Abstract
Poly(ADP-ribose) polymerase 1 (PARP1) inhibitors were recently shown to have clinical impact in a number of disease settings, particularly as related to cancer therapy, treatment for cardiovascular dysfunction, and suppression of inflammation. The molecular basis for PARP1 inhibitor function is complex, and appears to depend on the dual roles of PARP1 in DNA damage repair and transcriptional regulation. Here, the mechanisms by which PARP-1 inhibitors elicit clinical response are discussed, and strategies for translating the preclinical elucidation of PARP-1 function into advances in disease management are reviewed.
INTRODUCTION
Poly(ADP-ribose) polymerase 1, PARP1, is the founding member of an enzyme superfamily that serves to add PAR (poly(ADP) ribose) moieties onto target proteins, and in doing so to exerts powerful effects on a number of biological processes critical for cell growth and survival (Gibson and Kraus, 2012). At present, there are at least 17 members of the PARP (and related tankyrase) superfamily; these play important and varied roles in DNA repair, transcriptional regulation, chromatin dynamics, response to hypoxia, cell cycle control, oncogene activity, cell death, maintenance of genomic integrity, and spindle pole regulation (Lupo and Trusolino, 2014). PARP1 is the most abundant member of this family, and shares overlapping functions with the related protein PARP2. In the last decade, intensive focus has been placed on discerning the mechanisms that regulate PARP1 function and the downstream consequence of PARP1 biological activity, given provocative preclinical and clinical observations with regard to the promise of PARP1/2 inhibitors as a means to combat a subset of human malignancies.
From a structural standpoint, PARP1 is composed of six functional domains: there are two homologous zinc finger domains (Zn1 and Zn2) that are associated with detection of DNA damage; a third zinc finger domain that is responsible for coupling the DNA-binding and enzymatic functions of PARP1 (Zn3); a BRCT domain that controls protein – protein interactions, a WGR domain that promotes inter-domain communication, and a C-terminal catalytic domain that controls PAR catalysis (Steffen et al., 2013). Notably, PARP1 function is known to be induced in response to a wide arrays of cellular signals and stresses, including nucleosome conformational changes (Ji and Tulin, 2010; Thomas et al., 2014), altered interacting partners, and induction signaling pathways associated with oxidative, oncogenic, genomic, or inflammatory stress (Luo and Kraus, 2012), but has perhaps been most extensively characterized in the presence of DNA damage. In the context of DNA damage, PARP1 binds damage DNA dependent on the N-terminal domains; this event activates the C-terminal domain to hydrolyze NAD+ (nicotinamide adenine dinucleotide), a cofactor for redox reactions and effector of other cellular events including signal transduction and gene regulation; this then generates PAR chains. Through this mechanism, PARP1 covalently attaches PAR subunits to the Glu, Lys, or Asp residues of target proteins. Similar mechanisms of regulation have been ascribed to PARP2, but there are structural differences, and studies with Parp1−/− mice (Wang et al., 1995) demonstrate that PARP-1 accounts the majority of total cellular PARP activity. Thus, it is generally thought that the cellular consequence of PARylation is largely driven by PARP1, and that the therapeutic effects of PARP1/2 inhibitors are likely to be manifest through modulation of the PARP1 enzyme.
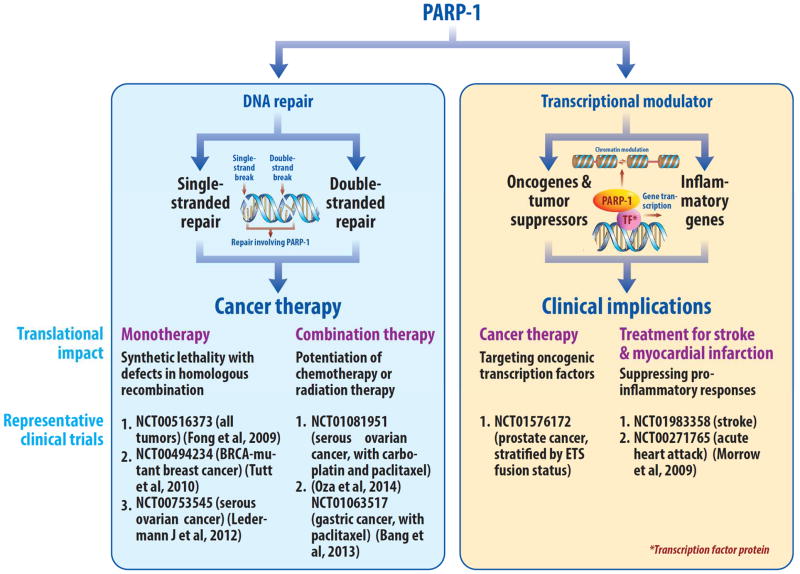
Recent studies have begun to reveal the mechanisms by which PAR exerts its biological effects (Gibson and Kraus, 2012; Lupo and Trusolino, 2014; Schiewer and Knudsen, 2014). PARylation is recognized by PAR-recognizing proteins (“readers”), that contain macrodomains, PAR-binding zinc-fingers (PBZFs), PAR-binding linear motifs (PBMs), and WWE-domains (Barkauskaite et al., 2013). Removal of PAR is also highly regulated, and can occur within minutes (Alvarez-Gonzalez and Althaus, 1989; Gagne et al., 2006). Specialized macrodomain proteins such as PARG (Poly ADP-ribose glycohydrolase) remove PAR and thereby also act as “erasers” (Slade et al., 2011), leaving behind mono-ADP ribosylation that is more stable but is ultimately resolved (Jankevicius et al., 2013). The majority of PARP1 activity is self-directed, and as such, auto-PARylation of PARP1 is a known readout to monitor PARP1 activity. However, proteomic analyses using various human cancer cell lines of demonstrated that outside of auto-modification, the substrates modified by PARP1 are divergent (dependent on cell context), and identify nuclear targets of PARP1 that include DNA repair factors, transcription factors, chromatin remodeling factors, and histones (Zhang et al., 2013). Specific targets of PARylation in the DNA damage response include ALC1 (also known as CHD1L), a macrodomain-containig ATPase and remodeling enzyme that is attracted to sites of PAR formation and therein is thought to facilitate chromatin remodeling at sites of DNA damage (Ahel et al., 2008; Gottschalk et al., 2009). The macrodomain proteins macroH2A.1 also responds to PAR formation and initiates chromatin compaction events that likely contribute to DNA repair (Timinszky et al., 2009). While these functions exemplify the role of PAR-binding proteins on DNA damage associated chromatin alterations, other PARylated proteins play more proximal roles in DNA repair and/or DNA damage checkpoints, including the PBZFs APLF (aprataxin PNK-like factor) and CHFR (checkpoint protein with FHA and RING domains) (Ahel et al., 2008; Rulten et al., 2011). Distinct from these downstream effects of PARP-1 are the PARylation events that modulate transcriptional regulation. Both PARP-1 and PARG play established roles in gene expression through via regulating PARylation and PAR-degradation at target gene regulatory loci (Frizzell et al., 2009; Krishnakumar et al., 2008). Modulation of the KDM5B (JARID1B) histone demethylase provides yet a different mechanism of transcriptional regulation by PARP-1, wherein PARylation of KDM5B suppresses chromatin interaction, thereby sustaining of histone modifications that facilitate gene expression (H2K4me3)(Krishnakumar and Kraus, 2010). Observations in lower eukaryotes have also provided insight into transcriptional regulation by PARP-1, wherein PARP-1 orchestrates chromatin alterations and engages differential transcriptional networks downstream of cellular stresses, such as in response to heat shock (Petesch and Lis, 2012; Sala et al., 2008). These collective observations further implicate PARP1 as a critical effector of DNA repair, transcriptional regulation, and chromatin dynamics, and underscore the need to discern which of the variant PARP1 functions underlie the molecular rationale for targeting PARP1 function in the clinical setting, and for determining which disease types might most benefit from treatment with a PARP1 inhibitor. This review will focus on the dual roles of PARP1 in DNA repair and transcriptional regulation and as relates to clinical utility (Figure 1), highlighting PARP1 functions that are being exploited in on-going clinical trials.
