of the bound protein. This resembles RNA crosslinking and immunoprecipitation based techniques (CLIP) that crosslink proteins to RNA in living cells to map RNA-binding sites throughout the transcriptome9–12.
Here, we show that certain anti-m6A antibodies induce specific mutational signatures that enable precise identification of m6A residues in RNA. In this approach, anti-m6A antibodies are crosslinked to RNA using UV light to create antibody-RNA crosslinks. Reverse transcription of crosslinked RNA then results in a highly specific pattern of mutations or truncations in the cDNA. These mutational signatures also enable identification of the related modified nucleotide, N6,2′-O-dimethyladenosine (m6Am), which is the first nucleotide following the 7-methylguanosine cap of certain mRNAs13. Using these signatures we map m6A and m6Am residues throughout the transcriptome at single-nucleotide resolution.
Results
Determining the mutational profile of anti-m6A antibodies
Because UV-induced RNA-protein crosslinks can be highly variable14, we first established the mutational signature of different commercially available anti-m6A antibodies. Since each antibody presumably has a different antigen-binding pocket for m6A, we reasoned that different antibodies would produce different types of mutations in cDNA, and that some antibodies might induce consistent mutations that can be used to locate m6A residues.
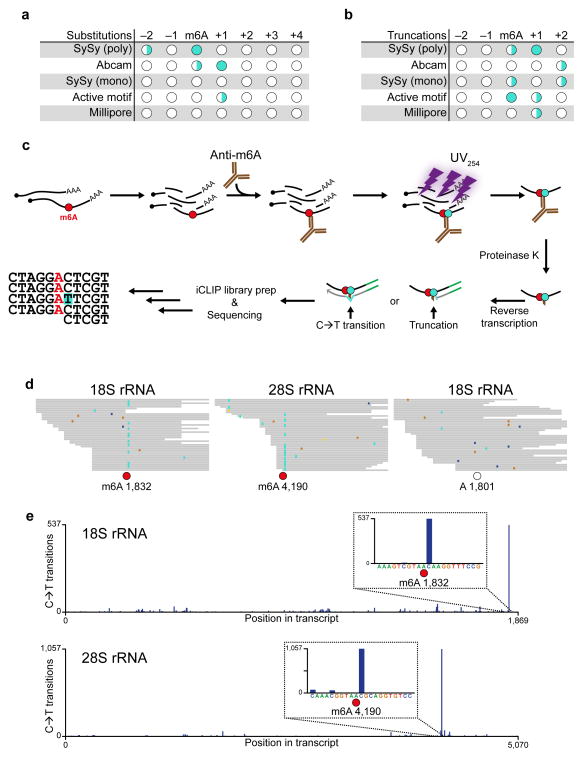
To test this, we crosslinked an in vitro-transcribed RNA containing a single m6A to different anti-m6A antibodies. The RNA was reverse transcribed, and the cDNA was sequenced to identify nucleotide substitutions and/or truncations created during reverse transcription10,15. Some antibodies induced an unpredictable pattern of substitutions at different positions relative to the m6A residue, making it difficult to unambiguously determine the position of the m6A (Fig. 1a,b). However, other antibodies resulted in more consistent patterns of substitutions. For example, the Abcam antibody induced a nucleotide substitution at the invariant cytosine residue adjacent to the m6A as well as at the m6A itself (Fig. 1a).
Figure 1. Anti-m6A antibodies induce signature mutations that directly indicate the location of m6A.
(a) In vitro transcribed RNA containing a single m6A was UV-crosslinked to different antibodies, purified, reverse transcribed and sequenced. Positions with crosslinking-induced single-nucleotide substitutions were identified and categorized into high (●), medium (◑) and low (○) frequency sites (>25%, 5–25%, and <5%, respectively).
(b) Similar to a, but with analysis of truncations instead of substitutions.
(c) Schematic of the miCLIP protocol. Purified cellular RNA is fragmented and incubated with an anti-m6A antibody. After crosslinking with UV light (254 nm), covalently bound antibody-RNA complexes are recovered by protein A/G-affinity purification, SDS-PAGE and nitrocellulose membrane transfer. RNA is then released from the membrane by proteinase K and reverse transcribed. Peptide fragments that remain on the RNA lead to nucleotide incorporation errors (indicated as C→T transition) and cDNA truncations.
(d) miCLIP reads from HEK293 total RNA were aligned to rRNA and analyzed for mismatches at the two most abundant cellular m6A residues (red circles) at positions 1,832 and 4,190 of 18S and 28S rRNA, respectively. As a control, a non-methylated DRACH consensus site at position 1,801 of 18S rRNA was analyzed (open circle). C→T transitions are shown in turquoise, other single-nucleotide mismatches are indicated by blue, brown and yellow colors.
(e) Quantitative representation of the C→T transitions in 18S (upper panel) and 28S (lower panel) rRNAs.
In addition to substitutions, we also analyzed antibodies for their ability to induce truncations during reverse transcription. We found that the SySy polyclonal antibody efficiently induced truncations at the +1 position relative to the m6A (Fig. 1b). Although other antibodies also induced specific patterns of mutations, the Abcam and SySy polyclonal antibodies were chosen for further investigation due to their high immunoprecipitation efficiency and predictable pattern of crosslink-induced substitutions and truncations.
Transcriptome-wide characterization of crosslinking sites
In order to exploit these mutational signatures for transcriptome-wide mapping of m6A, we developed “m6A individual-nucleotide resolution crosslinking and immunoprecipitation” (miCLIP). In miCLIP, cellular RNA is sheared and crosslinked to an anti-m6A antibody using UV-light (Fig. 1c). Antibody-crosslinked RNA fragments are then purified and converted into a cDNA library following the iCLIP protocol10. Then, crosslink-induced mutations and truncations introduced during reverse transcription are analyzed to determine precise positions of m6A throughout the transcriptome.
