Abstract
Integral to the discovery of new pharmaceutical entities is the ability to predict in vivo pharmacokinetic parameters from early stage in vitro data generated prior to the onset of clinical testing. Within the pharmaceutical industry, a whole host of assay methods and mathematical models exist to predict the in vivo pharmacokinetic parameters of drug candidates. One of the most important pharmacokinetic properties of new drug candidates predicted from these methods and models is the hepatic clearance. Current methods, while useful, are still limited in their predictive efficacy. In order to address this issue, we have established a novel microfluidic in vitro culture system, the patented HμREL® device. The device comprises multiple compartments that are designed to be proportional to the physiological architectures and enhanced with the consideration of flow. Here we demonstrate the functionality of the liver-relevant chamber in the HμREL® device, and the feasibility of utilizing our system for predicting hepatic clearance. Cryopreserved human hepatocytes from a single donor were seeded within the HμREL® device to predict the in vivo hepatic clearance (CLH) of six marketed model compounds (carbamazepine, caffeine, timolol, sildenafil, imipramine, and buspirone). The intrinsic clearance rates from static culture controls, as well as clearance rates from the HμREL® device were subsequently compared to in vivo data available from the literature.
Keywords: Microfluidic, Human hepatocyte, Hepatic clearance
1. Introduction
One of the fundamental challenges researchers face in drug discovery within the pharmaceutical industry is the extrapolation of metabolic data from in vitro systems and in vivo animal models to humans, for the purpose of risk assessment. Traditional methods of predicting human response to drugs utilize surrogates—typically either static, in vitro cell-based assays, or in vivo animal studies. Static in vitro cell-based assays are of limited value because they do not adequately mimic the complexity of the physiological environment to which a drug candidate is subjected within a human, and thus may not accurately predict human exposure. While in vivo animal testing can replicate some of the complex inter-cellular and inter-tissue effects, animal studies are expensive, labor-intensive, and time consuming; they also bear the ethical burden of requiring the sacrifice of large numbers of living creatures in the course of their effectuation. One of the most significant drawbacks of in vivo animal testing, due to the pharmacokinetic limitations inherent in the allometric scale-up and extrapolation of assay results from one species to another, is that animal studies are frequently of extremely limited predictive correlation when evaluating human risk [1–3]. Thus a need exists for the development of new, cell-based in vitro methods and devices that can improve the prediction of in vivo drug disposition.
Here we describe a microfluidic microscale cell culture analogue (CCA) system for culturing cellular materials and evaluating the pharmacodynamic and pharmacokinetic interactions between those materials and molecular entities that may be presented to them under conditions of perfusion (flow). The system comprises a biochip (HμREL® chip) on which reside one or more discrete but microfluidically interconnected compartments. The different compartments can house various cell types, thereby simulating, through microfluidic intermediation between them, the metabolic interaction between different human organs (Fig. 1A). The specific chamber geometry is a physical analogue to the concept of a physiologically based pharmacokinetic (PBPK) model—a mathematical model that represents the body as interconnected compartments specific for a particular organ [4–7]. The system also comprises a housing which encloses four HμREL® biochips, a fluid reservoir, and a peristaltic pump (Fig. 2). The elements of the system are interconnected with tubing to comprise, collectively, the HμREL® prototype instrument.
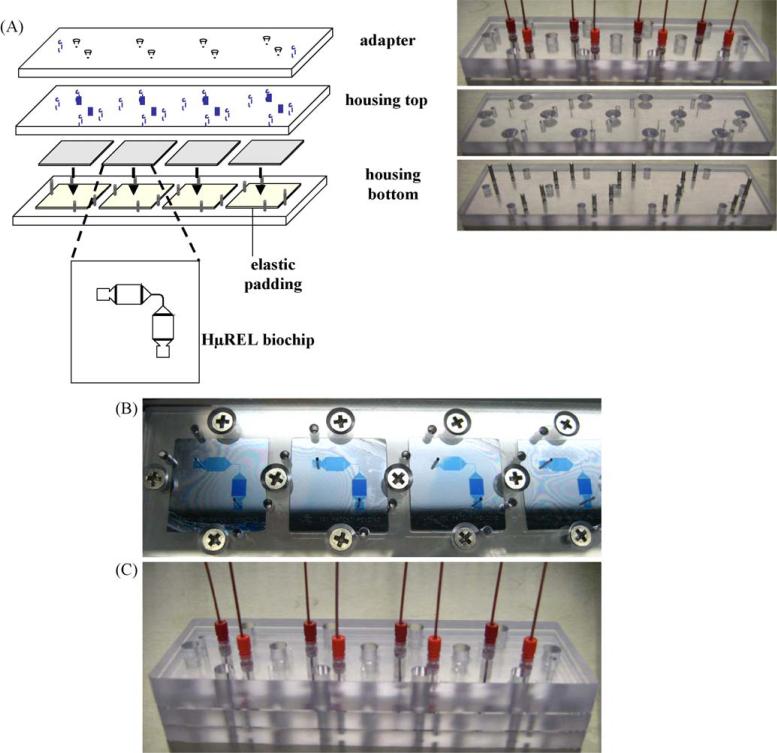
Fig. 1.
The assembly of HμREL® housing set with four biochips. (A) Schematic drawings and the corresponding pictures of the HμREL® housing set: from the top—adapter, housing top, HμREL® biochips, housing bottom with 4 elsatomeric pads. (B) The assembled HμREL® biochips inside the housing top and housing bottom. A blue dye was used to show the functional features on the HμREL® biochip. (C) The picture of a complete set with the adapter plate connected with the tubing.
