Abstract
Background
The early stages of ovarian follicle formation—beginning with the breakdown of germ cell cysts and continuing with the formation of primordial follicles and transition to primary and secondary follicles—are critical in determining reproductive life span and fertility. Previously, we discovered that global knockouts of germ cell-specific transcriptional co-regulators Sohlh1, Sohlh2, Lhx8, and Nobox, cause rapid oocyte loss and ovarian failure. Also factors such as Nobox and Sohlh1 are associated with human premature ovarian failure. In this study, we developed a conditional knockout of Lhx8 to study oocyte-specific pathways in postnatal folliculogenesis.
Results
The conditional deficiency of Lhx8 in the oocytes of primordial follicles leads to massive primordial oocyte activation, in part, by indirectly interacting with the PI3K-AKT pathway, as shown by synergistic effects on FOXO3 nucleocytoplasmic translocation and rpS6 activation. However, LHX8 does not directly regulate members of the PI3K-AKT pathway; instead, we show that LHX8 represses Lin28a expression, a known regulator of mammalian metabolism and of the AKT/mTOR pathway. LHX8 can bind to the Lin28a promoter, and the depletion of Lin28a in Lhx8-deficient oocytes partially suppresses primordial oocyte activation. Moreover, unlike the PI3K-AKT pathway, LHX8 is critical beyond primordial follicle activation, and blocks the primary to secondary follicle transition.
Conclusions
Our results indicate that the LHX8-LIN28A pathway is essential in the earliest stages of primordial follicle activation, and LHX8 is an important oocyte-specific transcription factor in the ovary for regulating postnatal folliculogenesis.
Electronic supplementary material
The online version of this article (doi:10.1186/s12915-015-0151-3) contains supplementary material, which is available to authorized users.
Keywords: Lhx8, Primordial follicle, Oocyte, Lin28a, Ovarian reserve
Background
Following the proliferation of primordial germ cells, oogonia enter meiosis circa embryonic day 13 (E13.5) and are then referred to as oocytes in the embryonic mouse ovary. Oocytes in the embryonic ovary exist in clusters, surrounded by epithelial pre-granulosa cells [1]. Germ cell cyst breakdown results in the envelopment of individual oocytes by flat granulosa cells to form primordial follicles. Primordial follicles contain small oocytes (<20 μm) surrounded by flat granulosa cells. These early steps in the formation of primordial follicles are critical, since primordial follicles constitute the fundamental reproductive units of the ovary and give rise to all dominant follicles. Primordial follicle activation (PFA) is a process by which primordial follicles are selected into the growing follicle pool [2]. Morphologically, PFA is characterized by oocyte growth to greater than 20 μm and transition of the flat epithelial layer to cuboidal granulosa cells. PFA is independent of gonadotropins [3], occurs prior to puberty, and is spontaneously observed when an ovary is transplanted or cultured in vitro [4]. Several groups have shown the importance of the ubiquitous PI3K-AKT-mTORC1 pathways within oocytes in regulating PFA [5–9]. Conditional ablation of Foxo3, Pten, and Tsc1/2 in oocytes triggers massive oocyte activation [5, 6, 8, 10]. Activated oocytes in these animals survive beyond 5 weeks, perhaps not surprising given the importance of this pathway in regulating apoptosis and cell death.
The role of oocyte-specific pathways in oocyte activation is unknown. Lhx8 encodes a highly conserved LIM homeodomain protein that is preferentially expressed in mammalian ovaries, including human ovaries [11, 12]. Female mice with global Lhx8 deficiency are infertile and rapidly lose oocytes after birth [13]; so great is the loss in Lhx8 global knockout that few oocytes remain by postnatal day 7. To study the postnatal role of Lhx8 in oocyte activation, we conditionally ablated Lhx8 in oocytes of primordial follicles (primordial oocytes). Our results indicate that Lhx8 represses oocyte activation and plays a dominant role over PTEN-led pathways in oocyte survival. Lhx8 depletion in primordial oocytes decouples oocyte activation from somatic differentiation. Moreover, Lhx8 directly regulates Lin28a expression and indirectly interacts with the PI3K-Akt pathway to effect repression of PFA. Furthermore, unlike the Pten pathway, we found that conditional deletion of Lhx8 from oocytes of primary follicles (primary oocytes) causes primary follicle death and depletion of the secondary/antral follicle pool.
Results
Conditional depletion of Lhx8 by Gdf9Cre causes massive primordial follicle activation
We previously reported that global knockout of Lhx8 causes infertility and loss of oocytes by postnatal day 7 (PD7) [14]. In the global knockout of Lhx8, primordial-like follicles form (oocytes less than 20 μm in diameter and surrounded by flat granulosa cells), but oocytes do not grow. Since Lhx8 is expressed in both embryonic and postnatal female germ cells, it is possible that global knockout of Lhx8 disrupts early embryonic pathways that lead to postnatal oocyte depletion. We therefore investigated the postnatal functions of Lhx8 by generating a conditional knockout mouse, using a floxed Lhx8 allele (Lhx8flx/flx) [15] and a Gdf9Cre transgenic mouse [16]. The Gdf9Cre transgene will inactivate Lhx8 specifically in primordial oocytes. Gdf9Cre is highly efficient in oocytes and, when present in either Lhx8flx/flx or Lhx8flx/- animals, displayed the same ovarian phenotype. We used Lhx8flx/flxGdf9Cre to study the effects of Lhx8 conditional deficiency in primordial follicles on ovarian development.
At PD7 and PD14, LHX8 protein was depleted in Lhx8flx/flxGdf9Cre ovaries and massive oocyte activation occurred in the primordial follicles, as manifested by oocytes reaching a diameter greater than 20 μm without significant transformation of the surrounding flat granulosa cells (Fig. 1a–f). At PD7, Lhx8flx/flxGdf9Cre mice had 398 ± 53 activated primordial follicles per ovary compared to 16 ± 2 per ovary in Lhx8flx/flx controls. The number of primordial follicles was 1917 ± 23 per ovary in Lhx8flx/flx controls and was significantly reduced to 1043 ± 119 per ovary in Lhx8flx/flxGdf9Cre mice (Fig. 1c). We detected a decline in primary follicles in Lhx8flx/flxGdf9Cre mice, implying a block in the transition from activated primordial follicles to primary follicles. There were a negligible number of advanced follicle types (oocytes surrounded by multiple layers of granulosa cells) in PD7 Lhx8flx/flxGdf9Cre ovaries, compared to the Lhx8flx/flx controls.
Fig. 1.