To test this, we generated miCLIP libraries from total cellular RNA crosslinked to the Abcam and SySy antibodies. Comparison of single-nucleotide transitions in the Abcam and SySy libraries revealed strong enrichment of C→T transitions in the Abcam library (Supplementary Fig. 1a,b). No other transitions were enriched in the Abcam library. Thus, C→T transitions may serve as a signature mutation to map m6A with miCLIP using the Abcam antibody.
To determine whether C→T transitions identify m6A in cellular RNA, we examined these mutations at known m6A residues in the human 18S and 28S rRNA16. At these m6A residues, mapped reads exhibited a high frequency of C→T transitions at the +1 position relative to the m6A, while reads covering a non-methylated site did not show such enrichment (Fig. 1d). Strikingly, quantitative analysis across the length of the 18S and 28S transcripts revealed that C→T transitions were enriched ~500 and ~1000 fold at m6A positions, respectively (Fig. 1e). Thus, C→T transitions induced by the Abcam antibody mark m6A with high specificity.
We next asked if the Abcam antibody induced other mutations near m6A residues. We therefore analyzed single-nucleotide substitutions around RAC triplets—the core m6A motif—throughout the transcriptome. The only mutation seen with high frequency at these triplets was the C→T transition at position +1 relative to the A (Supplementary Fig. 1c). Thus, the Abcam antibody induces C→T transitions at m6A in a highly position-specific manner.
We next asked if C→T transitions are a unique feature of the Abcam antibody. Notably, the SySy antibody, which primarily induces truncations, also induced C→T transitions at the +1 position. Thus, C→T transitions may be a common feature of anti-m6A antibodies. However, in the SySy library, other substitutions were present and the mutations occurred with lower positional accuracy (Supplementary Fig. 1d). Because in vitro analysis indicated that the SySy antibody efficiently induces cDNA truncations at the +1 position of m6A (Fig. 1b), truncations were used as this antibody’s mutational signature.
Identification of m6A using antibody-induced mutations
We next used miCLIP to generate two independent, transcriptome-wide maps of human m6A residues. For this, we crosslinked the Abcam and SySy antibodies to RNA purified from human embryonic kidney cells (HEK293).
We first analyzed C→T transitions induced by the Abcam antibody. We prepared short antibody-bound RNA fragments (~35 nt), and used paired-end sequencing to reduce mutation noise (see Methods). Mapping these reads to the reference genome resulted in ~40-nt wide peaks, unlike the ~200-nt wide peaks seen in MeRIP-Seq (Supplementary Fig. 2). Combining reads from four replicates resulted in identification of 80,774 peaks on mRNA and 92,068 C→T transitions at 48,694 unique genomic positions.
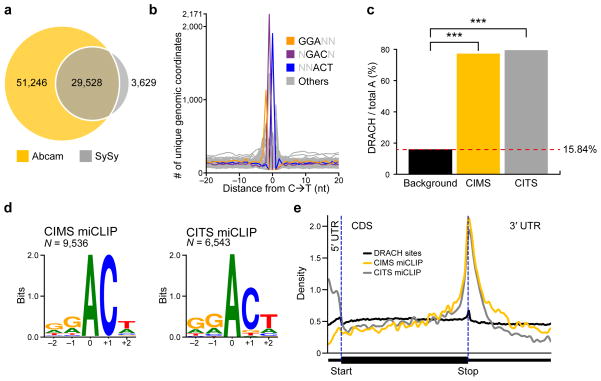
We then analyzed the miCLIP library prepared with the SySy antibody. Peaks were similar in width and overall shape to those produced by the Abcam antibody (Supplementary Fig. 2). We detected 33,157 peaks in this library that mapped to mRNAs. These peaks had a high degree of positional overlap (89.06%) with the Abcam peaks (Fig. 2a).
Figure 2. C→T transitions and truncations map m6A throughout the transcriptome.
(a) Overlap analysis of miCLIP peak clusters generated by the Abcam and SySy antibodies.
(b) Frequency plot of nucleotide triplets at C→T transitions. The most abundant 3-mers at positions −1, 0 and +1 are GGA, GAC and ACT, and are shown in color. These reconstitute the most common m6A consensus sequence GGACT. AAC, the other RAC motif known to be highly methylated, is the second most abundant 3-mer at the transition site (shown in grey here for clarity).
(c) Adenosines called by miCLIP are significantly more often in DRACH sequence context than expected by the background distribution of this motif. (P < 1 × 10−15, Fisher’s exact test).
(d) Transcripome-wide sequence logos of C→T transitions and truncations are identical to the m6A consensus motif. Weblogo analyses of the sequence environment of C→T transitions (left panel) or truncations (right panel) (m6A is at position 0).
(e) miCLIP-identified m6A residues show a metagene distribution profile that is typical for m6A. Metagene distribution plots of DRACH consensus site background (black) and the m6A residues predicted by the CIMS- (yellow) and CITS-based (grey) miCLIP.
We next sought to validate that C→T transitions induced by the Abcam antibody are found at m6A residues throughout the transcriptome. Because biochemical experiments have demonstrated that most m6A in mRNAs is located in either a GAC or AAC consensus sequence5, we analyzed whether these triplets occur in the vicinity of the transitions. Indeed, GAC and AAC were strongly enriched at transition sites (Fig. 2b). Furthermore, the triplets GGA and ACT were enriched at positions −1 and +1, respectively, recapitulating the most prevalent m6A consensus sequence, GGACT (Fig. 2b). Thus, C→T transitions predominantly occur at m6A consensus motifs.