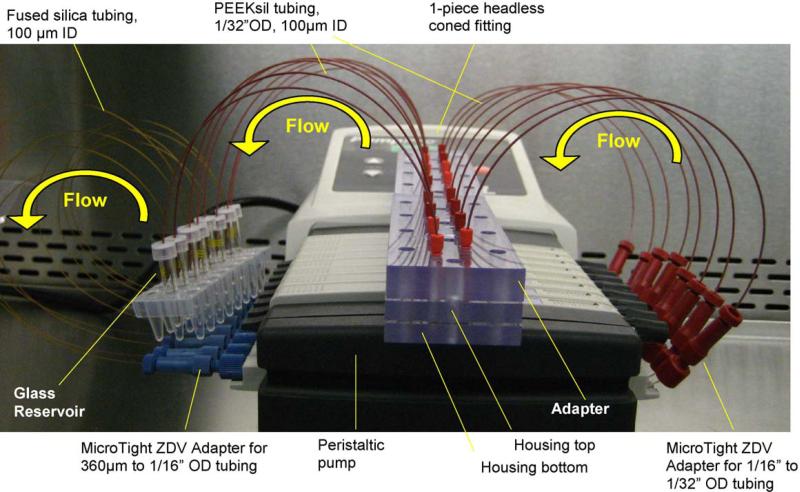
Fig. 2.
The complete setup of a HμREL® prototype instrument. A peristaltic pump was used to generate the flow of culture medium. Two HμREL® housing sets each containing four biochips with cells cultured upon them (for a total of eight microfluidic circuits configured to operate in parallel) were connected to the inlet and outlet PEEKsil tubing (I.D. 100 μm). The inlet PEEKsil tubing was connected to HμREL®tubing (I.D. 250 μm, 8 in. long), which was specially modified to be fitted inside the peristaltic pump, through MicroTight ZDV adapters. The other end of the HμREL® tubing was connected to the fused silica tubing(I.D. 100 μm), which was inserted inside the reservoirs, through MicroTight ZDV adapters. The outlet PEEKsil tubing was inserted in the reservoirs to complete recirculation loops. The glass vials were served as reservoirs and de-bubblers.
In these studies we evaluated six well-characterized drugs with known human in vivo clearance data to test the metabolic competency of human hepatocytes cultured in the HμREL® system. Metabolic clearance data obtained from the HμREL® device was incorporated into a clearance model modified to give effect to flow, the defining characteristic of the system, and was then compared to intrinsic clearance data obtained for the static culture system. These in vitro data were then compared to human in vivo data. We demonstrate significant correlation using the HμREL® device.
2. Materials and methods
2.1. Materials and reagents
Timolol, carbamazepine, buspirone, loperamide, imipramine, caffeine, and dimethyl sulfoxide (DMSO) were obtained from Sigma–Aldrich (St. Louis, MO). Sildenafil was a generous gift from Schering-Plough Research Institute (Kenilworth, NJ). Hanks’ balanced salt solution (HBSS), methanol, isopropanolol, and all other organic solvents were purchased from Fisher Scientific (Waltham, MA) unless otherwise specified. Cryopreserved human hepatocytes, InVitroGRO HI (incubation) medium, InVitroGRO CP (plating) medium and Torpedo Antibiotic were acquired from Celsis in vitro Technologies (Baltimore, MD). Rat Tail Collagen Type I and BD Biocoat™ collagen I 96-well microplates were obtained from BD Biosciences (Franklin Lakes, NJ). LIVE/DEAD viability/cytotoxicity kit for mammalian cells was purchased from Invitrogen (Eugene, OR).
Pharmed® and Tygon®-MHLL tubing were purchased from Cole-Parmer Instrument Company (Vernon Hills, IL). Polystyrene HμREL® biochips and polycarbonate housing sets, as well as HμREL® non-specific-binding peristaltic pump tubing, were obtained from Hurel Corp. (Beverly Hills, CA). All other parts for the microfluidic instrument were obtained from IDEX Health & Science Group (Oak Harbor, WA) unless specified separately.
2.2. Microfluidic device assembly
The details of the microfluidic instrument assembly are similar to those described previously by Sin et al. [8], with some modifications. In brief, a set of polycarbonate plates consisting of a housing bottom, a housing top and an adapter was used to enclose the open features of four HμREL® biochips (Fig. 1) after cells had been incubated upon them, thereby forming a micro-fluidically sealed, linear path for the recirculation of culture medium through each of the four devices under positive pressure actuated by an off-board pump. The setup shown in Fig. 1 provides for four microfluidic circuits, each of which utilizes one of the four HμREL® biochips operating in parallel with and separately from the other three; hence, in this configuration of the HμREL® instrument four separate microfluidic experiments could be run in parallel at one time. The adapter connects to four sets of PEEKsil tubing (100 μm inner diameter (I.D.) and 1/32 in. outer diameter (O.D.)) which serve as inlets to and outlets from the respective biochips through headless one-piece fittings. The inlet PEEKsil tubing in turn connects through MicroTight adapters to 1/16 in. high purity HμREL® tubing (250 μm I.D.) with absorption characteristics composed to minimize non-specific binding of test substrates; the HμREL® tubing conveys culture medium into the interior of a peristaltic pump (205 U, 8-channel; Watson-Marlow, Wilmington, MA). Inside the pump, the HμREL® tubing is inserted into the pump's cassette adapter (not visible in Fig. 2), which holds the tubing in position for exposure to the pump's peristaltic actuation rollers (not visible). The other end of the HμREL® tubing connects to fused silica tubing (100 μm I.D., 360 μm O.D.) through MicroTight adapters. The fused silica tubing is inserted into glass vials, which serve as reservoirs capped with pre-slit snap caps (Fisher Scientific, Pittsburgh, PA), to provide culture medium to the cells, and to which the outlet PEEKsil tubing was inserted to complete the four closed microfluidic circuits that afford recirculation of the culture medium.