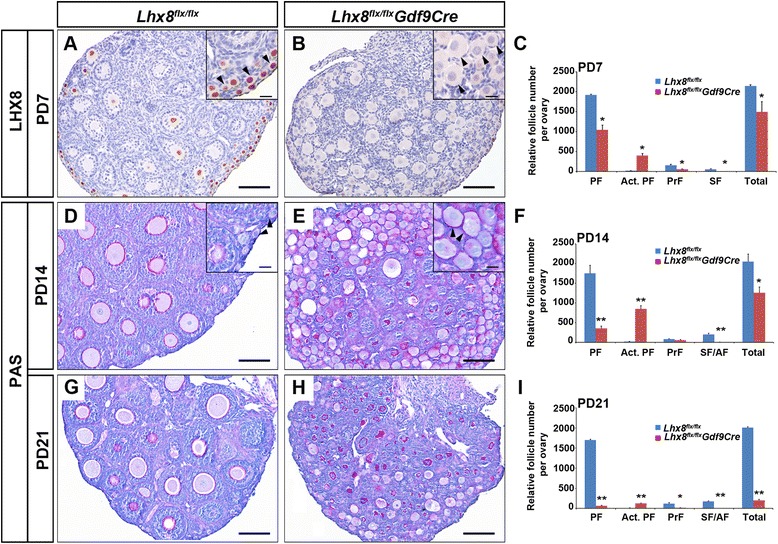
Postnatal inactivation of Lhx8 causes premature activation of primordial follicles and ovarian failure. a and b Anti-LHX8 antibodies were used to detect oocytes in paraformaldehyde-fixed and hematoxylin-counterstained ovaries taken at postnatal day 7 (PD7). Ovaries were derived from control (Lhx8 flx/flx, A) and Lhx8 conditional knockout (Lhx8 flx/flx Gdf9Cre, B) mice. Arrowheads in the inset of panel A indicate primordial follicles that stain with anti-LHX8 antibodies (brown signal). Arrowheads in the inset in panel B show activated primordial follicles (oocytes larger than 20 μm without cuboidal granulosa cells). d, e, g and h Periodic acid–Schiff (PAS) staining of PD14 (D and E) and PD21 (G and H) ovaries derived from control (D and G) and Lhx8 conditional knockout (E and H) mice. Arrowheads in the inset of panels D and E indicate primordial activated primordial follicles. c, f and i Quantification of ovarian follicle types in Lhx8 flx/flx and Lhx8 flx/flx Gdf9Cre mice. Five pairs of ovaries from PD7 (C), PD14 (F), and PD21 (I) were embedded in paraffin and serially sectioned at 5 μm thickness, and the follicles were counted. Anti-NOBOX antibodies stain oocyte nuclei throughout folliculogenesis and were used to identify oocytes in our counts. Every fifth section was counted. We scored primordial follicles (PF), activated primordial follicles (Act. PF, oocyte diameter greater than 20 μm without cuboidal granulosa cells), primary follicles (PrF) and secondary/antral follicles (SF/AF). *P < 0.05, **P < 0.01. Scale bars: 100 μm (A, B, D, E, G and H); 20 μm (inset in A, B, D and E)
At PD14, Lhx8flx/flxGdf9Cre ovaries contained 847 ± 82 activated primordial follicles per ovary, compared to 15 ± 7 per ovary in Lhx8flx/flx controls. The number of primordial follicles significantly diminished from 1753 ± 204 in Lhx8flx/flx controls to 353 ± 58 in Lhx8flx/flxGdf9Cre ovaries (Fig. 1f). By PD21, the total number of follicles from primordial to secondary was greatly diminished in the Lhx8flx/flxGdf9Cre ovary (Fig. 1g–i). By PD35, there were barely any oocytes and follicles detected in the Lhx8flx/flxGdf9Cre ovary (see Additional file 1: Figure S1C) and Lhx8flx/flxGdf9Cre females were sterile (see Additional file 2: Figure S2A). Our studies show that postnatal inactivation of Lhx8 within oocytes of primordial follicles leads to massive oocyte activation, decoupling of oocyte activation from somatic cell transformation, oocyte death, and infertility.
LHX8 interacts with PI3K-AKT pathway
We examined RNA expression of genes that encode important members of the PI3K-AKT-mTOR pathways in PD7 Lhx8flx/flxGdf9Cre oocytes. Real-time polymerase chain reaction (PCR) did not detect significant changes in the expression of Pten, Akt, Foxo3, Pdk1, Mtor, Rps6, Deptor, Rictor, or Tsc1/2 (some of these are shown in Fig. 2). These results indicate that LHX8 does not directly affect transcription of a subset of genes known to encode proteins in the PI3K-AKT-mTOR pathways. Then we assessed whether the PI3K-AKT pathway was activated at the protein level in Lhx8flx/flxGdf9Cre oocytes. AKT is a serine/threonine-specific protein kinase that plays a key role in apoptosis and PFA. Phosphorylated AKT activates multiple downstream pathways, including FOXO3 phosphorylation, resulting in oocyte activation [8]. Western blot analyses on oocytes from PD7 ovaries showed that phosphorylation of AKT at two sites, S473 and T308, was higher in Lhx8flx/flxGdf9Cre oocytes than in controls (Fig. 2h). But interestingly, only p-AKT (T308) was detected in the activated primordial follicles (see Additional file 3: Figure S3).
Fig. 2.
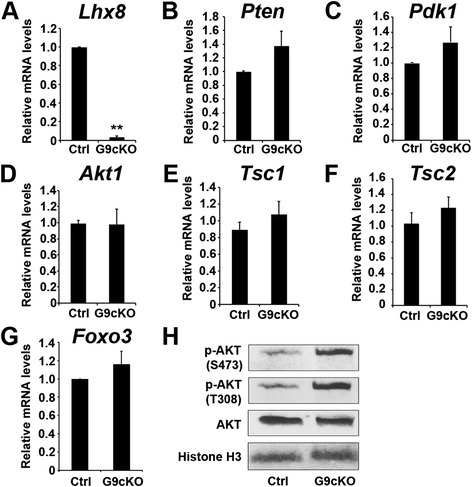
AKT is activated in Lhx8 flx/flx Gdf9Cre oocytes. a–g Oocytes were isolated from PD7 control (Lhx8 flx/flx, Ctrl) and Lhx8 flx/flx Gdf9Cre (G9cKO) mouse ovaries and RNA was extracted for cDNA conversion and real-time quantitative polymerase chain reaction (RT-qPCR). Data were normalized to Gapdh expression and are given as the mean relative quantity (compared with control), with error bars representing the standard error of the mean. Student’s t-test was used to calculate P values. The only significant difference was noted in the expression of Lhx8, as expected. ** P < 0.01. h Oocytes were isolated from PD7 control and Lhx8 flx/flx Gdf9Cre ovaries, protein was extracted, and a Western blot test was performed on three independent samples, using antibodies against AKT and its two phosphorylated forms (S473 and T308). Histone H3 immunoreactivity was used as a control
Previous studies have shown FOXO3 nucleocytoplasmic translocation and rpS6 phosphorylation via the PI3K-AKT-mTOR pathways to be associated with PFA [5, 6, 8]. We examined FOXO3 nucleocytoplasmic translocation and rpS6 phosphorylation in PD7 Pten and Lhx8 conditional knockouts (Fig. 3 and Additional file 4: Figure S4). FOXO3 nucleocytoplasmic translocation was prominent in oocytes larger than 30 μm but did not show obvious translocation among oocytes between 20 and 30 μm in Lhx8flx/flxGdf9Cre mice (Fig. 3d–f, f', p). Oocytes of primordial follicles in Pten and Lhx8 conditional single knockouts as well as corresponding controls did not show FOXO3 nucleocytoplasmic translocation. However, FOXO3 nucleocytoplasmic translocation was significantly induced in the primordial (<20 μm) oocytes (Fig. 3j–l, l', p) of the PD7 double Lhx8/Pten conditional knockouts (Lhx8flx/flxPtenflx/flxGdf9Cre). These data indicate a synergistic action of the LHX8 and PTEN proteins on FOXO3 nucleocytoplasmic translocation.
Fig. 3.