Figure 1.
Schematic highlighting the translational implications of the dual roles of PARP1 in DNA repair and transcriptional regulation.
THE ROLE OF PARP1 IN SENSING DNA DAMAGE AND FACILITATING DNA REPAIR
Of all the known molecular functions of PARP1, perhaps the best characterized is that associated with DNA damage and DNA repair. From an enzymatic standpoint, DNA damage-induced binding and activation of PARP1 is thought to promote a relaxed chromatin structure and to facilitate access of DNA repair enzymes to bind damaged lesions. PARP1 plays a well-established role in base excision repair (BER), and as part of a core complex composed of DNA ligase III, Pol-B, and XRCC1 (Beck et al., 2014). This DNA repair pathway resolves single base pair lesions that occur as a result of genetic insults, including deamination, oxidation, and alkylation. PARP1 senses these lesions, binds, synthesizes PAR at the lesion, and recruits the BER machinery to the site. Other pathways that resolve or generate single-strand breaks, such as nucleotide excision repair (NER) and mismatch repair (MMR) have also been suggested to invoke PARP1 activity (Liu et al., 2011; Robu et al., 2013), although the role of PARP1 in these processes is less well defined. It is clear that PARP1 activity is significantly enhanced in response to ultraviolet (UV) radiation and the generation of UV-induced thymine dimers. Consistent with these functions, PARP1/2 inhibitors sensitize cells to single-strand DNA breaks and base damage.
More recently, a role for PARP1 in double strand DNA break repair has emerged (Helleday et al., 2005; Wang et al., 2006). PARP1 has been found in association with double strand DNA breaks, and PARylation at double strand breaks is thought to facilitate recruitment of appropriate double strand DNA break repair factors. Moreover, PARP1 binding and function have been implicated in the resolution of stalled DNA replication forks (Bryant et al., 2009; Ying et al., 2012). Both double strand DNA break repair and stalled replication forks invoke homologous recombination (HR) and non-homologous end joining (NHEJ) pathways, and a potential role for PARP1 in these processes further substantiates the rationale for utilization of PARP1 inhibitors in the clinical setting. The functional role of PARP1 in double-stranded DNA break repair has been extensively reviewed elsewhere (Golia et al., 2015; Li and Yu, 2014); briefly, PARP1 binds to double-stranded DNA breaks within milliseconds of the damage event, induces PARylation, and attracts the MRN (Mre11, Rad50, NBS1) complex for homologous HR mediated repair. After double strand DNA break resection, resultant single-strand DNA is bound by Rad51, which facilitates template dependent DNA synthesis. Critical to this process are BRCA1 and BRCA2, which facilitate Rad51 loading, and resultant efficient completion of HR-mediated repair. Recent studies further implicate PARP1 in this process; BRCA1 is itself a substrate of PARP1, and the PARylation of BRCA1 DNA-binding domain attenuates its function. Suppression of BRCA1 PARylation resulted in hyperactive HR and genomic instability, demonstrating for the first time that PARP1 and BRCA1 might paradoxically both support and suppress HR. With regard to the more error-prone double-stranded DNA break repair pathway of NHEJ, a potential role for PARP1 was implicated by the observation that the enzyme is frequently found in complex with and to modify DNA-PK, a kinase whose function is critical for NHEJ-mediated DNA repair. However, the overall contribution of PARP1 to classical NHEJ remains uncertain. A potential role for PARP1 in alternative end joining (A-EJ) (Robert et al., 2009), a process that slowly resolves radiation-induced double strand DNA breaks, has also been proposed based on the requirement of PARP1 for A-EJ mediated repair in cells that lack the capacity for NHEJ. Moreover, substantial evidence implicates PARP-1 in backup end joining (B-NHEJ) (Iliakis, 2009). Collectively, the function of PARP1 in DNA repair has expanded from an established role in BER to multiple additional functions in both single and double strand DNA break repair processes. These properties of PARP1 are thought to play a major role in underpinning the function of PARP1 inhibitors in anticancer therapy, as evidenced by the initiation of multiple PARP1 inhibitor trials in this space.
PARP1 INHIBITION AS A STRATEGY FOR TARGETING DNA REPAIR IN CANCER
The concept of targeting PARP1 function in cancer therapy is not new––PARP1 inhibitors emerged in the 1980s, and were shown to both suppress DNA repair, and to enhance the response to DNA damaging agents (Durrant and Boyle, 1982; Nduka et al., 1980). PARP1 inhibitors generally function to suppress covalent attachment of ADP-ribose (monomers or polymers) to PARP1 substrates. With the development of more potent, specific, and effective PARP1 inhibitors, numerous clinical trials are now investigating these agents as an approach to target DNA repair in cancers (Figure 2 and www.clinicaltrials.gov).
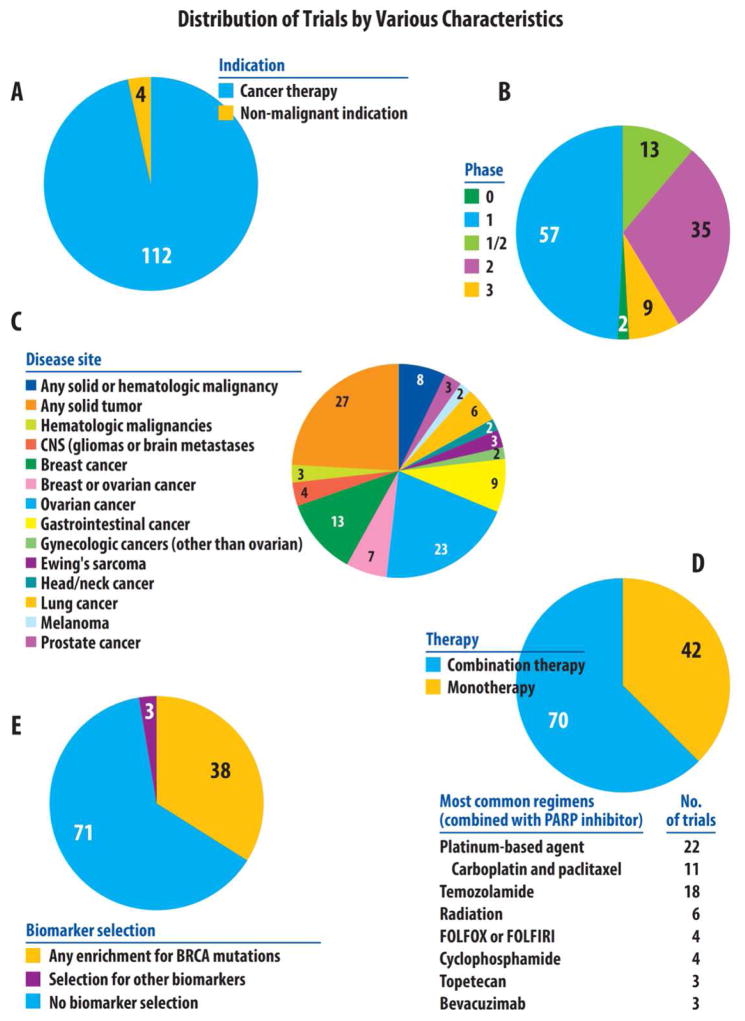
Figure 2.