2.3. Preparation of polystyrene HμREL® biochips
The biochips were sterilized by soaking in 70% isopropanol for an hour followed by rinsing with sterile distilled water. The biochips were then dried and subsequently treated with air plasma using a high frequency generator (Electro-Technic Products Inc., Chicago, IL) for 3–5 s to modify the surface properties of the biochips to be more hydrophilic, and to increase the collagen-coating efficiency. The “liver chamber” of the biochips was coated with rat tail type I collagen and the biochips were stored at 4 °C in sterile condition. The chips were rinsed with the plating medium 3 times before cell seeding.
2.4. Non-specific binding and stability assessment of drug molecules to the HμREL® device
All pharmaceutical compounds that were tested were diluted to a final concentration of 5 μM in incubation medium supplemented with 2% Torpedo Antibiotic. The drug solutions (100 μL) were placed in the glass vial reservoirs and circulated in the device without cells for 24 h at 37 °C in a humidified 5% CO2 incubator. Aliquots of 5 μL were taken at 0, 1, 2, 4 and 24 h from the reservoirs and were added to a 96-well plate, with each well containing 100 μL of methanol containing an internal standard (10 ng/ml of loperamide). Samples were stored at −20 °C until analyzed by LC–MS/MS.
2.5. Cell culture
2.5.1. Preparation of hepatocytes
In brief, cryopreserved human hepatocytes were removed from liquid nitrogen and thawed quickly in a water bath at 37 °C. The cells were transferred to a 50-mL conical tube containing 5 mL warm plating medium supplemented with 2% Torpedo Antibiotics. The cells were centrifuged at 45 × g (Beckman Coulter, TJ-25, Fullerton, CA) for 5 min at room temperature. After removing the supernatant, the cells were resuspended in plating medium with a cell density of 3 × 106 cells/mL for seeding. The cell viability and number were determined using trypan blue exclusion method in a hemacytometer.
2.5.2. Static culture in 96-well microplates
Hepatocytes suspended in the plating medium were seeded in BD Biocoat™ collagen I 96-well microplates with a seeding density of 30,000 cells per well. The cells were allowed to attach to the plate in a CO2 incubator at 37 °C for 4 h before the exposure of the cells to the drug solution. The drug solutions were prepared in the incubation medium and pre-warmed to 37 °C. The cell containing plates were incubated in a CO2 incubator at 37 °C on an orbital shaker with a shaking speed of 670 rpm. Aliquots of 5 μL were taken at pre-determined time points to 100 μL methanol containing 10 ng/mL loperamide as the internal standard. Samples were stored at −20 °C until analyzed by LC/MS/MS.
2.5.3. Hurel culture
Hepatocytes suspended in the plating medium were seeded in the “liver chamber” – one of the two cell culture compartments of the HμREL® biochips – with a seeding density of 30,000 cells per chamber (the second compartment of each biochip was maintained empty in these experiments, i.e., cell culture medium flowed through but was exposed to no cells in the second culture compartment of each biochip). The cells were allowed to attach to the biochips in a CO2 incubator at 37 °C for 4 h before assembling the biochips to the HμREL® housing sets and applying the flow of culture medium. Once the biochips were enclosed in the housing sets and connected to the tubing and the pump, the microfluidic device was transferred to a humidified CO2 incubator at 37 °C for 10 min to equilibrate the system. The exposure of the cells to the drugs was initiated by replacing the reservoirs to drug containing culture medium. Aliquots of 5 μL were taken from the reservoirs at pre-determined time points add to 100 μL methanol containing 10 ng/mL loperamide as the internal standard. Samples were stored at −20 °C until analyzed by LC–MS/MS.
2.6. Cell viability assay
The viability of human hepatocytes following 24 h flow experiments within the HμREL® device was determined using a LIVE/DEAD viability/cytotoxicity kit following the manufacturer's instruction with slight modifications. In brief, the HμREL® system was perfused with HBSS for 10 min, and followed by 2 μM calcein AM and 4 μM EthD-1 in HBSS solution for 30 min in dark at room temperature. The hepatocytes in the liver chambers of the biochips were observed under a Nikon eclipse TE200 fluorescence microscope. Digital images were acquired using Nikon ACT-1 (v. 2.70) software and a Nikon digital camera DXM1200 connected to the microscope.