To determine significant C→T transitions, we used a computational pipeline designed for the identification of crosslinking-induced mutation sites (CIMS) in HITS-CLIP data15 (see Methods). This resulted in a set of 11,832 called sites. This set was enriched in adenosines at the −1 position of the C→T transitions (80.66%), supporting that these transitions largely reflect m6A. Furthermore, 77.29% of these adenosines occurred in a DRACH consensus motif, a value that is significantly higher than expected by the background distribution of this motif in mRNA (Fig. 2c; P < 1 × 10−15; Fisher’s exact test). Thus, CIMS-based miCLIP (CIMS miCLIP) identified 9,536 putative m6A residues in the transcriptome (Supplementary Table 1; see also Supplementary Fig. 3b).
Identification of m6A using antibody-induced truncations
Next, we asked whether truncations induced by the SySy antibody could similarly map m6A residues in a transcriptome-wide manner. For this, we used a computational pipeline for detecting crosslinking-induced truncation sites (CITS) in CLIP data18. This resulted in 8,329 significant (P < 0.05) truncation sites that mapped to mRNAs. Most of these truncations occurred at adenosines (77.10%). Thus, CITS-based miCLIP (CITS miCLIP) identified 6,543 putative m6A sites (Supplementary Table 2). These were significantly enriched in DRACH consensus sites (Fig. 2c; 79.46%, P < 1 × 10−15; Fisher’s exact test).
Validation of m6A residues identified by miCLIP
Both, CIMS- and CITS-called sites localized predominantly in the coding sequence and the 3′UTR of mRNAs (Supplementary Fig. 3a), consistent with the known distribution of m6A1,2. Sequence logo analysis of both datasets confirmed that called sites occurred in the m6A consensus motif DRACH (Fig. 2d). Additionally, both metagene profiles followed the typical distribution of m6A with strong enrichment at the stop codon (Fig. 2e). These data suggest that miCLIP identifies true m6A residues.
Next, we examined the accuracy of m6A identification by miCLIP. We compared miCLIP sites to a control set of adenosines that were biochemically validated for their N6-methylation status using the thin-layer chromatography-based method SCARLET19. This dataset included a positive-control set of eight m6A residues in five transcripts, and 15 adenosine residues in DRACH sequence context that are not methylated.
To estimate the sensitivity and specificity of miCLIP, we determined the number of SCARLET-positive and SCARLET-negative sites that were called (Supplementary Fig. 3c). CIMS and CITS miCLIP identified six and five of the eight SCARLET-positive sites, respectively. Importantly, none of the 15 SCARLET-negative sites were called by CIMS miCLIP and only one by CITS miCLIP. Despite the small size of the SCARLET-derived set, this indicates that miCLIP detects m6A with high specificity and sensitivity.
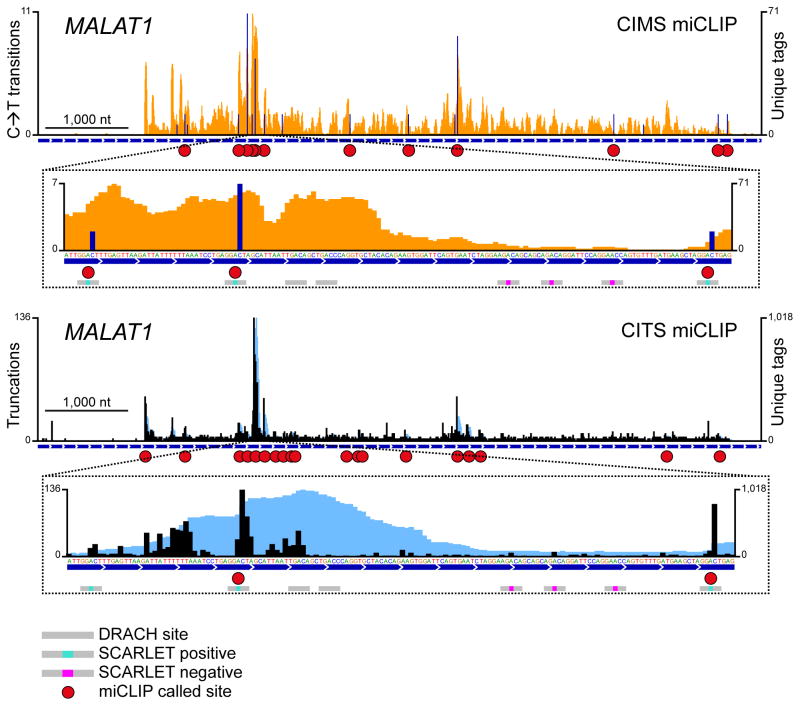
The accuracy of miCLIP can be seen on individual transcripts. For example, in MALAT1, three SCARLET-negative sites are located in a 110-nt window between two SCARLET-positive m6A residues (Fig. 3). While both SCARLET-positive sites were correctly identified by CIMS and CITS miCLIP, none of the interspersed negative sites were called. Thus, miCLIP has excellent spatial resolution and exhibits a low false-discovery rate.
Figure 3. miCLIP identifies m6A with single-nucleotide resolution.
m6A residues detected by CIMS and CITS miCLIP in the ncRNA MALAT1. Orange and dark blue tracks: CIMS miCLIP unique read coverage and C→T transitions, respectively. Light blue and black tracks: CITS miCLIP unique read coverage and unique read starts, respectively. Horizontal blue bars: Transcript models. Red circles: miCLIP called m6A. Small horizontal bars in insets: DRACH consensus sites with a methylation status that is undefined (grey), confirmed positive (turquoise) or confirmed negative (magenta) by SCARLET19.
m6A in clusters and infrequently methylated DRACH motifs
We next asked whether miCLIP can identify clustered m6As. Such regions of accumulated m6A residues have been predicted from the shape, size and distribution of MeRIP-Seq peaks1. We therefore used CIMS miCLIP to identify clustered m6A residues throughout the transcriptome (see Methods). We identified 958 clusters ranging in size from ~150 to ~500 nucleotides (Supplementary Fig. 4a) that contained up to 15 m6A residues (Supplementary Fig. 4b). The average distance between m6As in these clusters was 64 nucleotides.