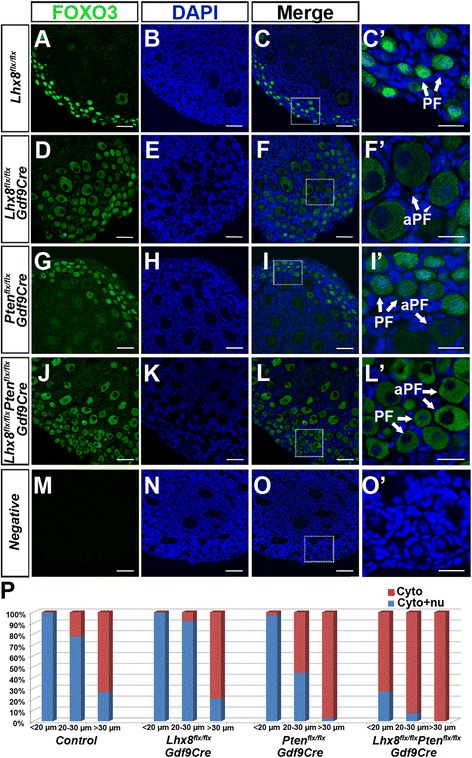
Lhx8 and Pten conditional knockout effects on FOXO3 localization. a–c In control mice (Lhx8 flx/flx), FOXO3 is expressed in the nucleus and cytoplasm of primordial oocytes (PF, arrows in c'). d–f In Lhx8 flx/flx Gdf9Cre mice, the extensive nucleocytoplasmic translocation is not observed in activated primordial follicles between 20 and 30 μm (aPF, arrow in f') but is noted in activated primordial follicles larger than 30 μm (aPF, arrow in F'). d–i A similar expression pattern of FOXO3 localization exists in Pten conditional knockout (Pten flx/flx Gdf9Cre) mice. The arrows in i' represent primordial follicles (PF) with both nuclear and cytoplasm expression of FOXO3 and cytoplasm expression of FOXO3 in activated primordial follicles (aPF) below 30 μm. j–l However, in mice that are conditionally deficient in both Lhx8 and Pten (Lhx8 flx/flx Pten flx/flx Gdf9Cre), FOXO3 nucleocytoplasmic translocation is present in primordial, activated, and primary oocytes. The negative control is immunofluorescence in the presence of secondary antibodies and is shown in m–o. The boxed areas in C, F, I, L, and O are shown magnified in C', F', I', L', and O'. p Graphic representation of FOXO3 distribution (cytoplasm only or nucleus and cytoplasm). Oocytes were grouped by size (diameter) as less than 20 μm, between 20 and 30 μm, and greater than 30 μm. Only oocytes with clear DAPI nuclear staining were counted. Scale bars: 50 μm (A–C, D–F, G–I, J–L, and M–O); 20 μm (C', F', I', L', and O')
We also examined the effects of double Pten and Lhx8 deficiency on rpS6. mTORC1 promotes protein translation and cell growth, in part, through activation of S6K1 (by phosphorylation of its threonine 389) and through phosphorylation and inactivation of eIF4E-binding proteins. S6K1 is responsible for phosphorylation and activation of rpS6, which leads to enhanced protein translation and ribosome biogenesis. In control, Lhx8flx/flxGdf9Cre, and Ptenflx/flxGdf9Cre ovaries, rpS6 was only significantly activated in growing follicles at PD7 (see Additional file 4: Figure S4). However, the PD7 Lhx8flx/flxPtenflx/flxGdf9Cre ovaries showed significant activation of rpS6 in primordial (<20 μm) oocytes (see Additional file 4: Figure S4D, D'). These results further indicate that the Lhx8 and Pten pathways synergistically interact to accelerate two events associated with PFA—FOXO3 nucleocytoplasmic translocation and phosphorylation of rpS6.
Lin28a RNA and protein expression are upregulated in Lhx8flx/flxGdf9Cre oocytes
LHX8 is a transcription factor, and we expect that its major effect will be at the RNA level. We therefore analyzed the transcriptome of Lhx8flx/flxGdf9Cre ovaries via high-throughput RNA sequencing (RNA-seq). We sequenced 180 million tags in each sample and compared the relative abundance of RNA tags encoded by genes in the PI3K-AKT-mTOR pathways derived from PD7 Lhx8flx/flxGdf9Cre and control ovaries. No significant differences in RNA expression of PI3K-AKT-mTOR pathway genes (Pik3r1, Pik3ca, Kit, Kitl, Gsk3b, Cdnd1, Wee1, Cdkn1a/b, Rheb, Mlst8, Mapkap1, Prr5, and Eif4ebp1) were evident in the RNA-seq experiment (see Additional file 5: Table S1).
In addition to analyzing the expression of known PI3K-AKT-mTOR genes, we studied global differences in the transcriptome of Lhx8flx/flxGdf9Cre and control ovaries. We detected a sixfold increase in Lin28a RNA transcripts in Lhx8flx/flxGdf9Cre compared to control ovaries (Fig. 4b and Additional file 5: Table S1). Immunofluorescence and Western blot analysis with anti-LIN28A antibodies showed that LIN28A protein was expressed at a significantly higher level in Lhx8flx/flxGdf9Cre oocytes compared to the controls (Fig. 4a, c).
Fig. 4.
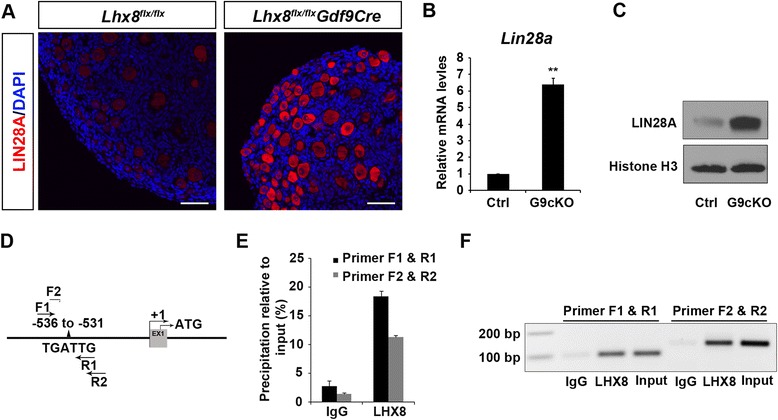
Lhx8 suppresses Lin28a expression. a Immunofluorescence with anti-LIN28A antibodies shows that LIN28A is preferentially expressed in oocytes within the ovary. LIN28A abundance is higher in Lhx8 conditionally deficient oocytes (Lhx8 flx/flx Gdf9Cre) compared to controls (Lhx8 flx/flx). b and c Lin28a transcripts and protein are significantly more highly expressed in Lhx8 flx/flx Gdf9Cre oocytes (G9cKO) unlike controls (Ctrl). a–f Chromatin immunoprecipitation (ChIP) assays with anti-LHX8 affinity purified antibodies on oocytes. d A putative LHX8 DNA binding site, TGATTG [22], which perfectly fits the LHX8 binding consensus sequence, was identified at position −536 to −531 relative to the Lin28a transcription initiation site. e Anti-LHX8 antibodies precipitate genomic DNA containing the TGATTG binding sequence from the Lin28a promoter region as shown by ChIP-quantitative PCR (qPCR). Immunoglobulin G (IgG) antibodies served as control. The percentage input method is used to analyze the qPCR data. “Input” is the PCR product from chromatin pellets before immunoprecipitation. A triplicate average Ct normalized to an adjusted input was used for the calculation of percentage input. Two sets of primers (F1 and R1) and (F2 and R2) were used to perform ChIP-qPCR. f PCR amplification of the oocyte input DNA, as well as DNA precipitated by normal guinea pig IgG and anti-LHX8 antibodies by the F1/R1 and F2/R2 primer sets. Scale bars: 50 μm (A)
LIN28A is an RNA-binding protein that blocks biogenesis of let-7 microRNAs and a known regulator of mammalian body size and metabolism, including onset of menarche [17, 18]. Lin28a is preferentially expressed in oocytes and embryo stem cells [19, 20]. Moreover, the PI3K-AKT-mTOR pathways can be activated by LIN28A [21]. LIN28A, therefore, may play a role in oocyte activation and growth.
LHX8 can directly bind to the LHX8 DNA binding motif in the Lin28a promoter
We tested whether LHX8 can bind to the Lin28a promoter. LHX8 is an oocyte-specific LIM homeodomain transcriptional regulator that is predicted to bind DNA. A previous study showed that the LHX8 homeodomain preferentially binds a TGATTG DNA motif [22]. We identified a single TGATTG DNA motif −531 to −536 bp upstream of the putative transcriptional initiation site in the Lin28a gene (Fig. 4d). The Lin28a TGATTG motif is conserved in other mammals, including humans. We performed a chromatin immunoprecipitation (ChIP) experiment on wild-type oocytes, using our highly specific and affinity purified anti-LHX8 antibodies [13]. Anti-LHX8 antibodies preferentially immunoprecipitated the Lin28a promoter DNA fragment containing the TGATTG motif (Fig. 4e, f). These data further suggest that LHX8 represses Lin28a expression by directly binding to the Lin28a promoter.
Lin28a deficiency partially rescues the Lhx8flx/flxGdf9Cre phenotype
Our data indicates that LHX8 suppresses Lin28a expression. Since LIN28A is a growth-promoting factor [17, 23] preferentially expressed in oocytes, we hypothesized that Lin28a deficiency will rescue Lhx8flx/flxGdf9Cre-induced PFA. We bred Lin28a and Lhx8 floxed mice with Gdf9Cre to generate Lhx8/Lin28a double conditional knockouts (Lhx8flx/flxLin28aflx/flxGdf9Cre).