Distribution of PARP inhibitor trials by various characteristics.
PARP inhibitor studies were identified on the clinical trial repository website (www.clinicaltrials.gov) using the keyword “PARP”. This search, completed in early March of 2015, identified 149 trials. Of these studies, 33 were excluded because they had been withdrawn prior to enrollment or because they did not utilize a true PARP inhibitor. Specifically, it should be noted that all studies including iniparib (BSI-201) were excluded, as it has been determined that this agent actually has poor selectivity toward PARP function, and does not meet the current criteria for a bona fide PARP1 inhibitor (Patel et al., 2012). Of the remaining 116 studies, the distribution of trials is shown by indication (cancer vs non-cancer indication) in Figure 2A and by phase (0–3) in Figure 2B. Of the 112 studies using PARP1 as cancer therapy, the distribution of these trials is then shown by disease site (Figure 2C), therapy (mono- vs combination therapy, Figure 2D), and by biomarker selection (Figure 2E).
The vast majority of current or completed PARP1 inhibitors clinical trials focus on the use of these agents in cancer patients (Figure 2A). Additionally, most of these studies are phase I trials (examining the safety and tolerability of these agents) or phase II trials (investigating the efficacy of these inhibitors in early studies) (Figure 2B). To date, there are 9 active phase III trials (which are large randomized studies definitively assessing the efficacy of PARP inhibitors compared to standard therapies, usually with a survival endpoint). In terms of disease sites, most of the studies targeting a particular cancer focus on either ovarian cancer (23 studies), breast cancer (13 studies), or breast and ovarian cancer (7 studies). It is also likely that many of the trials allowing for patients with any type of solid cancers (27 studies) have likely enrolled a disproportionately higher rate of patients with breast or ovarian cancer, compared to other cancer types. PARP1 inhibitors have been explored as either a monotherapy (42 trials) or in combination with conventional chemotherapy and/or radiotherapy (70 trials) (Curtin and Szabo, 2013) (Figure 2D).
Monotherapy strategies are based on the concept that PARP1 inhibitors may be effective in subsets of cancers that harbor defects in the HR pathways, based on the theoretically increased reliance on PARP1-dependent DNA repair mechanisms including BER (Farmer et al., 2005; McCabe et al., 2006). As predicted by preclinical studies, tumor cells with BRCA1/2 mutations proved highly sensitive to PARP1 inhibitors (Sandhu et al., 2010). In this situation, it is proposed that crippling PARP1 activity in the background of HR deficiency results in accumulation of unrepaired DNA breaks to the extent that the level of genomic instability achieved becomes non-viable. While initially conceived during the era wherein PARP1 was largely associated with BER, this hypothesis is increasingly attractive given the new state of knowledge regarding PARP1 function in double strand DNA break repair. Furthermore, it was posited that PARP1 and the HR pathway play distinct roles in restarting stalled DNA replication forks that occur in response to replication stress, therefore putting forth the provocative hypothesis that PARP1 inhibitor function could be at least partially attributed to suppressing DNA replication fork progression. Therefore in the setting of disabled HR though either somatic or germline alterations, the inhibitor of PARP1 activity leads an inability to repair DNA damage thus creating a synthetic lethal situation for the tumor cells.
To date, the use of synthetic lethality has been most effectively exploited in the context of BRCA1/2 deficient breast and ovarian cancers (Sonnenblick et al., 2015). Patients with germline mutations in BRCA1/2 are highly susceptible to the development of ovarian and breast cancers. In preclinical modeling in this disease type has confirmed that hypersensitivity to PARP1 inhibitors is observed when BRCA function is compromised. Building on these observations, a first in man, Phase I, clinical trial using the PARP1/2 inhibitor olaparib as a single agent in advanced cancers showed objective responses in patients carrying germline BRCA1/2 mutation (Fong et al., 2009). Similar results were observed in expansion cohorts assessing responses in ovarian cancer patients with BRCA mutations, and in Phase II trials that pre-selected for breast and ovarian cancer patients carrying BRCA mutations (Audeh et al., 2010; Tutt et al., 2010). Furthermore, a large randomized phase II study of olaparib versus placebo in patients with recurrent serous ovarian cancers has demonstrated significant improvements in progression-free survival in favor of olaparib treatment, with greater benefit seen in patients harboring a BRCA mutation (Ledermann et al., 2012; Ledermann et al., 2014). These trials have recently led to the regulatory approval of olaparib first by the European Medicines Agency (EMA), and then by the Food and Drug Administration (FDA). Currently, there are a large number of clinical trials for human malignancies using PARP1 inhibitors as a monotherapy, with a large number of these requiring or enriching for either germline BRCA1/2 mutation or other evidence of HR alterations as inclusion criteria (Figure 2E). However, variable response is observed even within published cohorts of patients with BRCA mutations, and PARP1 inhibition can result in improved outcomes even within cohorts with germline wild-type BRCA (Ledermann et al., 2014), thus indicating that effectors of the response to PARP1 inhibitors reach beyond that of BRCA/HR status.
In addition to studies assessing PARP1 inhibitor monotherapy, there are numerous clinical trials, across a range of cancer sites, which employ combination therapy strategies incorporating PARP1 inhibitors with cytotoxic therapies (Figure 2D). These trials are largely based on preclinical studies demonstrating that the suppression of DNA repair pathways by PARP1 inhibitors can potentiate the effects of conventional chemotherapy or radiation therapy (reviewed in (Curtin and Szabo, 2013)). Indeed, Parp1 deficient cells or cell treated with PARP-1 inhibitors are hypersensitive to DNA methylating agents, topoisomerase I inhibitors, and radiotherapy, albeit secondary to slightly differing mechanisms (Horton and Wilson, 2013; Liu et al., 1999; Masutani et al., 1999; Tentori et al., 2002; Wang et al., 1995). DNA methylating agents (e.g., temozolamide) methylate DNA at purine bases, and excision of the resultant N-methylpurines results in a DNA single strand break repaired by PARP1. Thus, by suppressing repair of these single strand breaks, PARP1 inhibition potentiates temozolamide effects. Similarly, topoisomerase I inhibitors (e.g., topotecan or irinotecan) result in DNA lesions repaired by BER, which is blocked by PARP1 inhibitors. Likewise, radiation therapy induces both DNA single-stranded breaks (SSB) and double-stranded breaks (DSB), and PARP1 inhibition can suppress the repair of SSBs that subsequently convert to DSBs upon collision with replication forks in S-phase. It is also likely that PARP1 inhibitors can directly inhibit the repair of DSBs generated by exposure to radiation. Additional in vitro and in vivo data support the potentiation of other cytotoxic agents in a context-specific manner, but the mechanisms underlying such synergy are still being explored. Notably, clinically utilized PARP1/2 inhibitors have also been reported to function in part by trapping PARP1 and PARP1 at sites of DNA damage, thus revealing important insight into the molecular basis by which suppression of PARP1/2 activity can cooperate with DNA damaging agents (Murai et al., 2012). As a consequence of these collective preclinical data, ongoing clinical trials have assessed the combination of PARP1 inhibitors with cytotoxic chemotherapy or radiation therapy, mostly in patient cohorts not screened for any particular DNA repair alterations. Ironically, the largest phase III study assessing combination therapy (with the addition of the presumed PARP1 inhibitor iniparib to gemcitabine and cisplatin) failed to show any additive anti-tumor effects with iniparib (O’Shaughnessy et al., 2014), but it was later determined that this agent actually has poor selectivity toward PARP function, and does not meet the current criteria for a bona fide PARP1 inhibitor (Patel et al., 2012). While this study temporarily dampened enthusiasm of the medical community for PARP1 inhibitor trials, continued reports of anti-tumor efficacy of combination therapy in both preclinical and clinical settings (Bang et al., 2013; Oza et al., 2014; Sonnenblick et al., 2015) has rekindled interest in PARP1 inhibitors, with over 110 clinical trials, most of them ongoing, employing combination therapies incorporating these agents (Figure 2 and www.clinicaltrials.gov).