Because MeRIP-Seq peaks are typically ~100–200 nucleotides wide and the bioinformatic prediction of m6A residues is limited to one site per peak6, this approach misses a substantial portion of clustered m6As. Indeed, while m6A clusters on average had 2.39 miCLIP-called m6As, they contained only 1.06 MeRIP-Seq-predicted m6As. This indicates that peak-based prediction algorithms may miss more than half of m6A residues occurring in clusters. In contrast, miCLIP identifies individual m6A residues that are separated by as little as three nucleotides (Supplementary Fig. 4c).
Bioinformatic methods utilize a predefined subset of consensus motifs to predict the m6A residue within a peak, based on the idea that some DRACH pentamers are preferred targets for methylation6. However, this approach misses m6As that occur in motifs outside of this predefined subset. As miCLIP does not require a priori assumptions about the sequence context of m6A (except for the invariant cytosine in CIMS miCLIP), it identifies m6A in all possible motifs. We determined the exact distribution of consensus sequences in which m6A occurs (Supplementary Fig. 5). Our findings confirm that most m6A residues reside in a subset of DRACH motifs6. In fact, 41% and 50% of m6A residues detected by CIMS and CITS miCLIP, respectively, reside in just four subtypes of the DRACH motif. However, a considerable portion of m6As (23% to 31% as determined by CITS and CIMS miCLIP, respectively) occur in DRACH motifs that would be missed by bioinformatic prediction.
CITS miCLIP identifies m6Am at the TSS
We next asked if miCLIP can identify N6,2′-O-dimethyladenosine (m6Am), a related base modification which is found in certain mRNAs at the first position following the 7-methylguanosine cap13. The function of m6Am is poorly understood. Thus, mapping m6Am is important for elucidating its functional role in vivo.
Unlike m6A, which is not detected at the first position of mRNAs, m6Am is limited to the first position in transcripts13. Recently, a MeRIP-Seq approach coupled with bioinformatic analysis that detects RNA 5′ ends was used to predict methylated adenosines at transcription start sites (TSSs)6. We reasoned that miCLIP can also be used to map m6Am at TSSs throughout the transcriptome. Indeed, the metagene profile of sites identified by CITS miCLIP showed an enrichment of called sites in the 5′UTR (Fig. 2e). This enrichment was absent in CIMS miCLIP (Fig. 2e), presumably because CIMS miCLIP requires a C at the +1 position of the modified A.
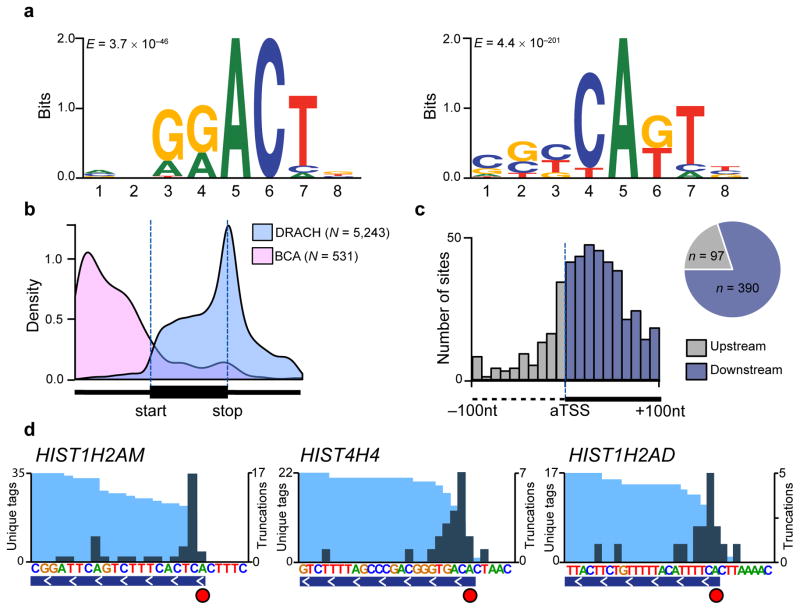
Because CITS miCLIP is not dependent on sequence context, we reasoned it could identify m6Am at the TSS. We analyzed 797 truncations localized to 5′UTRs and found that these occurred in the canonical m6A motif, DRACH, as well as a novel motif, BCA* (B=C, U, or G; A*=methylatable A) (Fig. 4a). 5′UTRs contained nearly three times as many methylated BCA motifs as DRACH motifs (434 sites vs. 151 sites, respectively). Interestingly, the extended form of the BCA motif (Fig. 4a) resembles the known pyrimidine-rich sequence at transcription start sites20, 24, further suggesting these sites are m6Am rather than internal m6A.
Figure 4. Antibody-induced truncations map m6Am throughout the transcriptome.
(a) MEME analysis of the sequence environment (−4 to +4) of 797 truncations localized to 5′UTRs identified two predominant motifs: The canonical m6A motif DRACH (left panel; E = 3.7 × 10−46), and a motif that is best described by the consensus sequence BCA (right panel; E = 4.4 × 10−201).
(b) Truncations in DRACH and BCA sequence context follow distinct metagene distribution profiles.
(c) Truncations in CITS miCLIP cluster around the annotated transcription start site (aTSS). The number of truncations in 10nt bins between −100nt and +100nt relative to the aTSS is plotted (upstream of aTSS, grey; downstream of aTSS, blue). The pie chart shows the proportion of truncations found upstream and downstream of the aTSS.
(d) Truncations identify m6Am with single-nucleotide resolution. Examples of known m6Am-containing 5′UTRs are shown (light blue and teal tracks, unique CITS miCLIP read coverage and unique read starts, respectively; horizontal blue bars, transcript models; red circles, called m6Am residues).
To further address the position of putative m6Am sites, we performed metagene analysis of truncations occurring in DRACH and e contexts (Fig. 4b). Whereas truncations in DRACH sites followed the canonical m6A distribution, truncations in BCA sequence context primarily localized to the 5′UTR (Fig. 4b), with highest enrichment near the annotated TSS (Fig. 4c).