We performed ovarian morphometric analyses to determine the effects of the double knockout on PFA. No morphometric difference was observed between Lin28aflx/flxGdf9Cre and control females at PD7 (Fig. 5a, c, e). As expected, we observed significantly fewer activated primordial follicles in Lhx8flx/flxLin28aflx/flxGdf9Cre compared to Lhx8flx/flxGdf9Cre ovaries, but they were not completely normal compared with the control (Fig. 5a, b, d, f).
Fig. 5.
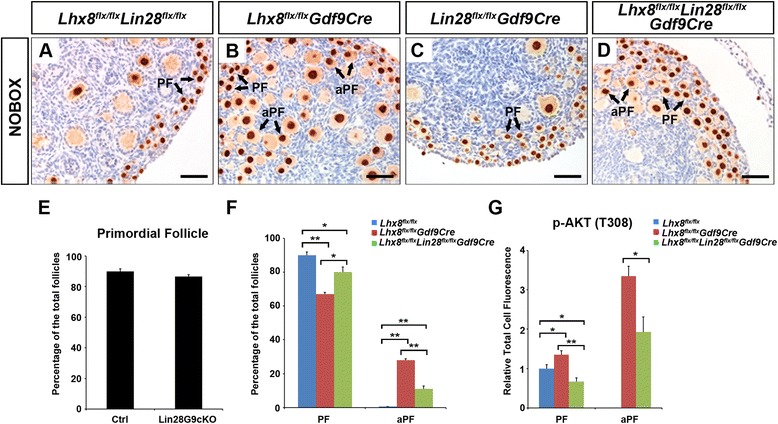
Lin28a deficiency rescues Lhx8 flx/flx Gdf9Cre-induced PFA. a–d Representative histology of control (Lhx8 flx/flx), Lhx8 conditional knockout (Lhx8 flx/flx Gdf9Cre), Lin28a conditional knockout (Lin28 flx/flx Gdf9Cre), and double Lhx8/Lin28a conditional knockout (Lhx8 flx/flx Lin28 flx/flx Gdf9Cre) ovaries. Double Lhx8/Lin28a conditional knockouts show a diminished number of activated primordial follicles. Anti-Nobox antibodies were used to label oocyte nuclei (dark brown). PF: primordial follicle; aPF: activated primordial follicle. e The Lin28 flx/flx Gdf9Cre (Lin28G9cKO) mice have a similar phenotype as control (Ctrl) mice and the primordial follicle count is expressed as the percentage of total follicles. f Quantitation of activated primordial follicles in control, Lhx8 conditional, and double Lhx8/Lin28a conditional knockouts. The percentage of primordial follicles (PF) and activated primordial follicles (aPF) is shown. Three pairs of ovaries from Lhx8 conditional knockout and double Lhx8/Lin28a conditional knockouts mice were serially sectioned, and every fifth section was counted. g AKT phosphorylation is diminished in double Lhx8/Lin28a conditional knockouts. We quantitated by fluorescence p-AKT (T308) expression in the primordial and activated primordial follicles of the Lhx8 flx/flx Gdf9Cre (G9cKO) and Lhx8 flx/flx Lin28a flx/flx Gdf9Cre (G9dKO) ovaries at PD7. There was a significant decrease in AKT phosphorylation in Lhx8 flx/flx Lin28a flx/flx Gdf9Cre primordial and activated primordial follicles compared to Lhx8 flx/flx Gdf9Cre. Student’s t-test was used to calculate P values. *P < 0.05, **P < 0.01. Scale bars: 50 μm (A–D)
The partial rescue of Lhx8flx/flxGdf9Cre-induced PFA by Lin28a deficiency argues that Lin28a is a regulator of oocyte growth. The diminished number of activated primordial follicles in Lhx8flx/flxLin28aflx/flxGdf9Cre ovaries suggests that AKT pathway activation is also diminished. We assayed the p-AKT (T308) signal in primordial and activated primordial follicles of Lhx8flx/flxLin28aflx/flxGdf9Cre and Lhx8flx/flxGdf9Cre ovaries (Fig. 5g) and found its expression was significantly reduced in the Lhx8flx/flxLin28aflx/flxGdf9Cre ovary compared to the ovaries of Lhx8flx/flxGdf9Cre mice.
Lhx8 regulates the primary to secondary follicle transition
Previous studies have shown that PTEN-regulated pathways are important in primordial oocyte activation, but not in primary oocytes [24]. We studied the role of Lhx8 in primary oocytes by generating Lhx8flx/flxZp3Cre mice. Zp3Cre is specifically expressed in primary oocytes, and Lhx8flx/flxZp3Cre ovaries continue to express LHX8 in primordial, but not primary, oocytes. Morphometric analyses revealed that the Lhx8flx/flxZp3Cre conditional knockout ovaries did not significantly differ from Lhx8flx/flx (control) mice at PD0 and PD7 (Fig. 6a–f). However, at PD14, we counted 159 ± 23 primary follicles and 29 ± 7 secondary/antral follicles per ovary in the Lhx8flx/flxZp3Cre mice, compared to 79 ± 6 primary follicles and 108 ± 7 secondary/antral follicles per ovary in the control mice (Fig. 6g–i). The relative increase of primary follicles at PD14 and the relative decrease of multilayer follicles in the Lhx8flx/flxZp3Cre ovary indicated that the transition from primary follicles to secondary follicles was blocked, which was consistent with the observation in Lhx8flx/flxGdf9Cre mice (Fig. 1f). At PD21 and PD30, we observed that many primary follicles were devoid of oocytes (Fig. 6k, n). We stained for LIN28A in PD21 ovaries and found that LIN28A was strongly expressed in Lhx8 deficient oocytes of the Lhx8flx/flxZp3Cre ovary, but no expression was detected in the empty follicles (see Additional file 6: Figure S5). However, excluding these empty primary follicles, the number of primary follicles between control and Lhx8flx/flxZp3Cre ovaries was not significantly different at PD21 or PD30 (Fig. 6l, o). For secondary/antral follicles, the number sharply dropped to 17 ± 3 at PD21 and to 2 ± 1 at PD30 in Lhx8flx/flxZp3Cre ovaries, compared to 170 ± 2 and 104 ± 4, respectively, in control mice. These findings imply that the growing follicle pool continued to be eliminated from the Lhx8flx/flxZp3Cre mice. Lhx8flx/flxZp3Cre mice were infertile (see Additional file 2: Figure S2A) and superovulation treatment of Lhx8flx/flxZp3Cre mice did not produce oocytes (see Additional file 2: Figure S2B). This result was in accord with the sharp fall of secondary/antral follicles in the Lhx8flx/flxZp3Cre ovary at PD21. Taken together, these data show that the folliculogenesis of Lhx8flx/flxZp3Cre mice is blocked in the transition from the primary to secondary follicle stage and results in primary oocyte death and infertility. The relative stability of the primordial follicle pool from PD14 to PD30 suggests that the PFA into primary follicles was not affected by the diminution in the number of secondary and more advanced ovarian follicles in Lhx8flx/flxZp3Cre ovaries.
Fig. 6.