TRANSCRIPTIONAL REGULATORY FUNCTIONS OF PARP1 AS ASSOCIATED WITH HUMAN DISEASE
In parallel to the realization that PARP1 encompasses a broad scope of DNA repair responses, it is evident that a major function of PARP1 in the absence of DNA damage is to serve as a potent modulator of gene transcription, through activities that include transcription factor regulation, chromatin regulation, and the ability of PARP1 to serve as a context specific transcriptional co-regulator and chromatin modifier (Gibson and Kraus, 2012; Kraus and Hottiger, 2013; Schiewer and Knudsen, 2014). Similar to DNA-PK (Goodwin and Knudsen, 2014), PARP1 interacts with RNA pol II complexes, and can both up- or down-regulate gene expression. Moreover, PARP1 can promote histone H1 exchange at the promoters of actively transcribed genes (Krishnakumar et al., 2008), thus facilitating an active chromatin state required for gene expression. In the context of cancer, PARP1-mediated transcriptional regulation is known to modulate transcriptional regulators whose functions are critical for tumor suppressor function (including p53), oncogene activity, effectors of metastases, chromatin modulators associated with human malignancy, maintenance of stemness/pluripotency, and numerous cell survival and adaptation pathways. HIF1-alpha function has also been shown to be sensitive to PARP1 (Martin-Oliva et al., 2006), therefore linking PARP1 activity to the response to hypoxia. Finally, a specialized role for PARP1 was revealed in hormone-dependent cancers (e.g., breast and prostate cancer), as PARP1 binds to and modulates the activity of a large number of nuclear receptors, including estrogen receptors alpha and beta, the progesterone receptor, and the androgen receptor (Schiewer and Knudsen, 2014). Moreover, PARP1 can modulate the transcription factor ERG activity and potentiate chromosomal rearrangements in prostate cancer (Brenner et al., 2011; Schiewer and Knudsen, 2014). Given the mounting evidence that PARP1 inhibitors show clinical efficacy in a subset of human malignancies, it has been hypothesized that these transcription regulatory functions of PARP1 significantly contribute to the observed anti-tumor activity. This hypothesis is further supported by evidence that PARP1 inhibitors confer significant benefit even in the absence of known BRCA mutations, although this may also in part be related to the loss of other DNA repair genes (Ledermann et al., 2014)
Importantly, PARP1-mediated transcriptional control appears to significantly impact processes with farreaching implications outside of cancer. Across a range of acute medical conditions, such as circulatory shock, myocardial infarction, and stroke, PARP1 has been demonstrated to promote the expression of pro-inflammatory genes which contribute to the pathology of these diseases (Curtin and Szabo, 2013). PARP1 interacts with a number of transcription factors and co-factors, including NFkB, NFAT, AP-1, YY1, sp1, and SIRT1, which have been associated with inflammatory gene expression (Bai and Virag, 2012). In fact, one of the best characterized interactions between PARP1 and a transcription factor is that with NFkB in cellular stress responses, such as inflammation (Curtin and Szabo, 2013; Hassa and Hottiger, 2002). Studies have shown that PARP-1 enzymatic activity directly affects NFkB-mediated transcriptional activity. In particular, PARP1 PARylates both subunits of NFkB, p50 and p65, in vitro and this PARylation inhibits the ability of NFkB to bind to DNA (Kameoka et al., 2000). Moreover, auto-PARylation of PARP1 also promotes the DNA binding ability of NFkB (Chang and Alvarez-Gonzalez, 2001) Coactivation of NFkB-mediated transcriptional programs by PARP1 can enhance expression of inducible nitric oxide synthase (iNOS), cell adhesion molecules (I-CAM, V-CAM, and L-CAM), and matrix metalloproteinases, all of which foster an inflammatory signaling cascade (Garcia Soriano et al., 2001; Ha et al., 2002; Hassa and Hottiger, 1999; Oliver et al., 1999). Pharmacologic inhibition or genetic knockout of PARP1 suppresses levels of inflammatory cytokines, such as TNF-α, IL-1β, IL-6, and IL-12, in animal models of inflammation (Shall and de Murcia, 2000). This may have relevance to the antitumor activity of PARP inhibitors. Moreover, in the context of circulatory shock, studies demonstrated that bacterial wall lipopolysaccharides (LPS, endotoxin) result in PARP1 activation in macrophages, which induces the expression of iNOS and results in the overproduction of nitric oxide, leading to development of vascular contractile failure. Inhibition or knockdown of PARP1 attenuates the tissue infiltration of inflammatory cells, improves vascular and organ function, and produces survival benefits in rodent models of circulatory shock (Curtin and Szabo, 2013; Szabo et al., 1996). Likewise, in the context of myocardial infarction or cardiac transplantation, PARP1 inhibition exerts a significant cardioprotective effect in rodent models, resulting in blunting of the inflammatory response, shielding from reperfusion injury, diminished infarct size, increased cardiac contractility and improved survival (Curtin and Szabo, 2013). In addition to cardiac ischemia, PARP1 inhibition also protects from neuronal injury in the context of ischemic strokes. In ischemic stroke models in monkeys, PARP1 inhibition significantly reduced cerebral infarct volumes and neurological deficits (Matsuura et al., 2011). It should be noted that while suppression of inflammation is a common theme across each of the highlighted medical conditions (shock, myocardial infarction, and stroke), the antiinflammatory properties of PARP1 inhibition only partially explain the efficacy of these agents in these situations. Other contributing mechanisms include inhibition of excitotoxicity that can trigger calcium overload resulting in cell necrosis in ischemic stroke, limitation of DNA strand breakage in the context of oxidative stress following myocardial infarction, and effects on cellular energetics and cell death signaling in all of these scenarios.