2.7. LC–MS/MS method
Samples were centrifuged at 1000 × g for 10 min, and an aliquot (10 μL) of the supernatant was analyzed by LC–MS/MS. The LC–MS/MS system comprised a Shimadzu LC-10ADvp pump (Shimadzu, Columbia, MD), HTS PAL CTC autosampler (Leap Technologies, Carboro, NC), and an API 4000 mass spectrometer with a Turbo Ion Spray probe (Applied Biosystems/MDS SCIEX, Ontario, Canada). The separation of compounds was achieved using a reversed phased stationary phase (Advantage ARMOR C-18, 5 μm, 30.0 mm × 2.1 mm, Analytical Sales and Services, Inc., Pompton Plains, NJ). The mobile phase was a gradient with 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) with a flow rate of 0.8 mL/min. The initial composition of the mobile phase was 5% of B for 0.1 min, followed by a linear gradient to 90% of B over 1.1 min, held at 90% B for 0.2 min, and back to 5% B in 0.1 min. All the samples were detected using multiple reaction monitoring (MRM) in positive ion mode. The area ratio of the analytes to the internal standard was calculated using the Analyst® software v. 1.4.1 (Applied Biosystems, Ontario, Canada).
2.8. Data analysis
2.8.1. Calculation of the CLintfor the static culture system
The in vitro human hepatocyte intrinsic clearances (CLint) are calculated from the substrate concentration profile in the hepatocyte incubation medium for static culture systems using Eq. (1) [9].
| (1) |
C0 and Ct are concentrations (μM) of compound at time 0 and t (min) respectively, AUC0–t is the area under the concentration–time curve from 0 to t with a unit of min μM, V is the volume of incubation solution (100 μL), and N is the number of hepatocytes (30,000 hepatocytes). The CLint is further normalized by 106 hepatocytes to have a unit of μL/min/106 hepatocytes.
A well-defined and widely used well-stirred model [10] is used to scale-up the in vitro intrinsic clearance obtained from the static culture conditions to the estimated hepatic clearance (CLH), assuming the drug is totally unbound in the serum-free culture medium:
| (2) |
where QH is the human hepatic blood flow (20.7 mL/min/kg) and SF is the scaling factor (2.3) from in vitro to in vivo [11].
2.8.2. Calculation of the CLhurel for the HμREL® device
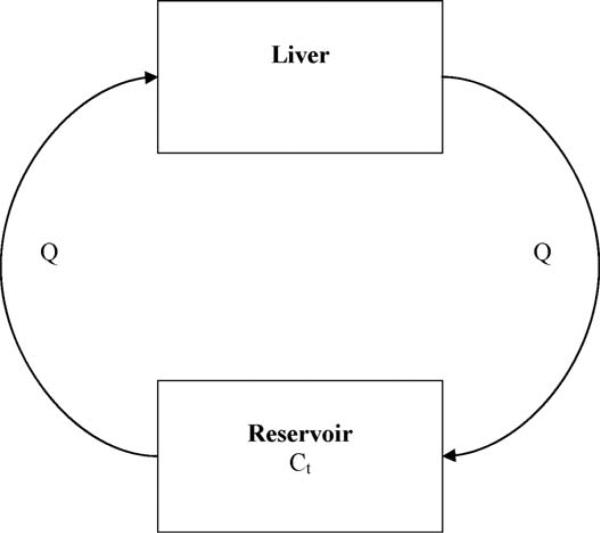
Fig. 3 illustrates the relationship between the clearances of drugs by the liver chamber of a HμREL® device, and the drug concentrations in the reservoir. The liver chamber in the system is the sole eliminating compartment, which is connected to a non-eliminating compartment, the reservoir. The liver compartment and the reservoir are connected via the medium flow with a flow rate of Qhurel (4.5 μL/min/chip) in a recirculation loop.
Fig. 3.
Diagrammatic representation of the liver and the reservoir of the HμREL® system. The arrow indicates the direction of flow (Q). The liver chamber represents the only eliminating compartment. Ct is the concentration of drug in the solution in the reservoir at time t.
The clearance of the HμREL® system (CLhurel, with a unit of μL/min/chip) by hepatocytes cultured in the device can be calculated from:
| (3) |
C0 and Ct are concentrations (μM) of compound in the reservoir at time 0 and t (min) respectively, AUC0–t is the area under the concentration–time curve from 0 to t with a unit of min μM, and V is the volume of incubation (100 μL). The extraction ratio of HμREL® system, Ehurel, is
| (4) |
where Qhurel is the flow rate of the HμREL® device, 4.5 μL/min. The predicted human hepatic clearance from the HμREL® device can be obtained by up-scaling the extraction ratio:
| (5) |
The data are presented in the format of average ± standard deviation with 3 or 4 replications. All figures were plotted using GraphPad Prism v. 4.02.