Finally, we sought to validate the idea that CITS miCLIP recognizes m6Am by comparing our data to a known set of m6Am-containing mRNAs25. We focused on histone mRNAs, which have been biochemically shown to start with m6Am25. Indeed, CITS miCLIP called truncation sites at the m6Am position of three histone mRNAs (Fig. 4d). Importantly, truncation efficiency at these sites was not 100%, indicating these truncations were induced by antibody crosslinking rather than the reverse transcriptase reaching the transcript end (Fig. 4d, Supplementary Figure 6). These data show that CITS miCLIP detects two distinct RNA modifications, m6A and m6Am, at single-nucleotide resolution throughout the transcriptome.
CIMS miCLIP identifies m6A residues in snoRNAs
An important criterion for calling peaks in MeRIP-Seq is that peaks must be higher than piled-up reads in adjacent areas within the same transcript, which define the background read level1. However, for small RNAs, the entire RNA can be covered with reads, making it difficult to establish the background. Thus, m6A residues in small RNAs are particularly difficult to identify with MeRIP-Seq. However, since CIMS miCLIP identifies C→T transitions, m6A residues can be readily detected in small RNAs.
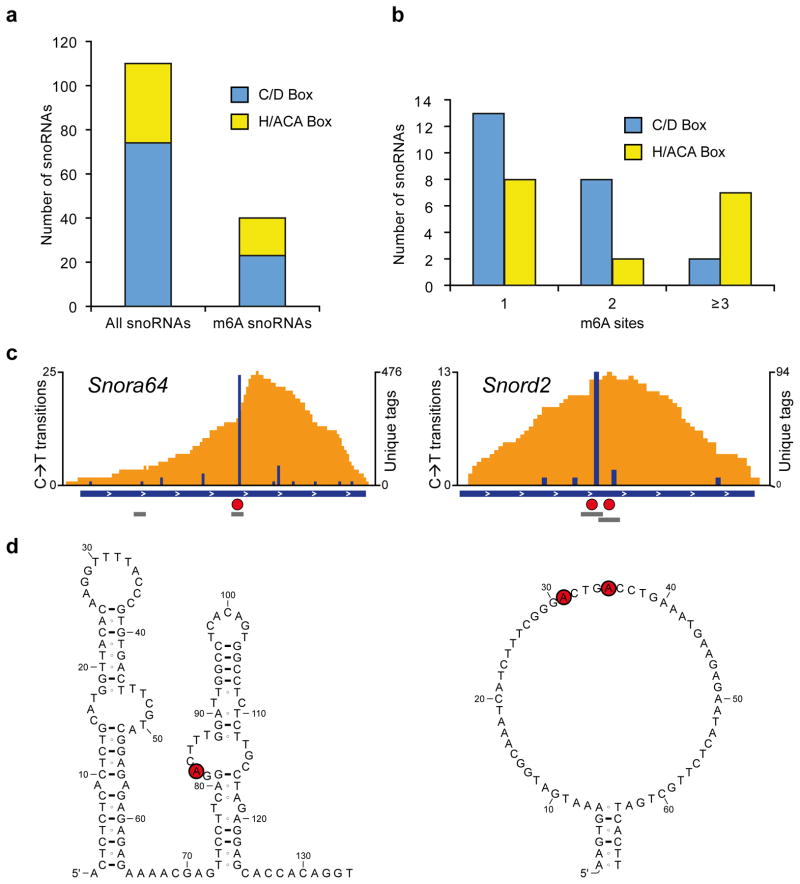
To validate that CIMS miCLIP can detect m6A residues in small RNAs, we focused on small nucleolar RNAs (snoRNAs). C→T transitions identified m6A residues in both H/ACA and C/D box snoRNA subclasses, with over 25% of both classes having at least one m6A (Fig. 5a,b). For example, we detected a high rate of C→T transitions in snoRNAs Snora64 and Snord2 (Fig. 5c). These transitions were found in typical DRACH motifs (Fig. 5d). The discovery of m6A in snoRNAs complements the finding of pseudouridine in snoRNAs26, suggesting this class of ncRNA is regulated by diverse RNA modifications.
Figure 5. m6As are abundant in mouse snoRNAs.
(a) Bargraph showing the number of snoRNAs analyzed and the number of snoRNAs with at least one m6A. Both, C/D-box and H/ACA-box snoRNAs are methylated.
(b) C/D-box and H/ACA-box snoRNAs can be methylated at multiple positions.
(c) Examples of m6A-modified snoRNAs. The H/ACA-box snoRNA Snora64 and the C/D-box snoRNA Snord2 contain m6A (red circles) in canonical DRACH consensus sites (grey bars). Orange and blue tracks: unique CIMS miCLIP read coverage and C→T transitions, respectively. Horizontal blue bars: transcript models. Grey bars: DRACH consensus sites. Red circles: called m6A residues.
(d) Methylation occurs in single-stranded regions of snoRNAs. Shown are secondary structures of Snora64 and Snord2 as predicted in the Ensembl database. Red circles: called m6A residues.
Discussion
m6A is the most widespread base modification in mRNA, but a method for identifying specific residues has been previously lacking. We show that m6A residues can be mapped by generating unique signature mutations with m6A-specific antibodies and UV crosslinking. We mapped m6A residues using two antibodies, each of which has specific advantages. One antibody produced a C→T transition that is limited to detecting m6A, but can readily detect clustered m6A residues. Another antibody produced truncations that can be used to simultaneously map m6A and m6Am residues. Although our experiments used specific antibodies, our approach to screen crosslink-induced mutations should make it straightforward to identify other anti-m6A antibodies suitable for miCLIP. This approach could additionally be used to map other RNA modifications for which antibodies exist.