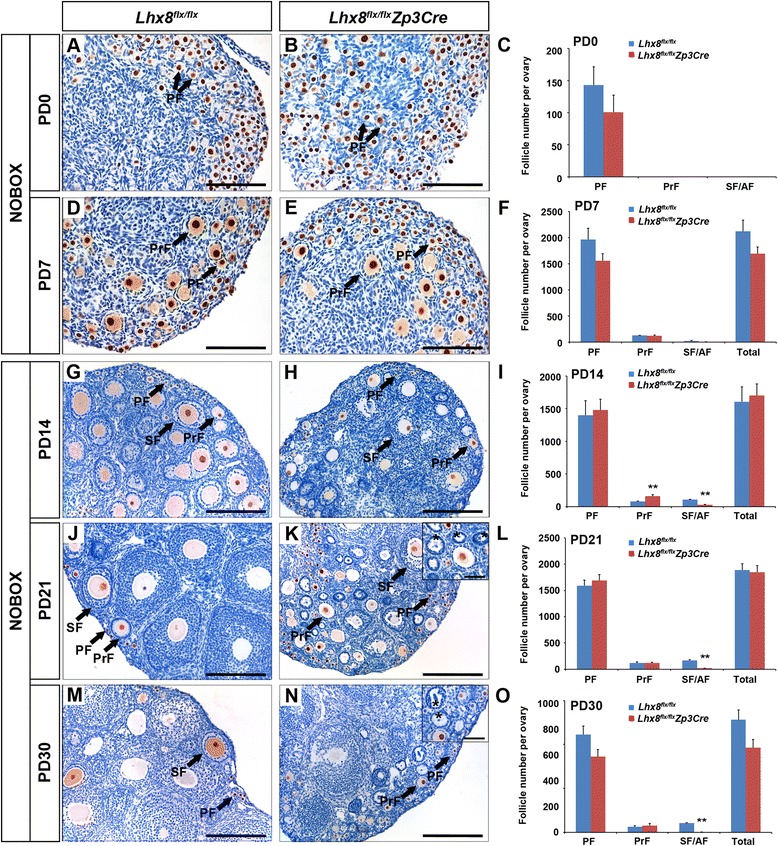
Lhx8 inactivation in primary follicles (Lhx8 flx/flx Zp3Cre) abolishes follicle growth. Histomorphological analysis was done on control (Lhx8 flx/flx) and Lhx8 deficient ovaries (Lhx8 flx/flx Zp3Cre) at various stages of postnatal ovarian development ranging from newborn (PD0) to postnatal day 30 (PD30). a–f Periodic acid–Schiff (PAS) staining and counting of different follicle types in the newborn and PD7 ovaries showed no significant differences between control and Lhx8 deficient ovaries. g–o Anti-NOBOX antibodies were used to stain oocytes (brown immunoreactivity) in ovaries from PD14 (G and H), PD21 (J and K), and PD30 (M and N) mice. At PD14, the Lhx8 flx/flx Zp3Cre ovaries showed a significantly higher number of primary follicles (PrF) and significantly diminished number of secondary/preantral (SF/AF) follicles characterized by two or more layers of granulosa cells. At PD21 and PD30, the primary follicle pool did not differ significantly between Lhx8 flx/flx Zp3Cre and control ovaries; however, there was a marked decrease in the number of secondary and more advanced ovarian follicles in conditional knockouts including degenerating follicles without oocytes (marked by asterisks in insets in K and N). The primordial follicle (PF) pool remained relatively stable between PD14 and PD30, with no significant difference between Lhx8 flx/flx Zp3Cre and control ovaries. **P < 0.01. Scale bars: 100 μm (A, B, D, and E); 200 μm (G, H, J, K, M, and N); 50 μm (insets of K and N)
Discussion
Early folliculogenesis in mammals is one of the least understood frontiers in ovarian follicle biology. Primordial follicles in mice form shortly after birth and remain dormant for prolonged intervals until individual follicles resume growth via little-understood mechanisms of PFA. Several breakthrough studies have highlighted the importance of the PI3K-AKT-mTORC pathways in PFA [5–8, 25, 26]. The Kit receptor mediates the action of the PI3K-AKT-mTORC pathways [27]. Moreover, recent studies show that the Hippo pathway is also involved in PFA [28]. These ubiquitous pathways clearly play important roles in many biological processes and tissues, including oocytes. The role of oocyte-specific pathways in PFA, however, is less clear.
Our previous studies on global knockouts of Lhx8 showed rapid loss of oocytes shortly after birth [13, 14, 29, 30]. The expression of Lhx8 is specifically confined to oocytes and commences during the meiotic prophase in the embryonic gonad and persists in primordial, primary, and antral oocytes during both early development and adulthood. To eliminate any effects of Lhx8 deficiency in the embryonic gonad, we utilized floxed Lhx8 mice and the efficient Gdf9Cre and Zp3Cre mice [16] to delete conditionally Lhx8 alleles in the oocytes of primordial and primary oocytes, respectively. Our results show that Lhx8 represses the earliest stage of PFA, oocyte activation, and is crucial for the differentiation of primary follicles.
The massive PFA caused by depletion of Lhx8 is somewhat reminiscent of the PI3K-Akt-mTORC1 activation caused by conditional deletion of its inhibitors, Pten [6] and Tsc1 [5]. However, unlike the PI3K-AKT-mTOR related experiments, LHX8 appears to act earlier as shown by the decoupling of oocyte activation and somatic cell transformation from flat to cuboidal granulosa cells, by earlier loss of oocytes from Lhx8flx/flxGdf9Cre ovaries, and by LHX8 being upstream of the KIT receptor [14]. PTEN antagonizes PI3K, and removal of PTEN activates AKT, which in turn, phosphorylates a wide range of intracellular targets, including FOXO3. FOXO3 is a specific target of the AKT pathway, and FOXO3 phosphorylation via AKT is thought to cause FOXO3 nuclear export and PFA. Our studies show that cross-talk exists between LHX8 and the PI3K-AKT-mTORC1 pathways. AKT phosphorylation is elevated in the activated oocytes of Lhx8flx/flxGdf9Cre ovaries and Lhx8/Pten double knockouts, which synergistically affects FOXO3 nucleocytoplasmic localization and rpS6 activation. The FOXO3 nucleocytoplasmic shuttling may not be the first step in initiating oocyte activation, because more than 70 % of unactivated primordial follicles (<20 μm) show FOXO3 nucleocytoplasmic translocation in the double Pten/Lhx8 conditionally deficient ovaries, and FOXO3 nucleocytoplasmic shuttling was mostly observed in oocytes greater than 30 μm among single Lhx8 conditional knockouts. Moreover, the Lhx8 conditional knockout phenotype is dominant to Pten as Pten/Lhx8 double conditional knockouts cannot rescue Lhx8 induced oocyte loss, and ovaries are indistinguishable from Lhx8 single conditional knockout ovaries (see Additional file 1: Figure S1).
The expression of genes that are part of the PI3K-AKT-mTORC1 pathways was not significantly affected by conditional inactivation of LHX8 in oocytes by both quantitative PCR and RNA-seq (Fig. 2 and Additional file 5: Table S1). These results indicate that LHX8 indirectly interacts with the members of the PI3K-AKT-mTORC1 pathways to affect PFA. Genes known to affect folliculogenesis, such as Gdf9, Bmp15, and Amh, were downregulated (see Additional file 5: Table S1). The lack of GDF9 and BMP15 is unlikely to explain the decoupling of oocyte activation and somatic cell differentiation as Gdf9 and Bmp15 knockouts form primary follicles [31]. It is likely that factors other than GDF9 and BMP15 are involved in primordial oocyte-soma communication. RNA-seq identified a sixfold upregulation of Lin28a in Lhx8flx/flxGdf9Cre ovaries. Real-time PCR confirmed the upregulation of Lin28a in Lhx8flx/flxGdf9Cre ovaries, but Lin28b was not significantly changed (see Additional file 7: Figure S6A). In addition, Lin28b was not modified in Lhx8flx/flxLin28aflx/flxGdf9Cre ovaries either (see Additional file 7: Figure S6B). LHX8 can bind to the conserved LHX8 DNA binding motif in the Lin28a promoter, and Lin28a deficiency suppresses primordial oocyte activation observed in Lhx8 deficient oocytes. Moreover, Lin28a deficiency reduced the AKT activation observed in LHX8 deficient oocytes. These experiments show that LHX8 indirect regulation of the AKT activation is mediated in part via LIN28A. Interestingly, LIN28A overexpression in myoblasts enhanced phosphorylation of AKT and mTORC1 signaling targets [21]. Lin28a is preferentially expressed in the gonads and germline [32]. In Caenorhabditis elegans, loss of lin-28 results in precocious vulva differentiation and premature developmental progression [33]. In mice, Lin28a gain-of-function leads to a delay in mouse puberty, with increased body size [17]. The mechanism behind LIN28A somatic growth-promoting functions in mammals involves increased glucose utilization in part via an increase in insulin-PI3K-mTOR signaling [17, 21]. Besides, LIN28A is also a well-known RNA-binding protein that blocks biogenesis of many microRNAs [34–36]. Although previous studies have shown that loss of LIN28A function in oocytes does not affect fertility or oogenesis [37], our results suggest that LIN28A overexpression, under the control of LHX8, regulates PFA.