Modulation of transcription by PARP1 extends beyond cancer drivers and inflammatory gene programs, often in a context-specific manner. For example, during neuronal differentiation, PARP1 was shown to be instrumental in promoting neurogenic gene expression events through modifying transcriptional co-repressors and displacing them from the promoters of pro-neurogenic genes (Ju et al., 2004). Similar hypotheses have emerged with regard to muscle differentiation gene programs, and the concept that PARP1 elicits a cell specific gene regulatory program is becoming evident. However, it is not clear that these gene regulatory events are universally sensitive to enzymatic inhibitors of PARP1. In some cases, PARP1 residence on chromatin is sufficient to modulate gene expression events, and to modulate chromatin function. In these situations it is postulated that PARP1 acts as a scaffold for recruitment of transcriptional modulators, including histone acetyltransferases. Furthermore, PARP1 activity has been shown to result in significant epigenetic alterations through modulation of CTCF (which regulates gene insulation) and Dnmt1 (a DNA methyltransferase). Finally, PARP1 hyperactivation has been linked to mitochondrial dysfunction (Bai et al., 2014) associated with neurodegeneration in aging, by virtue of modulating the NAD+-Sirt1-PGC1a axis. Taken together, it is apparent that a major cellular function of PARP1 on chromatin is to regulate gene expression, and that PARP1 sensitive transcriptional events can exert contextspecific biologic outcomes that are of relevance for translation of PARP1 inhibitors into the clinic.
PARP1 INHBITORS AS A STRATEGY FOR SELECTIVE MODULATION OF TRANSCRIPTIONAL PROGRAMS
The vast majority of PARP1 inhibitor-based clinical trials were designed and initiated to target DNA repair in cancer (including breast and ovarian cancer), using combinations with genotoxic stress and/or pre-selected populations known to harbor BRCA1/2 mutations. However, there is an emerging rationale for investigating PARP1 inhibitors as monotherapies, in part as a means to suppress transcriptional drivers of disease in the setting of cancer and non-cancer therapy. This rationale is supported by both preclinical data, as summarized above, as well as results from clinical studies suggesting that factors other than homologous recombination deficiencies can impact response to PARP1 inhibitors (Ledermann et al., 2014).
Within the arena of malignancies, PARP1 suppression as a single agent represents a particularly promising disease site for targeting both the DNA repair and transcriptional functions of PARP1 in prostate cancer, based on recent discoveries linking the hormone and DNA repair pathways. PARP1 is recruited to sites of AR (androgen receptor) function (Schiewer et al., 2012), and is it known that both early and late stage prostate cancers are dependent on AR function for growth and progression. Preclinical studies and ex vivo analyses of primary human tissues showed that PARP1 inhibitors suppress AR activity, AR dependent tumor growth, and progression to hormone therapy resistance in the absence of a DNA damaging agent or BRCA alterations. The rationale for utilizing PARP1 inhibitors was further enriched by the observation that AR promotes double-stranded DNA break repair through pathways that implicates PARP1, and invokes NHEJ through DNA-PK regulation (Goodwin et al., 2013; Polkinghorn et al., 2013). These findings provided the molecular basis for clinical observations which showed that anti-androgen therapy acts in concert with radiation in patients with locally advanced disease to improve overall survival and outcome, and suggest that PARP1 inhibitors as a monotherapy would serve to both suppress AR activity and hormone dependent DNA repair. Moreover, chromosomal translocations that result in hyper-expression of pro-oncogenic ERG transcription factors are common in prostate cancer, and PARP1 was shown in preclinical models to function as an ERG cofactor and to support ERG mediated transcriptional activity (Brenner et al., 2011). Thus, PARP1 inhibitors combinatorially suppress the function of the major transcription factors that drive prostate cancer growth and progression. Finally, while BRCA1/2 mutations are thought to be infrequent in prostate cancer, recent observations point toward other alterations that compromise HR and induce sporadic “BRCAness” in this tumor type, including ATM loss. Emerging data from the genomic study of advanced prostate cancers suggest that homozygous mutations in DNA repair genes including ATM in up to 15% of the cases (Beltran et al., 2013). Moreover, the lncRNA PCAT-1 is induced in this disease type and serves to repress BRCA2, thereby conferring marked sensitivity to PARP1 inhibitors as single agents (Prensner et al., 2014). Consistent with these preclinical findings, phase 1 studies were recently completed using the PARP1 inhibitors as single agents in two separate trials and shown significant clinical antitumour activity in patients with advanced sporadic prostate cancers that had progressed despite most available hormone treatments and chemotherapy. More recently, an abstract, presented at the 2014 ESMO meeting, reported preliminary results from an adaptive multi-part Phase II trial (TOPARP; CR-UK/11/029) investigating the activity of the PARP1 inhibitor olaparib in 30 patients with end-stage prostate cancer (Mateo et al., 2014). In this study, olaparib resulted in a 33% response rate, which is quite promising for this patient population. While whole exome sequencing revealed alterations in some DNA repair genes (eg BRCA2 and ATM) enriched among responding patients, a significant percentage of responders did not harbor defects in these DNA repair genes, indicating that the interaction with these genes may not explain all the PARP inhibitor sensitivity in this disease. It has been hypothesized that the effects of PARP1 on gene transcription may contribute to the antitumor activity of PARP inhibitors against metastatic prostate cancer, although more data are now required to interrogate these findings. Currently, there is an ongoing study (NCT01576172, www.clinicaltrials.gov), which stratifies castration resistant prostate cancer patients based on ETS rearrangement status, as ascertained by biopsy of a metastatic lesion, and then randomizes patients to abiraterone (an agent targeting androgen synthesis) alone or in combination with the PARP inhibitor veliparib. This study may provide additional insights on the efficacy of targeting oncogenic transcriptional drivers in prostate cancer with PARP1 inhibitors.
Outside of cancer, while there is an abundance of preclinical data supporting the investigation of PARP1 inhibitors as potential transcriptional regulators in non-oncological indications such as circulatory shock, myocardial infarction, or stroke, there has been, to date, only one completed clinical trial. This single randomized trial assessed the effect of the PARP1 inhibitor INO-1001 in 40 patients with myocardial infarction, undergoing percutaneous coronary revascularization (Morrow et al., 2009). While this study was not powered to assess clinical efficacy, there was a trend towards blunting of inflammatory response, as assessed by serial plasma c-reactive protein and IL-6 levels, with the addition of the PARP1 inhibitor. The only other active non-oncologic PARP1 inhibitor trial is a phase I study assessing safety of the “stroke-targeting” PARP1 inhibitor JPI-289 in healthy volunteers. One may wonder – why are there >140 clinical trials of PARP1 inhibitors in the oncological space, but only 2 studies of PARP1 inhibitors in the non-oncological space? The answer is relatively straightforward—the oncology arena likely represents a more acceptable space for testing drugs which inhibit DNA repair. Because many cancers are aggressive with limited therapeutic options, cancer patients and their physicians are willing to test out novel drugs with “potentially riskier” side-effect profiles (such as an extremely small risk of carcinogenesis from a DNA repair inhibitor). However, these same side-effect profiles are less acceptable to patients with less life-threatening illnesses and longer life expectancies. It is notable that mice lacking the PARP-1 (ADPRT) gene develop normally, and show no evidence of tumor formation, but do show some incidence of epidermal hyperplasic with aging (Wang et al., 1995). While improvements in drug formulation and delivery can help shorten the duration of treatment and further decrease side effects, it is unlikely that PARP1 inhibitors will be widely introduced into clinical trials assessing non-oncological indications, unless better function selectivity (for transcriptional regulation over DNA repair) can be achieved, until these agents gain greater traction with in the oncological space, or until more information can be gleaned about the long-term effects of PARP1 suppression. Nevertheless, the increasing evidence that these drugs are very well tolerated with very limited toxicity to date in oncology trials support the further evaluation of these agents in non-cancer indications.