3. Results
3.1. Non-specific binding and stability studies
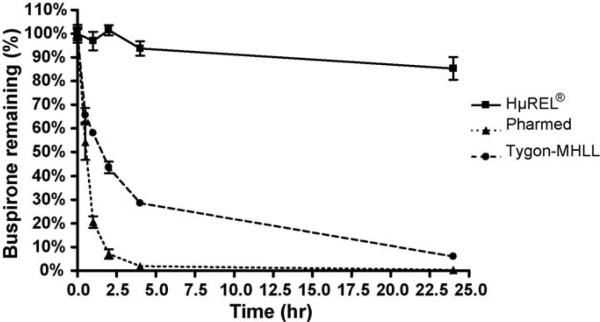
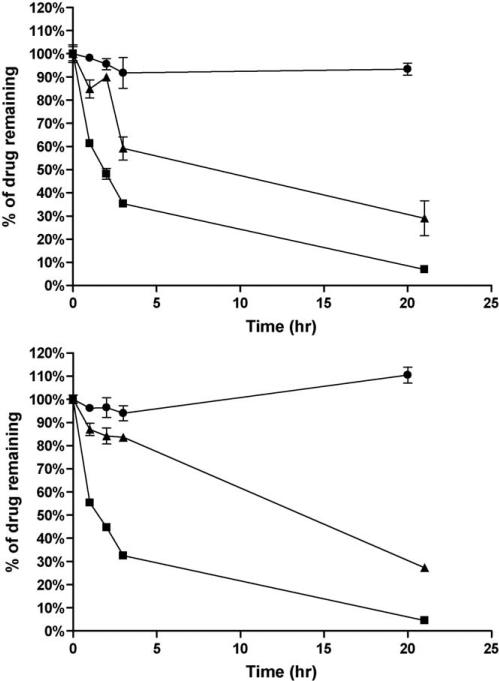
In the first Hurel experiment, it was important to ensure that non-specific binding and stability of our target pharmaceutical compounds with the chip, tubing, and reservoir was not an issue. Drug solutions were made using incubation media and subsequently perfused through the HμREL® device continuously for 24 h in four channels at a flow rate of 4.5 μL/min. Several different tubing types were tested with the target compounds. Non-specific binding was observed when using Pharmed and Tygon-MHLL tubing in the device (Fig. 4) especially when using buspirone, with significant improvement when HμREL® tubing was used within the device (Table 1). The result also indicated that the tested compounds were stable in the flow device under the conditions used throughout the drug clearance study when HμREL® tubing was used.
Fig. 4.
Non-specific binding of a model compound, buspirone, to different tubing used in the HμREL® device. Percentage of remaining buspirone (■, HμREL® tubing; ▲, Pharmed tubing; ●, Tygon-MHLL tubing) was plotted as a function of time. Significant loss of the parent compound was observed when the Pharmed and Tygon MHLL tubing was used due to non-specific binding of the drug molecules to the tubing. No apparent loss of the parent compound was observed when using HμREL® tubing in the HμREL® device.
Table 1.
Non-specific binding of various drug molecules to different peristaltic pump tubing.
| Compound | % Parent compound remaining |
||
|---|---|---|---|
| HμREL® | Pharmed | Tygon-MHLL | |
| Buspirone | 85.3 ± 9.6 | 0.3 ± 0.0 | 6.0 ± 1.3 |
| Carbamazepine | 113.3 ± 8.2 | 18.5 ± 1.3 | N/A |
| Timolol | 99.6 ± 9.6 | 62.7 ± 5.6 | N/A |
| Imipramine | 89.8 ± 4.0 | 0.1 ± 0.0 | N/A |
| Sildenafil | 100.0 ± 6.9 | N/A | N/A |
Solutions containing 5 μM compounds were perfused through the HμREL® system for 24 h at 37 °C. The concentration of drug remained in the system after 24-h recirculation was compared to the initial concentration. Limited amount non-specific binding was found when using HμREL® tubing, whereas significant non-specific binding was observed when using elastic tubing like Pharmed and Tygon-MHLL tubing. The data were expressed as average ± standard deviation in percentage. N/A: data not available.
3.2. Evaluation of cell seeding in the HμREL® device
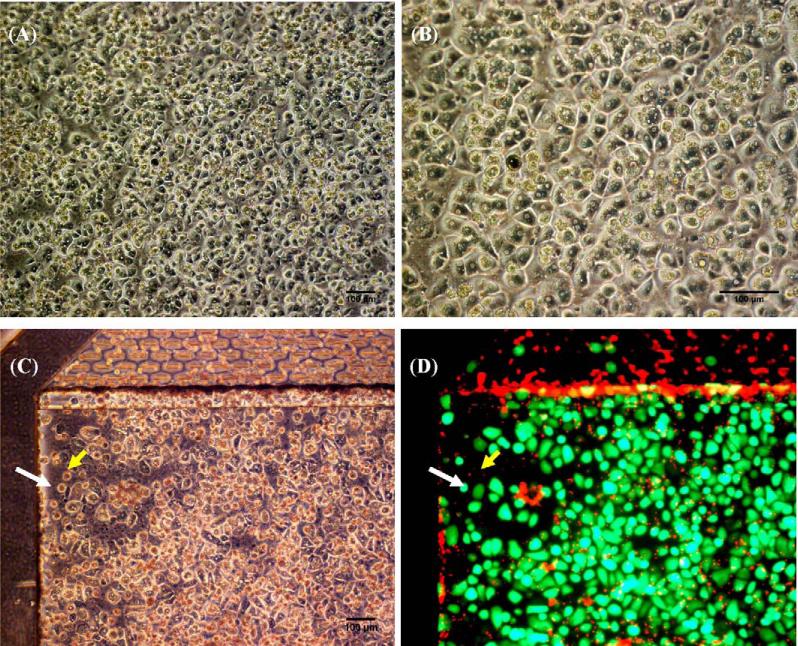
After ensuring that non-specific binding was not present within the system, the viability of cryopreserved human hepatocytes seeded within the device was evaluated. The morphology of the plated cryopreserved human hepatocytes on the HμREL® biochips was examined using a phase contrast microscope prior to applying flow to the cells (Fig. 5A and B). The human hepatocytes were able to attach to the liver chamber of the HμREL® biochips after seeding, with >90% confluency. The collagen-coated biochips showed equal cell seeding efficiency after 3 months storage at 4 °C (data not shown). The viability of cryopreserved human hepatocytes cultured under flow in the device for 24 h was determined (>95%) using a LIVE/DEAD stain, and a characteristic image is shown in Fig. 5D. The morphology of the cells under optical microscope of the same field is also shown for comparison. The superimposed green and red fluorescence image showed that the red fluorescent nuclei of dead cells did not overlay with the green fluorescent live cells. The result suggested that the human hepatocytes cultured inside the HμREL® biochips were viable under flow in the device for at least 24 h.