An important feature of miCLIP is that it can be used without pretreatment of cells with modified nucleotides such as 4-thiouracil (4SU), which is used in PAR-CLIP11. Indeed, a recent study crosslinked anti-m6A antibodies to 4SU-labeled RNA27. RNA fragments that crosslinked to the antibody contain characteristic T→C mutations that arise from 4SU crosslinks. This strategy is valuable since it reduces noise by identifying and narrowing PAR-CLIP peak clusters. However, this study did not use the T→C mutations to map specific m6A residues throughout the transcriptome. Unlike the signature mutations in miCLIP, PAR-CLIP experiments can generate multiple 4SU transitions at protein-binding sites in transcripts11. Because of the nonconsistent number and position of transitions relative to each m6A residue, it is challenging to use T→C mutations for m6A identification. In contrast, miCLIP relies on unique mutational signatures that are highly predictive of m6A residues.
Direct identification of m6A provides advantages over bioinformatic prediction of m6A residues from MeRIP-Seq peaks. Bioinformatic prediction is reliable if the m6A peak has a single clear summit, is caused by a single m6A residue, and has a single centrally-located DRACH motif. However, m6A residues are often clustered in mRNAs1. As a result, MeRIP-Seq peaks are often broad with diverse shapes. Thus, the summit of these peaks may not reflect the position of the m6A residues that account for the peak shape. In miCLIP, peak shape does not influence m6A identification. Furthermore, miCLIP does not constrain m6A identification to m6A residues that fall within a specified subset of DRACH motifs. Thus, miCLIP enables unbiased identification of m6A residues.
The inability to identify specific methylated adenosines in mRNAs at single nucleotide resolution has hampered functional studies of m6A. Indeed, recent studies proposing functions for m6A have not mutated specific adenosine residues to unequivocally determine the effect of m6A on a specific transcript. The single-nucleotide dataset presented here will make it straightforward to test the role of any m6A on the fate or function of any transcript.
METHODS
Antibodies
Anti-m6A antibodies from the following manufacturers were used: Synaptic Systems rabbit polyclonal (SySy; cat. no. 202 003), SySy mouse monoclonal (202 011), Abcam rabbit polyclonal (ab151230), Active Motif (61495), Millipore (ABE572).
In vitro mutagenesis assay
A 1,502 nt long RNA containing a single adenosine nucleotide at position 966 was transcribed in the presence of GTP, CTP, UTP and either ATP or m6ATP using the Ampliscribe in vitro transcription kit (Epicentre). Then, 6 μg of the fragmented transcript were incubated with 4μg of each of the anti-m6A antibodies tested. After UV-crosslinking with 0.15 J cm−2 UV light (254 nm), antibody-RNA complexes were processed as described for cellular RNA and a library was prepared for each antibody. Libraries were then sequenced on a MiSEQ instrument and reads covering the m6A position with single mismatches at positions −2 to +4 were quantified. For each nucleotide at positions −2 to +4 of the m6A, the frequency of truncation and transition events was determined. For this, reads terminating at that position (for truncations) and single-nucleotide mismatches at that position (for transitions) were counted and normalized to the total number of reads covering the m6A residue.
Cell lines and animals
For characterizing m6A residues in human, we used HEK293 cells (ATCC; CRL-1573). For profiling m6A residues in mouse, we used mouse liver (Charles River; CD-1/ICR, female, 8–10 weeks old). All experiments involving mice were approved by the Institutional Animal Care and Use Committee at Weill Cornell Medical College.
Preparation of mouse liver nuclei
For identification of m6A residues in small nucleolar RNAs (snoRNAs), intact nuclei were collected from mouse liver using an iodixanol gradient as described previously28. All steps were performed at 4°C. In brief, liver was homogenized in hypoosmotic medium (250 mM sucrose, 25 mM KCl, 5 mM MgCl2, 10 mM Tris-HCl, pH 7.4) with a dounce homogenizer. The homogenate was then mixed with 50% iodixanol (Sigma-Aldrich D1556; diluted with 250 mM sucrose, 150 mM KCl, 30 mM MgCl2, 60 mM Tris-HCl, pH 7.4) to a final concentration of 25% iodixanol. This mixture was then underlayered with 30% and 35% iodixanol and centrifuged in a swinging-bucket rotor for 20 minutes at 10,000 × g. Nuclear bands were collected, and nuclear RNA was extracted with Trizol (Life Technologies).
Crosslinking of cellular RNA
For initial miCLIP libraries, total RNA was purified from HEK293 cells. Poly(A)+ RNA was prepared from four biological replicates of HEK293 cells and processed in parallel for miCLIP. Mouse liver RNA was depleted of ribosomal RNA by Ribominus (Life Technologies).
RNA was fragmented using fragmentation reagent (Life Technologies) to a size between 30 and 130 nt. After stopping the reaction, 20 μg fragmented RNA was directly diluted in 450μl IP buffer (50 mM Tris pH 7.4, 100 mM NaCl, 0.05% NP40) and incubated with 1–5 μg anti-m6A antibody for 1–2 h at 4°C rotating head over tail. The solution was then transferred into a 3cm cell culture dish and crosslinked twice with 0.15 J cm−2 UV light (254 nm) in a Stratalinker (Agilent).
After crosslinking, the solution was transferred into Eppendorf tubes and incubated with 30 μl Protein A/G beads (Thermo Scientific) for 1–2 h at 4°C, rotating. Bead-bound antibody-RNA complexes were then recovered on a magnetic stand (Life technologies) and washed twice with high-salt buffer (50 mM Tris pH7.4, 1M NaCl, 1 mM EDTA, 1% NP40, 0.1% SDS), twice with IP buffer, and twice with PNK wash buffer (20 mM Tris, 10 mM MgCl2, 0.2% Tween20).