We examined expression of let-7 microRNAs in Lhx8flx/flxGdf9Cre and Lhx8flx/flxLin28aflx/flxGdf9Cre oocytes. Let-7 transcript levels were not changed in either mouse model (see Additional file 8: Figure S7). These results are consistent with a previous report showing that Lin28a and Lin28b do not regulate let-7a in oocytes [37]. Given that Lin28a deficiency reduced the AKT activation observed in LHX8 deficient oocytes, we consider that LIN28A regulates AKT activation through a way independent of let-7. LIN28A binds many RNA species, and further investigations are needed to identify LIN28A’s mechanisms of action in oocytes [38, 39].
Unlike PTEN-mediated pathways, whose actions are stage-specific and confined to PFA [24], Lhx8 deletion in primary oocytes disrupts follicle growth beyond primary oocytes and results in infertility. The initial rise in primary follicles circa PD14 is likely due to the onset of the Zp3Cre-mediated inactivation of Lhx8 that blocks the transition from primary to more advanced follicles. At PD21 and PD30, oocyte death in primary follicles is evident. Interestingly, we did not observe a significant decline in primordial follicles from PD0 to PD30, which argues against the notion that secondary and more advanced follicles inhibit PFA.
Conclusions
Our study shows that Lhx8 plays a critical role in primordial oocyte activation and somatic differentiation and proliferation. Lhx8-regulated pathways are dominant over PTEN pathways, are upstream of KIT, and unlike PTEN pathways, are required for later stages of oogenesis. Multiple pathways, both ubiquitous (PI3K-AKT and Hippo) and oocyte-specific (LHX8), play important roles in oocyte activation and survival. These models will allow further molecular dissection of oocyte-specific activation and signaling that allows the somatic component to differentiate from pre-granulosa cells to cuboidal and multi-layered follicles.
Methods
Mice
The conditional knockout of Lhx8 was established by mating Lhx8flx/flx (C57BL/6 J-129/Ola) mice and Gdf9Cre (C57-B6/SJL) or Zp3Cre mice (C57-BL/6 J) [15, 16, 40], to create Lhx8flx/flxGdf9Cre mice. The Ptenflx mice were obtained from the Jackson Laboratory (Bar Harbor, MN) [41]. Homozygous floxed B6;129-Lin28atm1Egm mice were purchased from Jackson Laboratory [42].
Histology, immunohistochemistry, and immunofluorescence analysis
Ovaries were fixed in 10 % buffered formalin or 4 % paraformaldehyde at room temperature for less than 24 h, processed, embedded in paraffin, serially sectioned (5 μm), and stained by periodic acid–Schiff (PAS) reagent (#26052-05, Electron Microscopy Sciences, Hatfield, PA) and Mayer’s hematoxylin counterstain for histological analysis. The immunohistochemistry was studied as previously described [13]. Affinity purified anti-LHX8 antibodies were generated as previously described [13]. Antibodies against p-rpS6 (S235/236, #2211) were purchased from Cell Signaling Technology (Danvers, MA). Immunofluorescence was performed as previously described [43]. Antibodies against FOXO3, LIN28A, p-AKT (S473), and p-AKT (T308) were purchased from Cell Signaling Technology (Danvers, MA) and Millipore (Temecula, CA). The immunofluorescence intensity of p-AKT (T308) was measured using ImageJ software [44, 45]. Corrected total cell fluorescence (CTCF) was used for statistical analysis [44, 45]: CTCF = Integrated density – (Area of selected cell × Mean fluorescence of background readings).
RNA isolation, reverse transcription, RT-qPCR, and RNA-seq analysis
RNA from PD7 ovaries was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA) and RNA from PD7 oocytes was extracted by the ARCTURUS PicoPure RNA Isolation Kit (Life Technologies, Grand Island, NY), then further subjected to DNase digestion with the RNase-Free DNase Set (Qiagen) and RNA cleanup. RT-qPCR was performed with the CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA). Each experiment was repeated with a minimum of three independent samples, and results were normalized to Gapdh. The relative amount of transcripts was calculated by the ΔΔCT method. Significance was calculated using Student’s t-test with Excel (Microsoft).
For RNA-seq, ten pairs of control and Lhx8 conditionally deficient PD7 ovaries were used to extract RNA; a TruSeq RNA sample prep kit (Illumina, San Diego, CA) was used to generate a cDNA library. Approximately 180 million reads were generated per sample (control and cKO cDNA) on Hi-Seq 2000 (Illumina, San Diego). The RNA-seq reads were analyzed using TopHat [46], Cufflinks [47], and Cuffdiff [48]. The raw RNA-seq data from this study have been submitted to the Sequence Read Archive of the National Center for Biotechnology Information with accession number SRP047051.
Oocyte isolation and Western blots
Oocytes were isolated according to the method described by Reddy et al. [26]. In total, 400 oocytes from control or Lhx8flx/flxGdf9Cre ovaries were isolated and used for Western blots. Primary antibodies of LIN28A (#8641), p-AKT (S473, #9271), p-AKT (T308, #2965), AKT (#9272), and histone H3 (#4499) were purchased from Cell Signaling.
Oocyte ChIP and ChIP-qPCR
Oocyte ChIP was performed following a micro-chromatin immunoprecipitation protocol [49] with some modification. Oocytes were isolated from five PD5 wild-type mice and fixed with 37 % formalin in a 1 % final concentration for 10 min. Chromatin fragmentation was conducted using 1 U micrococcal nuclease for 15 min at 37 °C, following the manufacturer’s manual (Pierce Chromatin Prep Module, #26158, Pierce Biotechnology, Rockford, IL). Chromatin was dissolved in 100 μl nuclear extraction buffer, and affinity purified LHX8 antibody [13] or normal guinea pig IgG (sc-2711) were used for immunoprecipitation. Immunoprecipitated DNA was subjected to real-time PCR analysis using the primers corresponding to the Lin28a promoter region that contains the putative LHX8 DNA binding element [22]. Primer set 1 forward: 5′-gagggggagggctcatccatttct-3′, reverse: 5′-ttccagagagttgggggagggagg-3′ (−584 to −485); and primer set 2 forward: 5′-gctcatccatttctcccttgacagg-3′, reverse: 5′-acacacacacacacacacaccacc-3′ (−574 to −434).
Statistics
Student’s t-test was performed for comparative studies. P < 0.05 was considered statistically significant.
Acknowledgements
We would like to thank Huaiyang Jiang for his assistance in the RNA-seq data analyses. This study was supported by Magee-Womens Research Foundation and R01 HD044858 funding.
Abbreviations
- bp
base pair
- ChIP
Chromatin immunoprecipitation
- IgG
Immunoglobulin G
- PAS
Periodic acid–Schiff
- PCR
Polymerase chain reaction
- PD
postnatal day
- PFA
Primordial follicle activation
- qPCR
Quantitative polymerase chain reaction
- RNA-seq
RNA sequencing
- RT-qPCR
real-time quantitative polymerase chain reaction
Additional files
AKT pathway activation does not rescue oocyte death in Lhx8 conditionally deficient ovaries. Ovaries were harvested at PD35 and examined for the presence of follicles in control (Lhx8 flx/flx, A), Pten conditional knockout (Pten flx/flx Gdf9Cre, B), Lhx8 conditional knockout (Lhx8 flx/flx Gdf9Cre, C), and double Lhx8/Pten conditional knockout (Lhx8 flx/flx Pten flx/flx Gdf9Cre, D) ovaries. Lhx8 flx/flx Gdf9Cre mice lose oocytes rapidly, so that by PD35 few remain. This is distinctly different from Pten flx/flx Gdf9Cre mice, whose activated oocytes persist beyond 5 weeks [6], as PTEN deletion activates the AKT pathway by increased AKT phosphorylation, which results in increased cell survival. The double knockout histology was not significantly different from Lhx8 conditional knockout and indicates that the Lhx8 pathway is dominant to the Pten pathway in controlling oocyte survival. Scale bars: 100 μm.