Fig. 5.
(A and B) Phase-contrast images of human hepatocytes 4 h after seeded on plastic HμREL® biochips and prior to applying flow to the cells. (C) Phase contrast image of human hepatocytes cultured on plastic HμREL® chips under flow for 24 h. (D) Fluorescence image (superimposed live and dead fluorescence images) of the same field of (C). The white arrow indicates a live cell and the yellow arrow indicates a dying or dead cell. Scale bars are equivalent 100 μm in the pictures.
3.3. Evaluation of parent compound clearance
To assess the metabolic competency of human hepatocytes cultured in the HμREL® biochips, 6 compounds with known in vivo clearance values covering a wide clearance range (from 1.3 to 28 mL/min/kg), were selected to be tested in the experiment (Table 2). The clearance of the same compounds was evaluated using human hepatocyte static cultures in a commercially available collagen-coated 96-well plate with the same cell seeding density (i.e., 30,000 hepatocytes/well) for comparison. In general, for compounds with medium or high clearance values, the compounds were cleared (Fig. 6) by hepatocytes cultured in both conditions tested. Since control samples, i.e., drug solutions recirculating within the HμREL® device with the absence of hepatocytes, were always conducted side-by-side with the HμREL® device with the presence of hepatocytes, the model compounds were all stable throughout the duration of the study (data not shown). The data from control samples were consistent with the stability study performed earlier (Table 1). Therefore, the result suggested that the disappearance of the model compounds in the HμREL® device with the presence of hepatocytes was due to the metabolic competence of the cultured hepatocytes under flow condition.
Table 2.
The in vivo clearance data and the hepatic extract ratios of the 6 tested compounds. The hepatic extract ratio is calculated by dividing the calculated hepatic clearance (CLin vivo × (100% – urinary excretion in %)) by the hepatic flow (QH, 20.7 mL/min/kg). The estimated hepatic clearance data of 6 compounds by human hepatocytes cultured in traditional 96-well Biocoat plates are obtained by using Eq. (1): , and Eq. (2): . The estimated hepatic clearance data by using the HμREL® device are calculated using. Eq. (1): , Eq. (2): , and Eq. (5): CLH = Qh × Ehurel.
| Chemical | In vivo |
HμREL® (flow) |
96-Well (shaking) | |||
|---|---|---|---|---|---|---|
| In vivo clearance (mL/min/kg) | Hepatic extraction ratio (E) | HμREL® clearance CLhurel (μL/min/chip) | HμREL® extraction ratio Ehurel | Estimated hepatic clearance CLH (mL/min/kg) | Estimated hepatic clearance CLH (mL/min/kg) | |
| Buspirone | 28.3 ± 10.3 | 1.00 | 3.13 | 0.69 | 14.4 | 10.0 |
| Imipramine | 13 ± 1.7 | 0.62 | 6.10 | 1.00 | 20.7 | 14.8 |
| Timolol | 7.7 ± 1.2 | 0.34 | 0.35 | 0.08 | 1.6 | 3.5 |
| Sildenafil | 6.0 ± 1.1 | 0.29 | 1.13 | 0.25 | 5.2 | 5.4 |
| Carbamazepine | 1.3 ± 0.5 | 0.06 | 0.01a | 0.01a | 1a | 1a |
| Caffeine | 1.4 ± 0.5 | 0.07 | 0.01a | 0.01a | 1a | 1a |
The difference in the drug concentrations between time 0 and time t (C0 – Ct) was not significant when compared to the standard deviations; therefore the CLhurel and CLint were too small to be determined.
Fig. 6.
Metabolic profiles of model compounds by human hepatocytes cultured under flow condition (HμREL® device, top) and traditional shaking plate (96-well collagen-coated plate, bottom). Percentage of remaining imipramine (■, high clearance), sildenafil (▲, medium clearance), and carbamazepine (●, low clearance) with initial concentration of 1 μM was plotted as a function of time. The concentrations of the compounds were monitored and expressed as percentage of the initial concentrations. The data were presented as a mean ± standard deviation with at least 3 replicates.
The kinetics of the model compounds were determined based on the depletion of the substrates in the incubation medium with human hepatocytes. The intrinsic clearance and estimated hepatic clearance of the tested compounds using plated hepatocyte in multi-titer plates were calculated using Eq. (1), and the intrinsic clearance was scaled-up using Eq. (2) to the estimated human hepatic clearance. The CLhurel was calculated using Eq. (4), and the estimated human hepatic clearance from the extraction ratio was calculated using Eq. (4). The Ehurel then was further scaled-up using Eq. (5) to the estimated human hepatic clearance (results presented in Table 2).
4. Discussion
In vivo–in vitro correlation approaches are often limited by assays and methodologies which, while providing some correlative potential, do not represent human in vivo conditions, particularly with respect to geometrical and flow properties. In order to provide more significant correlative data, we have developed the HμREL® system which is constructed as an analogue of the PBPK modeling approach to drug metabolism and pharmacokinetics (Fig. 1A). These chips, designed based on physiological geometries, are then incorporated into the overall flow housing (Fig. 1B and C), which allows the interface with a peristaltic pump (Fig. 2).