3′-dephosphorylation, linker ligation & labeling
The protocol for these steps was similar to the protocol described for iCLIP10. In brief, RNA 3′-ends were dephosphorylated on beads with PNK (NEB; M0201S) for 30 min in dephosphorylation buffer (70 mM Tris pH 6.5, 10 mM MgCl2, 1 mM DTT). After another round of extensive washing (twice with PNK wash buffer, once with IP buffer, once with high salt buffer, and twice with PNK wash buffer), the 3′-adaptor was ligated with T4 RNA ligase (NEB) overnight.
Antibody-RNA complexes were then eluted from beads with 1X NuPage sample buffer (Life Technologies) containing 50mM DTT, subjected to NuPage gel electrophoresis and transferred onto 0.45 μm nitrocellulose membranes (BioRad). After autoradiography (2 h to overnight), membrane regions containing RNA-crosslinked antibody heavy and light chains were excised and the RNA was released from the membrane by treatment with proteinase K (Life Technologies).
Library preparation
After phenol-chloroform extraction and precipitation, the RNA was reverse transcribed with Superscript III reverse transcriptase (Life Technologies) according to the manufacturer’s protocol. First strand cDNA was size-selected on a 6% TBE-Urea gel (Life Technologies) and regions corresponding to 80–120 nt (for the total RNA dataset) or 70–100 nt (for the mRNA dataset) were used for further analysis. Circularization and re-linearization of cDNA were performed as described in the iCLIP protocol10. Libraries were PCR amplified for 18 to 21 cycles and sequenced on an Illumina HiSeq 2500 instrument.
Read pre-processing
Fastq files were adapter trimmed using flexbar29 and demultiplexed based on their experimental barcode using the pyBarcodeFilter.py script of the pyCRAC tool suite30. The second part of the iCLIP random barcode was then moved to the read headers with the Unix tool awk (awk -F “##” ‘{sub(/..../, “##”$2, $2); getline($3); $4 = substr($3,1,2); $5 = substr($3,3); print $1 $2 $4 “\n”$5}’). For the HEK293 mRNA dataset (CIMS miCLIP), reads from the four replicates were combined at this point. Sequence-based removal of PCR duplicates was then performed with the pyFastqDuplicateRemover.py script30. Awk was then used to generate read headers compatible with downstream processing by the CIMS pipeline (awk -F ‘[_/]’ ‘/^>/{print $1“_”$2“_”$3“/”$4“#”$3“#”$2; getline($9); print $9}’).
Processing of paired-end data
In order to increase base coverage and thus facilitate the discrimination of mutations (which should be at the same position in both reads) from random sequencing errors (which should be evenly distributed in the reads), short insert libraries (~30–40 nt) were generated and subjected to paired-end sequencing. For this paired-end data, reverse reads were reverse complemented and processed like their forward mates. However, to distinguish the forward and reverse mates as distinct reads and prevent their collapsing during PCR duplicate removal, the reverse reads were assigned the reverse complement of the random barcode using a custom perl script (available in the Supplementary data section).
Read mapping
Reads were mapped with Novoalign (Novocraft Technologies Sdn Bhd) to a custom build of rRNA (28S, acc.no NR_003287.2; 18S acc.no. NR_003286.2; and 5.8S, NR_003285.2) or to the human (hg19) and mouse (mm10) genomes. A maximum alignment score of 85 was allowed, iterative trimming was used and parameters were adjusted to allow mapping of short reads (Novoalign -t 85 -s 1 -l 16 -F FA -o Native -r None).
Mutation calling
Positions of C→T transitions and the coordinates of mapped reads were extracted from native Novoalign output files using the novoalign2bed.pl script of the CIMS software package15. Reads that mapped to the same start and end coordinates and that shared the same random barcode were then collapsed into unique tags using the CIMS script tag2collapse.pl. For each mismatch position, unique tag coverage (k) and the number of C→T transitions (m) was determined using the CIMS.pl program15. Because the cluster permutation approach of the CIMS algorithm15 is not suited for assigning a false discovery rate to a specific transition subtype (i.e. the C→T transitions analyzed here), a filtering approach was used to minimize calling of false positive sites. In a first step, known SNPs (dbSNP 138) and regions marked as repetitive sequence in the respective genome were removed. Then, we filtered sites based on their number of C→T transitions (m) and the ratio of C→T transitions to unique tags covering that position (m/k): First, to reduce mismatches caused by random errors introduced during reverse transcription, PCR amplification and library sequencing, each transition had to be called at least twice (m2). Second, to further reduce noise and to exclude sites that have a very high mismatch rate (caused for example by unknown SNPs and read mismapping artifacts), m/k was required to be between 1% and 50%. Then, two filters were applied to reduce the number of false positive sites. First, to reduce mismatches caused by random errors introduced during reverse transcription, PCR amplification and library sequencing, each transition had to be called at least twice (m2). Second, the ratio of mutant reads to total reads covering a position was restricted to be between 1% and 50% (m/k1–50). This further reduces noise and simultaneously removes sites with very high mismatch rate such as produced by SNPs and mismapping artifacts. Together, these filters (m2 m/k1–50) reduced the signal to noise ratio more than twofold and calling of non-A positions to less than 20%. This resulted in a final dataset of 9,536 m6A residues in HEK293 cells and 780 m6A residues in mouse liver nuclei.
Truncation calling
Coordinates of mapped reads were extracted, and reads carrying the same random barcode that mapped to identical coordinates were collapsed as for mutation analysis (see above). Then, we used the CITS software package to call crosslink-sites18. In brief, coverage of unique reads was calculated with the tag2profile.pl script. Then, to detect potential crosslink-induced truncation sites, we used the bedExt.pl script of the CITS software package to map positions immediately upstream of the first nucleotide of each unique read. To differentiate reads that represent truncations from readthrough reads, we used the CIMS algorithm to identify unique reads carrying crosslink-induced deletions. Then, we used a bash script and joinWrapper.py of the CITS package to remove unique reads carrying deletions at potential truncation sites. Remaining reads were then clustered into peaks with tag2cluster.pl. Potential truncation positions (read start “pileups”) were then shuffled within their respective peak clusters using tag2peak.pl to identify truncation events of statistical significance. Events with a significance value of p≤0.05 that occurred at adenosines were retained to yield a list of 12,051 m6A residues in HEK293 cells.