Fertility test of Lhx8 flx/flx Zp3Cre mice. A Lhx8 flx/flx Gdf9Cre (G9cKO) mice and Lhx8 flx/flx Zp3Cre (Zp3cKO) mice were infertile. B To test the ovulation ability of the Lhx8 flx/flx Zp3Cre mice, 3-week-old mice were intraperitoneally injected with 5 IU pregnant mare serum gonadotropin (PMSG). After 48 hours, 5 IU human chorionic gonadotropin (HCG) was injected. Then 24 hours later, the ovary and oviduct were collected together to flush oocytes.
AKT expression in Lhx8 conditional knockout ovaries. A–F Anti-p-AKT (S473) antibodies show p-AKT(S473) expression mainly in the cytoplasm of primary oocytes in the control (Lhx8 flx/flx) and Lhx8 conditional knockout (Lhx8 flx/flx Gdf9Cre) ovaries, but were not significantly detected in activated primordial follicles (aPF, arrows in F'). G–L p-AKT (T308) was firmly located in the primary follicles (PrF, arrows) or the transitional follicles (PF/PrF, arrow in I'), both in control (Lhx8 flx/flx) and Lhx8 conditional knockout (Lhx8 flx/flx Gdf9Cre) ovaries. Unlike p-AKT (S473), activated primordial follicles in Lhx8 conditional knockouts express p-AKT (T308) (aPF, arrows in L'). The boxed areas in panels C, F , I, and L are shown magnified in C', F', I', and L'. Scale bars: 50 μm (A–L); 20 μm (C', F', I' and L').
Lhx8 and Pten conditional knockouts show synergism in p-rpS6 activation. Ovaries from control (Lhx8 flx/flx, A and A'), Lhx8 conditional knockout (Lhx8 flx/flx Gdf9Cre, B and B'), Pten conditional knockout (Pten flx/flx Gdf9Cre, C and C'), and double conditional knockout (Lhx8 flx/flx Pten flx/flx Gdf9Cre, D and D') mice, were subjected to immunohistochemistry using anti-p-rpS6 antibodies. In the control ovaries and single conditional knockouts, p-rpS6 is expressed primarily in primary or secondary follicles, but not in the vast majority of primordial follicles (PF) or activated primordial follicles (aPF), except for occasional cells (arrowhead in B'). However, in the double conditional knockout mice, the majority of activated primordial follicles are p-rpS6 positive (arrows). Scale bars: 100 μm (A–D); 20 μm (A'–D').
RNA-seq data derived from PD7 control (Lhx8 flx/flx, Ctrl) and Lhx8 flx/flx Gdf9Cre (G9cKO) ovaries. FPKM: Fragments per kilobase of transcript per million mapped reads. The P value is calculated by Cuffdiff software; a P value of less than 0.05 is taken as significant.
LIN28A expression in PD21 Lhx8 flx/flx and Lhx8 flx/flx Zp3Cre ovaries. In PD21 Lhx8 flx/flx control ovaries, LIN28A has weak expression in big follicles (arrows) (A–D), but it was still overexpressed in LHX8 depletion oocytes of Lhx8 flx/flx Zp3Cre mice (arrows) (E–H). However, follicles containing dead oocytes had no LIN28A expression (asterisk). Scale bars: 50 μm.
Lin28a and Lin28b expression in PD7 control (Lhx8 flx/flx and Lhx8 flx/flx Lin28a flx/flx), Lhx8 flx/flx Gdf9Cre, and Lhx8 flx/flx Lin28a flx/flx Gdf9Cre ovaries. A Lin28a and Lin28b transcription was examined by real-time PCR in PD7 control (Lhx8 flx/flx) and Lhx8 flx/flx Gdf9Cre mouse ovaries. B Lin28a and Lin28b transcription was examined by real-time PCR in PD7 control (Lhx8 flx/flx Lin28a flx/flx) and Lhx8 flx/flx Lin28a flx/flx Gdf9Cre mouse ovaries. Data were normalized to Gapdh expression and are given as the mean relative quantity (compared with control), with error bars representing the standard error of the mean. Student’s t-test was used to calculate P values. ** P< 0.01.
Expression of let-7 in Lhx8 flx/flx, Lhx8 flx/flx Gdf9Cre, Lhx8 flx/flx Lin28a flx/flx Gdf9Cre, and Lhx8 flx/flx Lin28a flx/flx Gdf9Cre oocytes. Total RNA from PD7 oocytes was extracted using Trizol. The NCode VILO miRNA cDNA Synthesis Kit (Life Technologies, Grand Island, NY) was used for the reverse transcription of all the small RNAs. Real-time PCR showed that let-7 was not changed in Lhx8 flx/flx Gdf9Cre or Lhx8 flx/flx Lin28a flx/flx Gdf9Cre oocytes. Data were normalized to 5s rRNA expression and are given as the mean relative quantity (compared with control), with error bars representing the standard error of the mean.
Footnotes
Yu Ren, Hitomi Suzuki and Krishna Jagarlamudi contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YR, HS, KJ, and AR designed the research. YR, HS, KJ, KG, and MM performed the research. RL and VP contributed Lhx8 flx mice. YR, HS, and AR analyzed the data. YR and AR wrote the paper. All authors read and approved the final manuscript.
Contributor Information
Yu Ren, Email: yur4@pitt.edu.
Hitomi Suzuki, Email: suzukih.arc@cmn.tmd.ac.jp.
Krishna Jagarlamudi, Email: Jagarlamudikr@mwri.magee.edu.
Kayla Golnoski, Email: mcgeekj@mwri.magee.edu.
Megan McGuire, Email: mmcguire887@gmail.com.
Rita Lopes, Email: rlopes@nimr.mrc.ac.uk.
Vassilis Pachnis, Email: vpachni@nimr.mrc.ac.uk.
Aleksandar Rajkovic, Email: rajkovic@upmc.edu.