In our first phase of optimization, we sought to eliminate nonspecific binding of the target pharmaceutical compounds to the HμREL® biochips as well as the tubing. After demonstrating that non-specific binding was not occurring within our system (Fig. 4) we next evaluated the viability of human hepatocytes culture within the HμREL® device. Prior to applying flow, cryopreserved hepatocytes seeded onto the chips were viewed under phase contrast; and it was observed that the cells exhibited >90% confluency, indicating that the surface properties of the treated HμREL® biochips provided adequate affinity for human hepatocytes. We then showed that the hepatocytes cultured under flow in the HμREL® device were viable for at least 24 h (Fig. 5). While the viability of cell lines cultured in microfluidic devices has been shown previously [5], the viability of primary human cells cultured in a microfluidic system was demonstrated here for the first time (Fig. 5).
There is a body of evidence demonstrating the beneficial effects of applying hydrodynamics to various cultural systems, including maintaining stronger enzymatic activities for long-term culture of primary hepatocytes [12,13] and of cell lines [14], preserving the viability and morphology of liver tissue slices [15], and the ability to perform cellular/genomic analysis of toxicity of organ tissues due to the metabolism of xenobiotics by hepatocytes [5,6,16,17]. Bader et al. [18] first reported that primary rat hepatocytes in a small-scale flat-membrane bioreactor maintained drug biotrans-formation capacity of uripidil for at least 14 days. A similar study utilizing porcine hepatocytes showed that cells cultured in a flat-membrane bioreactor maintained their phase I and phase II activities and responded to inducing drugs over a 3-week period [18,19]. Studies done with hepatocellular cell lines have shown similar results indicating that this model system is useful in studying drug metabolism [20]. One major goal of the current studies was thus to investigate the metabolic competency of human hepatocytes cultured in the HμREL® biochips under flow condition and compare it to a static hepatocyte culture approach. In the HμREL® device, the application of flow to cultured cells not only shows a beneficial effect on viability and cellular morphology but it is also advantageous in maintaining cellular metabolic competency for longer periods of time. For high and medium clearance compounds, hepatocytes cultured in the HμREL® device had similar metabolic activities as the ones cultured in the static condition (Fig. 6). The result indicates that human hepatocytes cultured under flow condition are at least as metabolically active as the ones cultured under a traditional static surrogate system. For the low clearance compounds carbamazepine, and caffeine, the HμREL® device and the static culture system did not show significant depletion of the parent compounds at 24 h. Future experiments will evaluate long-term culture within the HμREL® device, and the capacity to clear low clearance compounds due to the system's ability to maintain hepatic viability and metabolic function for longer periods of time under conditions of flow.
After establishing viability and metabolic functionality within the HμREL® device, our final major goal was to evaluate the ability to generate in vivo–in vitro correlations using data generated with the HμREL® device, and to compare predictability versus the static culture approach. For static culture systems, the intrinsic clearance was calculated, and a well-stirred model [10] was applied to obtain the predictive hepatic clearance. Since a flow component is incorporated in the HμREL® device, the device itself represents a well-stirred model; clearance data obtained using the HμREL® device can, therefore, scale-up directly to predict the hepatic clearance. Here we proposed mathematic calculations utilizing the extraction ratio concept to scale-up the clearance data obtained using the flow device to the estimated human hepatic clearance. The predicted data are comparable to the conventional intrinsic clearance calculation [9] with the model compounds tested (Table 2). The device thus provides the apparent benefit of avoiding mathematical modeling to predict in vivo parameter value(s).
In summary, we have demonstrated that the HμREL® device is capable of metabolizing drug molecules by human hepatocytes cultured under a flow (perfusion) condition. While we have not yet evaluated a large panel of pharmaceutical compounds, we have verified that cultured human hepatocytes are viable and metabolically competent, and that reference compounds exhibiting different degrees of in vivo clearance are metabolized at similarly differential clearance rates within the HμREL® device. In addition, the flow device may be a useful tool to investigate drug protein binding under flow conditions, drug–drug interactions, and interactions between different organs, as well as varying metabolic disposition of pharmaceutical compounds depending upon the underlying genetic makeup of the populations to which they are exposed.