Peak calling
To determine the number of peaks generated by miCLIP, unique reads from each dataset were clustered into peaks using the tag2cluster.pl script of the CIMS software package. The resulting list of peak clusters was filtered for those with at least four stacked reads to generate 80,774 HEK293 and 14,055 mouse liver nuclei peaks for the CIMS miCLIP and 33,157 HEK293 peaks for the CITS miCLIP.
Analysis of clustered m6A
To define sites of clustered methylation, sliding windows (length 100 nt, step size 25 nt) that overlapped with annotated RefSeq transcripts were analyzed for m6A site coverage. Windows with two or more m6As were then merged into a cluster. Finally, the number of miCLIP-called m6A residues in each cluster was determined. In addition, we counted the number of MeRIP-Seq coverage-predicted m6As6 in each cluster.
Annotation of m6A residues
Called sites were annotated with the annotatePeaks.pl script from the Homer software suite31. To compare the genomic distribution of m6A residues with the distribution of meRIP-Seq peaks, a previously published HEK293 MeRIP-Seq dataset1 was analyzed the same way.
Supplementary Material
Acknowledgments
We thank K. Meyer and D. Patil for useful comments and suggestions. This work was supported by NIH grants NIDA DA037150 (S.R.J.), NS076465 (C.E.M), T32 HD060600 (A.G.), T32 CA062948 (A.O.-G.), a German Research Foundation (DFG) fellowship (B.L.), the Irma T. Hirschl and Monique Weill-Caulier Charitable Trusts, the STARR Consortium (I7-A765, C.M.), the Vallee Foundation (C.M), and the WorldQuant Foundation (C.E.M.).
Footnotes
Accession Codes
Data were deposited in NCBI’s Gene Expression Omnibus (GEO) under accession number GSE63753.
AUTHOR CONTRIBUTIONS
B.L., A.G., A.O.-G., and S.R.J. conceived and designed the experiments and analyzed data, C.M. and C.E.M. analyzed mutational profiles of initial miCLIP libraries, and B.L., A.G., A.O.-G., and S.R.J. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Meyer Kate D, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 3.Perry RP, Kelley DE, Friderici K, Rottman F. The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5′ terminus. Cell. 1975;4:387–394. doi: 10.1016/0092-8674(75)90159-2. [DOI] [PubMed] [Google Scholar]
- 4.Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proceedings of the National Academy of Sciences of the United States of America. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schibler U, Kelley DE, Perry RP. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. Journal of Molecular Biology. 1977;115:695–714. doi: 10.1016/0022-2836(77)90110-3. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz S, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Reports. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Squires JE, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Research. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugimoto Y, et al. Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions. Genome Biology. 2012;13:R67. doi: 10.1186/gb-2012-13-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.König J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nature Structural & Molecular Biology. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ule J, et al. CLIP Identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 12.Schibler U, Perry RP. The 5′-termini of heterogeneous nuclear RNA: a comparison among molecules of different sizes and ages. Nucleic Acids Research. 1977;4:4133–4149. doi: 10.1093/nar/4.12.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kramer K, et al. Photo-cross-linking and high-resolution mass spectrometry for assignment of RNA-binding sites in RNA-binding proteins. Nature Methods. 2014;11:1064–1070. doi: 10.1038/nmeth.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnology. 2011;29:607–614. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piekna-Przybylska D, Decatur WA, Fournier MJ. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Research. 2007;36:D178–D183. doi: 10.1093/nar/gkm855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore MJ, et al. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nature Protocols. 2014;9:263–293. doi: 10.1038/nprot.2014.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weyn-Vanhentenryck SM, et al. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Reports. 2014;6:1139–1152. doi: 10.1016/j.celrep.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu N, et al. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA. 2013;19:1848–1856. doi: 10.1261/rna.041178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carninci P, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nature Genetics. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 20.Ni T, et al. A paired-end sequencing strategy to map the complex landscape of transcription initiation. Nature Methods. 2010;7:521–527. doi: 10.1038/nmeth.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plessy C, et al. Linking promoters to functional transcripts in small samples with nanoCAGE and CAGEscan. Nature Methods. 2010;7:528–534. doi: 10.1038/nmeth.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frith MC, et al. A code for transcription initiation in mammalian genomes. Genome Research. 2008;18:1–12. doi: 10.1101/gr.6831208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moss B, Gershowitz A, Weber LA, Baglioni C. Histone mRNAs contain blocked and methylated 5′ terminal sequences but lack methylated nucleosides at internal positions. Cell. 1977;10:113–120. doi: 10.1016/0092-8674(77)90145-3. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz S, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen K, et al. High-resolution N6-methyladenosine (m6A) map using photo-crosslinking-assisted m6A sequencing. Angewandte Chemie International Edition. 2014;54:1587–1590. doi: 10.1002/anie.201410647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graham JM. Isolation of nuclei and nuclear membranes from animal tissues. Current Protocols in Cell Biology. 2001:3–10. doi: 10.1002/0471143030.cb0310s12. [DOI] [PubMed] [Google Scholar]
- 29.Dodt M, Roehr JT, Ahmed R, Dieterich C. FLEXBAR—flexible barcode and adapter processing for next-generation sequencing platforms. Biology. 2012;1:895–905. doi: 10.3390/biology1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webb S, Hector RD, Kudla G, Granneman S. PAR-CLIP data indicate that Nrd1-Nab3-dependent transcription termination regulates expression of hundreds of protein coding genes in yeast. Genome Biology. 2014;15:R8. doi: 10.1186/gb-2014-15-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.