CONCLUSIONS AND FUTURE DIRECTIONS
Biochemical investigation, preclinical discovery, and clinical analyses have not only nominated PARP1 as a viable therapeutic target for human malignancies, but suggest that PARP1 inhibitors may be effective in cardiovascular dysfunction and inflammatory syndromes. While recent advances suggest that the molecular basis of PARP1 inhibitor function likely depends on the pleiotropic roles of the enzyme in DNA repair, transcriptional regulation, and modulation of chromatin dynamics, significant gaps in understanding remain which limit translation of these findings into the clinical setting. First, which function(s) of PARP1 are critical for observed clinical responses? Although early studies suggest that the transcription regulation and DNA repair associated functions of PARP1 can be functionally segregated through mutational analyses, the molecular underpinnings of these divergent functions remain poorly understood and are closely interlinked. Second, what molecular markers can be identified that may predict sensitivity to PARP1 inhibitors? While the observation that patients with germline or somatic mutation in BRCA1/2 can show exceptional responses to PARP1 inhibitors in the oncology setting, not all patients show this profile, or in other known DNA repair pathways. Studies should be prioritized that will allow for a full molecular dissection of tumors that show heightened sensitivity to PARP1 inhibitors, and these observations challenged in the clinical setting. These studies should include assessment of not only DNA repair factors that might modify response to PARP1 inhibitors, but should include investigation of up- and downstream effectors of PARP1 function (including readers, writers, and erasers of PARylation). Third, as the transcriptional regulatory roles of PARP1 are thought to contribute to the function of PARP1 inhibitors as single agents, what is the genomic profile of PARP1 binding to chromatin, and the basis of selectivity for controlling disease-relevant transcriptional programs? Defining the PARP1 cistrome and transcriptome in human tissues and/or preclinical models that mimic human disease would be of potentially high translational and clinical value. Fourth, the role of other PARP family members should be considered. For example, PARP16 has been shown to contribute to cell stress and unfolded protein responses (Jwa and Chang, 2012), pathways known to be altered in cancers. Finally, what is the contribution of PARP2 to clinical responses to PARP inhibitors? Most agents that have been utilized in clinical trials effectively suppress the activity of both PARP1 and PARP2; while preclinical studies point to PARP1 as the critical therapeutic target, the contribution of PARP2 to clinical responses cannot be dismissed, and a richer understanding of PARP2 mediated molecular and cellular activities would be of likely benefit for interpreting clinical data. Overall, recent findings have clearly invigorated translational and clinical interest in PARP1 inhibitors for use in oncology care, cardiovascular dysfunction, and inflammatory diseases, while biochemical dissection and preclinical modeling has potentiated our understanding of PARP1 inhibitor function from chromatin to the clinic. The molecular basis of PARP1 inhibitor function remains fertile ground for translational discovery.
Acknowledgments
The authors regret omission of meritorious citations due to page constraints. We would also like to thank the Knudsen, de Bono, Rubin, and Feng laboratories for ongoing discussions and intellectual input, most especially Dr. M. Schiewer, G. Goodwin, J. Dean, V. Kothari and J. Evans. Additional advice and input from the following collaborators and colleagues: Arul Chinnaiyan, Maha Hussain, W. Kevin Kelly, Adam Dicker, Leonard Gomella, Scott Tomlins, and Robert Den were greatly appreciated. This work was supported by a PCF Challenge Award (to KEK, JSdeB, MAR and FYF).
Footnotes
Author Contributions: FYF and KEK were the lead authors, but all authors contributed to the content and scope of the study
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- Alvarez-Gonzalez R, Althaus FR. Poly(ADP-ribose) catabolism in mammalian cells exposed to DNA-damaging agents. Mutat Res. 1989;218:67–74. doi: 10.1016/0921-8777(89)90012-8. [DOI] [PubMed] [Google Scholar]
- Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, Scott C, Weitzel JN, Oaknin A, Loman N, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- Bai P, Nagy L, Fodor T, Liaudet L, Pacher P. Poly(ADP-ribose) polymerases as modulators of mitochondrial activity. Trends Endocrinol Metab. 2014 doi: 10.1016/j.tem.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Bai P, Virag L. Role of poly(ADP-ribose) polymerases in the regulation of inflammatory processes. FEBS Lett. 2012;586:3771–3777. doi: 10.1016/j.febslet.2012.09.026. [DOI] [PubMed] [Google Scholar]
- Bang YJ, et al. Olaparib plus paclitaxel in patients with recurrent or metastatic gastric cancer: a randomized, double-blind phase II study [abstract] Journal of Clinical Oncology. 2013;31(Suppl):a4013. doi: 10.1200/JCO.2014.60.0320. [DOI] [PubMed] [Google Scholar]
- Barkauskaite E, Jankevicius G, Ladurner AG, Ahel I, Timinszky G. The recognition and removal of cellular poly(ADP-ribose) signals. FEBS J. 2013;280:3491–3507. doi: 10.1111/febs.12358. [DOI] [PubMed] [Google Scholar]
- Beck C, Robert I, Reina-San-Martin B, Schreiber V, Dantzer F. Poly(ADP-ribose) polymerases in double-strand break repair: Focus on PARP1, PARP2 and PARP3. Exp Cell Res. 2014;329:18–25. doi: 10.1016/j.yexcr.2014.07.003. [DOI] [PubMed] [Google Scholar]
- Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, Patel S, Wang X, Liang H, Yu J, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28:2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambon P, Weill JD, Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem Biophys Res Commun. 1963;11:39–43. doi: 10.1016/0006-291x(63)90024-x. [DOI] [PubMed] [Google Scholar]
- Chang WJ, Alvarez-Gonzalez R. The sequence-specific DNA binding of NF-kappa B is reversibly regulated by the automodification reaction of poly (ADP-ribose) polymerase 1. The Journal of biological chemistry. 2001;276:47664–47670. doi: 10.1074/jbc.M104666200. [DOI] [PubMed] [Google Scholar]
- Curtin NJ, Szabo C. Therapeutic applications of PARP inhibitors: anticancer therapy and beyond. Molecular aspects of medicine. 2013;34:1217–1256. doi: 10.1016/j.mam.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant LG, Boyle JM. Potentiation of cell killing by inhibitors of poly(ADP-ribose) polymerase in four rodent cell lines exposed to N-methyl-N-nitrosourea or UV light. Chem Biol Interact. 1982;38:325–338. doi: 10.1016/0009-2797(82)90062-x. [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- Frizzell KM, Gamble MJ, Berrocal JG, Zhang T, Krishnakumar R, Cen Y, Sauve AA, Kraus WL. Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J Biol Chem. 2009;284:33926–33938. doi: 10.1074/jbc.M109.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JP, Hendzel MJ, Droit A, Poirier GG. The expanding role of poly(ADP-ribose) metabolism: current challenges and new perspectives. Curr Opin Cell Biol. 2006;18:145–151. doi: 10.1016/j.ceb.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Garcia Soriano F, Virag L, Jagtap P, Szabo E, Mabley JG, Liaudet L, Marton A, Hoyt DG, Murthy KG, Salzman AL, et al. Diabetic endothelial dysfunction: the role of poly(ADP-ribose) polymerase activation. Nature medicine. 2001;7:108–113. doi: 10.1038/83241. [DOI] [PubMed] [Google Scholar]
- Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nature reviews Molecular cell biology. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- Golia B, Singh HR, Timinszky G. Poly-ADP-ribosylation signaling during DNA damage repair. Front Biosci (Landmark Ed) 2015;20:440–457. doi: 10.2741/4318. [DOI] [PubMed] [Google Scholar]
- Goodwin JF, Knudsen KE. Beyond DNA repair: DNA-PK function in cancer. Cancer Discov. 2014;4:1126–1139. doi: 10.1158/2159-8290.CD-14-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin JF, Schiewer MJ, Dean JL, Schrecengost RS, de Leeuw R, Han S, Ma T, Den RB, Dicker AP, Feng FY, et al. A hormone-DNA repair circuit governs the response to genotoxic insult. Cancer Discov. 2013;3:1254–1271. doi: 10.1158/2159-8290.CD-13-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci U S A. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha HC, Hester LD, Snyder SH. Poly(ADP-ribose) polymerase-1 dependence of stress-induced transcription factors and associated gene expression in glia. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:3270–3275. doi: 10.1073/pnas.052712399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassa PO, Hottiger MO. A role of poly (ADP-ribose) polymerase in NF-kappaB transcriptional activation. Biological chemistry. 1999;380:953–959. doi: 10.1515/BC.1999.118. [DOI] [PubMed] [Google Scholar]
- Hassa PO, Hottiger MO. The functional role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-kappaB in inflammatory disorders. Cellular and molecular life sciences : CMLS. 2002;59:1534–1553. doi: 10.1007/s00018-002-8527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T, Bryant HE, Schultz N. Poly(ADP-ribose) polymerase (PARP-1) in homologous recombination and as a target for cancer therapy. Cell Cycle. 2005;4:1176–1178. doi: 10.4161/cc.4.9.2031. [DOI] [PubMed] [Google Scholar]
- Horton JK, Wilson SH. Strategic Combination of DNA-Damaging Agent and PARP Inhibitor Results in Enhanced Cytotoxicity. Front Oncol. 2013;3:257. doi: 10.3389/fonc.2013.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliakis G. Backup pathways of NHEJ in cells of higher eukaryotes: cell cycle dependence. Radiother Oncol. 2009;92:310–315. doi: 10.1016/j.radonc.2009.06.024. [DOI] [PubMed] [Google Scholar]
- Jankevicius G, Hassler M, Golia B, Rybin V, Zacharias M, Timinszky G, Ladurner AG. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat Struct Mol Biol. 2013;20:508–514. doi: 10.1038/nsmb.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Tulin AV. The roles of PARP1 in gene control and cell differentiation. Current opinion in genetics & development. 2010;20:512–518. doi: 10.1016/j.gde.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju BG, Solum D, Song EJ, Lee KJ, Rose DW, Glass CK, Rosenfeld MG. Activating the PARP-1 sensor component of the groucho/ TLE1 corepressor complex mediates a CaMKinase IIdelta-dependent neurogenic gene activation pathway. Cell. 2004;119:815–829. doi: 10.1016/j.cell.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Jwa M, Chang P. PARP16 is a tail-anchored endoplasmic reticulum protein required for the PERK- and IRE1alpha-mediated unfolded protein response. Nat Cell Biol. 2012;14:1223–1230. doi: 10.1038/ncb2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameoka M, Ota K, Tetsuka T, Tanaka Y, Itaya A, Okamoto T, Yoshihara K. Evidence for regulation of NF-kappaB by poly(ADP-ribose) polymerase. The Biochemical journal. 2000;346(Pt 3):641–649. [PMC free article] [PubMed] [Google Scholar]
- Kraus WL, Hottiger MO. PARP-1 and gene regulation: progress and puzzles. Mol Aspects Med. 2013;34:1109–1123. doi: 10.1016/j.mam.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell. 2010;39:736–749. doi: 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, Scott C, Meier W, Shapira-Frommer R, Safra T, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med. 2012;366:1382–1392. doi: 10.1056/NEJMoa1105535. [DOI] [PubMed] [Google Scholar]
- Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, Scott CL, Meier W, Shapira-Frommer R, Safra T, et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15:852–861. doi: 10.1016/S1470-2045(14)70228-1. [DOI] [PubMed] [Google Scholar]
- Li M, Yu X. The role of poly(ADP-ribosyl)ation in DNA damage response and cancer chemotherapy. Oncogene. 2014 doi: 10.1038/onc.2014.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Taverna P, Whitacre CM, Chatterjee S, Gerson SL. Pharmacologic disruption of base excision repair sensitizes mismatch repair-deficient and -proficient colon cancer cells to methylating agents. Clin Cancer Res. 1999;5:2908–2917. [PubMed] [Google Scholar]
- Liu Y, Kadyrov FA, Modrich P. PARP-1 enhances the mismatch-dependence of 5′-directed excision in human mismatch repair in vitro. DNA Repair (Amst) 2011;10:1145–1153. doi: 10.1016/j.dnarep.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26:417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo B, Trusolino L. Inhibition of poly(ADP-ribosyl)ation in cancer: old and new paradigms revisited. Biochim Biophys Acta. 2014;1846:201–215. doi: 10.1016/j.bbcan.2014.07.004. [DOI] [PubMed] [Google Scholar]
- Martin-Oliva D, Aguilar-Quesada R, O’Valle F, Munoz-Gamez JA, Martinez-Romero R, Garcia Del Moral R, Ruiz de Almodovar JM, Villuendas R, Piris MA, Oliver FJ. Inhibition of poly(ADP-ribose) polymerase modulates tumor-related gene expression, including hypoxiainducible factor-1 activation, during skin carcinogenesis. Cancer Res. 2006;66:5744–5756. doi: 10.1158/0008-5472.CAN-05-3050. [DOI] [PubMed] [Google Scholar]
- Masutani M, Nozaki T, Nishiyama E, Shimokawa T, Tachi Y, Suzuki H, Nakagama H, Wakabayashi K, Sugimura T. Function of poly(ADP-ribose) polymerase in response to DNA damage: gene-disruption study in mice. Mol Cell Biochem. 1999;193:149–152. [PubMed] [Google Scholar]
- Mateo J, Hall E, Sandhu SK, Omlin AG, Miranda S, Carreira S, Goodall J, Gillman A, Mossop H, Ralph C, et al. Antitumour activity of the PARP inhibitor olaparib in unselected sporadic castration-resistant prostate cancer (CRPC) in the TOPARP trial. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2014;25:1–41. [Google Scholar]
- Matsuura S, Egi Y, Yuki S, Horikawa T, Satoh H, Akira T. MP-124, a novel poly(ADP-ribose) polymerase-1 (PARP-1) inhibitor, ameliorates ischemic brain damage in a non-human primate model. Brain Res. 2011;1410:122–131. doi: 10.1016/j.brainres.2011.05.069. [DOI] [PubMed] [Google Scholar]
- McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, Giavara S, O’Connor MJ, Tutt AN, Zdzienicka MZ, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- Morrow DA, Brickman CM, Murphy SA, Baran K, Krakover R, Dauerman H, Kumar S, Slomowitz N, Grip L, McCabe CH, et al. A randomized, placebo-controlled trial to evaluate the tolerability, safety, pharmacokinetics, and pharmacodynamics of a potent inhibitor of poly(ADP-ribose) polymerase (INO-1001) in patients with ST-elevation myocardial infarction undergoing primary percutaneous coronary intervention: results of the TIMI 37 trial. J Thromb Thrombolysis. 2009;27:359–364. doi: 10.1007/s11239-008-0230-1. [DOI] [PubMed] [Google Scholar]
- Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, Ji J, Takeda S, Pommier Y. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nduka N, Skidmore CJ, Shall S. The enhancement of cytotoxicity of N-methyl-N-nitrosourea and of gamma-radiation by inhibitors of poly(ADP-ribose) polymerase. Eur J Biochem. 1980;105:525–530. doi: 10.1111/j.1432-1033.1980.tb04528.x. [DOI] [PubMed] [Google Scholar]
- O’Shaughnessy J, Schwartzberg L, Danso MA, Miller KD, Rugo HS, Neubauer M, Robert N, Hellerstedt B, Saleh M, Richards P, et al. Phase III Study of Iniparib Plus Gemcitabine and Carboplatin Versus Gemcitabine and Carboplatin in Patients With Metastatic Triple-Negative Breast Cancer. J Clin Oncol. 2014;32:3840–3847. doi: 10.1200/JCO.2014.55.2984. [DOI] [PubMed] [Google Scholar]
- Oliver FJ, Menissier-de Murcia J, Nacci C, Decker P, Andriantsitohaina R, Muller S, de la Rubia G, Stoclet JC, de Murcia G. Resistance to endotoxic shock as a consequence of defective NF-kappaB activation in poly (ADP-ribose) polymerase-1 deficient mice. The EMBO journal. 1999;18:4446–4454. doi: 10.1093/emboj/18.16.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza AM, Cibula D, Benzaquen AO, Poole C, Mathijssen RH, Sonke GS, Colombo N, Spacek J, Vuylsteke P, Hirte H, et al. Olaparib combined with chemotherapy for recurrent platinum-sensitive ovarian cancer: a randomised phase 2 trial. Lancet Oncol. 2014 doi: 10.1016/S1470-2045(14)71135-0. [DOI] [PubMed] [Google Scholar]
- Patel AG, De Lorenzo SB, Flatten KS, Poirier GG, Kaufmann SH. Failure of iniparib to inhibit poly(ADP-Ribose) polymerase in vitro. Clin Cancer Res. 2012;18:1655–1662. doi: 10.1158/1078-0432.CCR-11-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT. Activator-induced spread of poly(ADP-ribose) polymerase promotes nucleosome loss at Hsp70. Mol Cell. 2012;45:64–74. doi: 10.1016/j.molcel.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polkinghorn WR, Parker JS, Lee MX, Kass EM, Spratt DE, Iaquinta PJ, Arora VK, Yen WF, Cai L, Zheng D, et al. Androgen receptor signaling regulates DNA repair in prostate cancers. Cancer Discov. 2013;3:1245–1253. doi: 10.1158/2159-8290.CD-13-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner JR, Chen W, Iyer MK, Cao Q, Ma T, Han S, Sahu A, Malik R, Wilder-Romans K, Navone N, et al. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 2014;74:1651–1660. doi: 10.1158/0008-5472.CAN-13-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert I, Dantzer F, Reina-San-Martin B. Parp1 facilitates alternative NHEJ, whereas Parp2 suppresses IgH/c-myc translocations during immunoglobulin class switch recombination. J Exp Med. 2009;206:1047–1056. doi: 10.1084/jem.20082468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu M, Shah RG, Petitclerc N, Brind’Amour J, Kandan-Kulangara F, Shah GM. Role of poly(ADP-ribose) polymerase-1 in the removal of UV-induced DNA lesions by nucleotide excision repair. Proc Natl Acad Sci U S A. 2013;110:1658–1663. doi: 10.1073/pnas.1209507110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulten SL, Fisher AE, Robert I, Zuma MC, Rouleau M, Ju L, Poirier G, Reina-San-Martin B, Caldecott KW. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol Cell. 2011;41:33–45. doi: 10.1016/j.molcel.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Sala A, La Rocca G, Burgio G, Kotova E, Di Gesu D, Collesano M, Ingrassia AM, Tulin AV, Corona DF. The nucleosome-remodeling ATPase ISWI is regulated by poly-ADP-ribosylation. PLoS Biol. 2008;6:e252. doi: 10.1371/journal.pbio.0060252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu SK, Yap TA, de Bono JS. Poly(ADP-ribose) polymerase inhibitors in cancer treatment: a clinical perspective. Eur J Cancer. 2010;46:9–20. doi: 10.1016/j.ejca.2009.10.021. [DOI] [PubMed] [Google Scholar]
- Schiewer MJ, Goodwin JF, Han S, Brenner JC, Augello MA, Dean JL, Liu F, Planck JL, Ravindranathan P, Chinnaiyan AM, et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012;2:1134–1149. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiewer MJ, Knudsen KE. Transcriptional roles of PARP1 in cancer. Mol Cancer Res. 2014;12:1069–1080. doi: 10.1158/1541-7786.MCR-13-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shall S, de Murcia G. Poly(ADP-ribose) polymerase-1: what have we learned from the deficient mouse model? Mutation research. 2000;460:1–15. doi: 10.1016/s0921-8777(00)00016-1. [DOI] [PubMed] [Google Scholar]
- Slade D, Dunstan MS, Barkauskaite E, Weston R, Lafite P, Dixon N, Ahel M, Leys D, Ahel I. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 2011;477:616–620. doi: 10.1038/nature10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenblick A, de Azambuja E, Azim HA, Jr, Piccart M. An update on PARP inhibitors-moving to the adjuvant setting. Nat Rev Clin Oncol. 2015;12:27–41. doi: 10.1038/nrclinonc.2014.163. [DOI] [PubMed] [Google Scholar]
- Steffen JD, Brody JR, Armen RS, Pascal JM. Structural Implications for Selective Targeting of PARPs. Front Oncol. 2013;3:301. doi: 10.3389/fonc.2013.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo C, Zingarelli B, O’Connor M, Salzman AL. DNA strand breakage, activation of poly (ADP-ribose) synthetase, and cellular energy depletion are involved in the cytotoxicity of macrophages and smooth muscle cells exposed to peroxynitrite. Proc Natl Acad Sci U S A. 1996;93:1753–1758. doi: 10.1073/pnas.93.5.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tentori L, Leonetti C, Scarsella M, d’Amati G, Portarena I, Zupi G, Bonmassar E, Graziani G. Combined treatment with temozolomide and poly(ADP-ribose) polymerase inhibitor enhances survival of mice bearing hematologic malignancy at the central nervous system site. Blood. 2002;99:2241–2244. doi: 10.1182/blood.v99.6.2241. [DOI] [PubMed] [Google Scholar]
- Thomas CJ, Kotova E, Andrake M, Adolf-Bryfogle J, Glaser R, Regnard C, Tulin AV. Kinase-mediated changes in nucleosome conformation trigger chromatin decondensation via poly(ADP-ribosyl)ation. Molecular cell. 2014;53:831–842. doi: 10.1016/j.molcel.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, Friedlander M, Arun B, Loman N, Schmutzler RK, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- Wang M, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZQ, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner EF. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- Ying S, Hamdy FC, Helleday T. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res. 2012;72:2814–2821. doi: 10.1158/0008-5472.CAN-11-3417. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang J, Ding M, Yu Y. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nature methods. 2013;10:981–984. doi: 10.1038/nmeth.2603. [DOI] [PubMed] [Google Scholar]