References
- 1.Pepling ME, Spradling AC. Mouse ovarian germ cell cysts undergo programmed breakdown to form primordial follicles. Dev Biol. 2001;234:339–51. doi: 10.1006/dbio.2001.0269. [DOI] [PubMed] [Google Scholar]
- 2.McGee EA, Hsueh AJ. Initial and cyclic recruitment of ovarian follicles. Endocr Rev. 2000;21:200–14. doi: 10.1210/edrv.21.2.0394. [DOI] [PubMed] [Google Scholar]
- 3.Mason AJ, Hayflick JS, Zoeller RT, Young WS, Phillips HS, Nikolics K, et al. A deletion truncating the gonadotropin-releasing-hormone gene is responsible for hypogonadism in the Hpg mouse. Science. 1986;234:1366–71. doi: 10.1126/science.3024317. [DOI] [PubMed] [Google Scholar]
- 4.Wandji SA, Srsen V, Voss AK, Eppig JJ, Fortune JE. Initiation in vitro of growth of bovine primordial follicles. Biol Reprod. 1996;55:942–8. doi: 10.1095/biolreprod55.5.942. [DOI] [PubMed] [Google Scholar]
- 5.Adhikari D, Zheng W, Shen Y, Gorre N, Hamalainen T, Cooney AJ, et al. Tsc/mTORC1 signaling in oocytes governs the quiescence and activation of primordial follicles. Hum Mol Genet. 2010;19:397–410. doi: 10.1093/hmg/ddp483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddy P, Liu L, Adhikari D, Jagarlamudi K, Rajareddy S, Shen Y, et al. Oocyte-specific deletion of Pten causes premature activation of the primordial follicle pool. Science. 2008;319:611–3. doi: 10.1126/science.1152257. [DOI] [PubMed] [Google Scholar]
- 7.Castrillon DH, Miao L, Kollipara R, Horner JW, DePinho RA. Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science. 2003;301:215–18. doi: 10.1126/science.1086336. [DOI] [PubMed] [Google Scholar]
- 8.John GB, Gallardo TD, Shirley LJ, Castrillon DH. Foxo3 is a PI3K-dependent molecular switch controlling the initiation of oocyte growth. Dev Biol. 2008;321:197–204. doi: 10.1016/j.ydbio.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Kawamura K, Cheng Y, Liu S, Klein C, Liu S, et al. Activation of dormant ovarian follicles to generate mature eggs. Proc Natl Acad Sci USA. 2010;107:10280–4. doi: 10.1073/pnas.1001198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adhikari D, Flohr G, Gorre N, Shen Y, Yang H, Lundin E, et al. Disruption of Tsc2 in oocytes leads to overactivation of the entire pool of primordial follicles. Mol Hum Reprod. 2009;15:765–70. doi: 10.1093/molehr/gap092. [DOI] [PubMed] [Google Scholar]
- 11.Moriguchi H, Zhang Y, Mihara M, Sato C. Successful cryopreservation of human ovarian cortex tissues using supercooling. Sci Rep. 2012;2:537. doi: 10.1038/srep00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White YAR, Woods DC, Takai Y, Ishihara O, Seki H, Tilly JL. Oocyte formation by mitotically active germ cells purified from ovaries of reproductive-age women. Nat Med. 2012;18:413–21. doi: 10.1038/nm.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pangas SA, Choi Y, Ballow DJ, Zhao Y, Westphal H, Matzuk MM, et al. Oogenesis requires germ cell-specific transcriptional regulators Sohlh1 and Lhx8. Proc Natl Acad Sci USA. 2006;103:8090–5. doi: 10.1073/pnas.0601083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi Y, Ballow DJ, Xin Y, Rajkovic A. Lim homeobox gene, Lhx8, is essential for mouse oocyte differentiation and survival. Biol Reprod. 2008;79:442–9. doi: 10.1095/biolreprod.108.069393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopes R, van Wijk NV, Neves G, Pachnis V. Transcription factor LIM homeobox 7 (Lhx7) maintains subtype identity of cholinergic interneurons in the mammalian striatum. Proc Natl Acad Sci USA. 2012;109:3119–24. doi: 10.1073/pnas.1109251109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lan ZJ, Xu XP, Cooney AJ. Differential oocyte-specific expression of Cre recombinase activity in GDF-9-iCre, Zp3cre, and Msx2Cre transgenic mice. Biol Reprod. 2004;71:1469–74. doi: 10.1095/biolreprod.104.031757. [DOI] [PubMed] [Google Scholar]
- 17.Zhu H, Shah S, Shyh-Chang N, Shinoda G, Einhorn WS, Viswanathan SR, et al. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat Genet. 2010;42:626–30. doi: 10.1038/ng.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elks CE, Ong KK, Scott RA, van der Schouw YT, Brand JS, Wark PA, et al. Age at menarche and type 2 diabetes risk: The EPIC-InterAct study. Diabetes Care. 2013;36:3526–34. doi: 10.2337/dc13-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA. 2002;99:4465–70. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi Y. s. Dev Reprod. 2012;16:379. [DOI] [PMC free article] [PubMed]
- 23.Shyh-Chang N, Daley GQ. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell. 2013;12:395–406. doi: 10.1016/j.stem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jagarlamudi K, Liu L, Adhikari D, Reddy P, Idahl A, Ottander U, et al. Oocyte-specific deletion of Pten in mice reveals a stage-specific function of PTEN/PI3K signaling in oocytes in controlling follicular activation. Plos One. 2009;4 doi: 10.1371/journal.pone.0006186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adhikari D, Liu K. Molecular mechanisms underlying the activation of mammalian primordial follicles. Endocr Rev. 2009;30:438–64. doi: 10.1210/er.2008-0048. [DOI] [PubMed] [Google Scholar]
- 26.Reddy P, Adhikari D, Zheng W, Liang S, Hamalainen T, Tohonen V, et al. PDK1 signaling in oocytes controls reproductive aging and lifespan by manipulating the survival of primordial follicles. Hum Mol Genet. 2009;18:2813–24. doi: 10.1093/hmg/ddp217. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Risal S, Gorre N, Busayavalasa K, Li X, Shen Y, et al. Somatic cells initiate primordial follicle activation and govern the development of dormant oocytes in mice. Curr Biol. 2014;24:2501–8. doi: 10.1016/j.cub.2014.09.023. [DOI] [PubMed] [Google Scholar]
- 28.Kawamura K, Cheng Y, Suzuki N, Deguchi M, Sato Y, Takae S, et al. Hippo signaling disruption and Akt stimulation of ovarian follicles for infertility treatment. Proc Natl Acad Sci USA. 2013;110:17474–9. doi: 10.1073/pnas.1312830110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajkovic A, Pangas SA, Ballow D, Suzumori N, Matzuk MM. NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science. 2004;305:1157–9. doi: 10.1126/science.1099755. [DOI] [PubMed] [Google Scholar]
- 30.Choi YS, Ballow D, Zhao YG, Westphal H, Rajkovic A. Lhx8 deficiency disrupts early folliculogenesis and oocyte-specific gene expression in the mouse ovary. Biol Reprod. 2007;77:91. doi: 10.1095/biolreprod.107.060459. [DOI] [Google Scholar]
- 31.Yan CN, Wang P, DeMayo J, DeMayo FJ, Elvin JA, Carino C, et al. Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function. Mol Endocrinol. 2001;15:854–66. doi: 10.1210/mend.15.6.0662. [DOI] [PubMed] [Google Scholar]
- 32.Childs AJ, Kinnell HL, He J, Anderson RA. LIN28 is selectively expressed by primordial and pre-meiotic germ cells in the human fetal ovary. Stem Cells Dev. 2012;21:2343–9. doi: 10.1089/scd.2011.0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–46. doi: 10.1016/S0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 34.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heo I, Joo C, Cho J, Ha M, Han JJ, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell. 2008;32:276–84. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 36.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–49. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flemr M, Moravec M, Libova V, Sedlacek R, Svoboda P. Lin28a is dormant, functional, and dispensable during mouse oocyte-to-embryo transition. Biol Reprod. 2014;90:131. doi: 10.1095/biolreprod.114.118703. [DOI] [PubMed] [Google Scholar]
- 38.Hafner M, Max KEA, Bandaru P, Morozov P, Gerstberger S, Brown M, et al. Identification of mRNAs bound and regulated by human LIN28 proteins and molecular requirements for RNA recognition. RNA. 2013;19:613–26. doi: 10.1261/rna.036491.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilbert ML, Huelga SC, Kapeli K, Stark TJ, Liang TY, Chen SX, et al. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol Cell. 2012;48:195–206. doi: 10.1016/j.molcel.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Vries WN, Binns LT, Fancher KS, Dean J, Moore R, Kemler R, et al. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis. 2000;26:110–12. doi: 10.1002/(SICI)1526-968X(200002)26:2<110::AID-GENE2>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 41.Lesche R, Groszer M, Gao J, Wang Y, Messing A, Sun H, et al. Cre/loxP-mediated inactivation of the murine Pten tumor suppressor gene. Genesis. 2002;32:148–9. doi: 10.1002/gene.10036. [DOI] [PubMed] [Google Scholar]
- 42.Chakraborty P, Buaas FW, Sharma M, Snyder E, de Rooij DG, Braun RE. LIN28A marks the spermatogonial progenitor population and regulates its cyclic expansion. Stem Cells. 2014;32:860–73. doi: 10.1002/stem.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki H, Ahn HW, Chu T, Bowden W, Gassei K, Orwig K, et al. SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev Biol. 2012;361:301–12. doi: 10.1016/j.ydbio.2011.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gavet O, Pines J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev Cell. 2010;18:533–43. doi: 10.1016/j.devcel.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burgess A, Vigneron S, Brioudes E, Labbe JC, Lorca T, Castro A. Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc Natl Acad Sci USA. 2010;107:12564–9. doi: 10.1073/pnas.0914191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–15. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dahl JA, Collas P. A rapid micro chromatin immunoprecipitation assay (mu ChIP) Nat Protoc. 2008;3:1032–45. doi: 10.1038/nprot.2008.68. [DOI] [PubMed] [Google Scholar]