References
- 1.Marathe PH, Rodrigues AD. In vivo animal models for investigating potential CYP3A- and Pgp-mediated drug–drug interactions. Curr Drug Metab. 2006;7:687–704. doi: 10.2174/138920006778520598. [DOI] [PubMed] [Google Scholar]
- 2.Kramer JA, Sagartz JE, Morris DL. The application of discovery toxicology and pathology towards the design of safer pharmaceutical lead candidates. Nat Rev Drug Discov. 2007;6:636–49. doi: 10.1038/nrd2378. [DOI] [PubMed] [Google Scholar]
- 3.Olson H, Betton G, Robinson D, Thomas K, Monro A, Kolaja G, et al. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul Toxicol Pharmacol. 2000;32:56–67. doi: 10.1006/rtph.2000.1399. [DOI] [PubMed] [Google Scholar]
- 4.Oh TI, Sung JH, Tatosian DA, Shuler ML, Kim D. Real-time fluorescence detection of multiple microscale cell culture analog devices in situ. Cytometry A. 2007;71:857–65. doi: 10.1002/cyto.a.20427. [DOI] [PubMed] [Google Scholar]
- 5.Viravaidya K, Sin A, Shuler ML. Development of a microscale cell culture analog to probe naphthalene toxicity. Biotechnol Prog. 2004;20:316–23. doi: 10.1021/bp0341996. [DOI] [PubMed] [Google Scholar]
- 6.Viravaidya K, Shuler ML. Incorporation of 3T3-L1 cells to mimic bioaccumulation in a microscale cell culture analog device for toxicity studies. Biotechnol Prog. 2004;20:590–7. doi: 10.1021/bp034238d. [DOI] [PubMed] [Google Scholar]
- 7.Sin A, Chin KC, Jamil MF, Kostov Y, Rao G, Shuler ML. The design and fabrication of three-chamber microscale cell culture analog devices with integrated dissolved oxygen sensors. Biotechnol Prog. 2004;20:338–45. doi: 10.1021/bp034077d. [DOI] [PubMed] [Google Scholar]
- 8.Sin A, Chin KC, Jamil MF, Kostov Y, Rao G, Shuler ML. The design and fabrication of three-chamber microscale cell culture analog devices with integrated dissolved oxygen sensors. Biotechnol Prog. 2004:338–45. doi: 10.1021/bp034077d. [DOI] [PubMed] [Google Scholar]
- 9.Lau YY, Sapidou E, Cui X, White RE, Cheng K-C. Development of a novel in vitro model to predict hepatic clearance using fresh, cryopreserved, and sandwich-cultured hepatocytes. Drug Metab Dispos. 2002;30:1446–54. doi: 10.1124/dmd.30.12.1446. [DOI] [PubMed] [Google Scholar]
- 10.Pang KS, Rowland M. Hepatic clearance of drugs. I. Theoretical considerations of a “well-stirred” model and a “parallel tube” model. Influence of hepatic blood flow, plasma and blood cell binding, and the hepatocellular enzymatic activity on hepatic drug clearance. J Pharmacokinet Biopharm. 1977;5:625–53. doi: 10.1007/BF01059688. [DOI] [PubMed] [Google Scholar]
- 11.Li C, Liu T, Cui X, Uss AS, Cheng KC. Development of in vitro pharmacokinetic screens using Caco-2, human hepatocyte, and Caco-2/human hepatocyte hybrid systems for the prediction of oral bioavailability in humans. J Biomol Screen. 2007;12:1084–91. doi: 10.1177/1087057107308892. [DOI] [PubMed] [Google Scholar]
- 12.Schmitmeier S, Langsch A, Jasmund I, Bader A. Development and characterization of a small-scale bioreactor based on a bioartificial hepatic culture model for predictive pharmacological in vitro screenings. Biotechnol Bioeng. 2006;95:1198–206. doi: 10.1002/bit.21089. [DOI] [PubMed] [Google Scholar]
- 13.Zeilinger K, Sauer IM, Pless G, Strobel C, Rudzitis J, Wang A, et al. Three-dimensional co-culture of primary human liver cells in bioreactors for in vitro drug studies: effects of the initial cell quality on the long-term maintenance of hepatocyte-specific functions. Altern Lab Anim. 2002;30:525–38. doi: 10.1177/026119290203000506. [DOI] [PubMed] [Google Scholar]
- 14.Gebhardt R, Lippert C, Schneider A, Doehmer J. Improved determination of drug metabolism by perfusion of recombinant V79 cells carrying human CYP3A4. Toxicol In Vitro. 1999;13:639–43. doi: 10.1016/s0887-2333(99)00020-x. [DOI] [PubMed] [Google Scholar]
- 15.Schumacher K, Khong YM, Chang S, Ni J, Sun W, Yu H. Perfusion culture improves the maintenance of cultured liver tissue slices. Tissue Eng. 2007;13:197–205. doi: 10.1089/ten.2006.0046. [DOI] [PubMed] [Google Scholar]
- 16.Kane BJ, Zinner MJ, Yarmush ML, Toner M. Liver-specific functional studies in a microfluidic array of primary mammalian hepatocytes. Anal Chem. 2006;78:4291–8. doi: 10.1021/ac051856v. [DOI] [PubMed] [Google Scholar]
- 17.Gebhardt R, Wegner H, Alber J. Perfusion of co-cultured hepatocytes: optimization of studies on drug metabolism and cytotoxicity in vitro. Cell Biol Toxicol. 1996;12:57–68. doi: 10.1007/BF00143356. [DOI] [PubMed] [Google Scholar]
- 18.Bader A, Fruhauf N, Zech K, Haverich A, Borlak JT. Development of a small-scale bioreactor for drug metabolism studies maintaining hepatospecific functions. Xenobiotica. 1998;28:815–25. doi: 10.1080/004982598239074. [DOI] [PubMed] [Google Scholar]
- 19.Langsch A, Bader A. Longterm stability of phase I and phase II enzymes of porcine liver cells in flat membrane bioreactors. Biotechnol Bioeng. 2001;76:115–25. doi: 10.1002/bit.1151. [DOI] [PubMed] [Google Scholar]
- 20.Iwahori T, Matsuura T, Maehashi H, Sugo K, Saito M, Hosokawa M, et al. CYP3A4 inducible model for in vitro analysis of human drug metabolism using a bioartificial liver. Hepatology. 2003;37:665–73. doi: 10.1053/jhep.2003.50094. [DOI] [PubMed] [Google Scholar]