Abstract
Aims: The Wnt planar cell polarity (PCP) pathway is one of the Wnt pathways which plays a critical role in cell proliferation and fate. The VANGL1 protein is one of Wnt-PCP pathway components. It is known that Wnt-PCP pathway has major roles in cell motility but its role in hepatocellular carcinoma (HCC) progression through invasion and metastasis needs to be clarified. Methods: We silenced VANGL1 gene expression in the HepG2 HCC cell line by stable transfection with a vector containing siRNA template for VANGL1 and investigated the change in cell invasion and motility. Results: Transfected cells with the siRNA template showed significantly suppressed invasive capacity when compared to controls although cellular motility was only slightly affected. Conclusion: Our study showed a basal role for VANGL1 with respect to the invasive capacity of HCC cells. This suggests that the Wnt-PCP pathway may play a role in progression of HCC through cellular invasion but further studies are needed to clarify its role in cell motility.
Introduction
Hepatocellular carcinoma (HCC) is the fifth most frequent cancer worldwide and is responsible for an estimated 600,000 deaths annually (Llovet et al., 2003). There is no effective treatment for this malignancy and a number of molecular changes, including hypermethylation or hypomethylation changes, mutations, and loss of heterozygosity of some genes, were thought to be associated with its development; however, there is no specific mechanism implicated in HCC (Moeini et al., 2012).
The Wnt proteins are a large family of highly conserved glycoproteins that have multiple roles in the cell fate and development of several organisms. Wnt pathways regulate different cellular functions, such as proliferation, differentiation, polarity, survival, and migration, and affect the organization of body plan, organogenesis, and tissue patterning during development (Gómez-Orte et al., 2013; Rosenbluh et al., 2014). The activation of Wnt signaling contributes to the etiology of several human cancers and may participate in malignant metastasis (An et al., 2013; Gao et al., 2014). While the Wnt/β-catenin pathway (known as canonical) plays a major role in Wnt signaling, there are also alternative (noncanonical) pathways, including the Wnt-planar cell polarity (PCP) and Wnt-Ca+2 pathways (Takahashi-Yanaga and Kahn, 2010). The Wnt system regulates motility and invasion not only by the canonical pathway but also by the PCP pathway in neoplasia as well as in physiologic processes. Although alterations in the Wnt/β-catenin pathway have been reported to be involved in hepatocarcinogenesis, there are limited data related to the Wnt/PCP pathway in HCC (Ikenoue et al., 2002; Pez et al., 2013).
It is well known that the components of the noncanonical Wnt pathway are essential for the regulation of PCP during embryonic differentiation. The core components of this pathway are Frizzled (Fz), Van Gogh (Vang), (Strabismus [Stbm]), Flamingo (Fmi), Disheveled (Dsh), Prickle (Pk), and Diego proteins. The PCP is established by both the Fz gradient and the asymmetric distribution of core PCP proteins in the surface of cells (Rosso and Inestrosa, 2013). Convergent extension movement of embryonic cells is also mediated by the noncanonical Wnt pathway (Walck-Shannon et al., 2014). Although various other signaling pathways besides the Wnt pathway have been implicated in cell motility, several data indicate that the noncanonical Wnt signaling pathway has the major role in migration during embryogenesis (Walck-Shannon and Hardin, 2014).
The VANGL1 gene is located on human chromosome 1p13. It encodes a transmembrane protein that interacts with PCP core proteins Disheveled (Dvl), Prickle (Pk), and Frizzled (Fz), a tumor metastasis suppressor KAI1 protein, and an intestinal epithelial restitution factor ITF protein (Jenny et al., 2003). The role of VANGL1 in normal and malignant human cells needs to be further investigated to better understand the mechanisms of cell invasion and motility. In this study, we investigated the effects of the inhibition of VANGL1 gene expression on cellular motility and invasion on human HCC cells by the siRNA method.
Materials and Methods
Cell lines and cell culture
Human HCC cell lines HEP-G2, HEP-3B, and SK-HEP-1, the colon adenocarcinoma cell line SW-480, and the chronic myeloid leukemia cell line K-562 were grown in Dulbecco's Modified Eagle's Medium (DMEM; Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich), 2 mM L-glutamine (Sigma-Aldrich), and 100 IU/mL penicillin (Sigma-Aldrich), and 100 μg/mL streptomycin (Sigma-Aldrich) and cultured at 37°C humid air containing 5% CO2.
Real-time reverse transcriptase-polymerase chain reaction
Total RNA was extracted from the cell lines with the Nucleospin® RNA II kit (Macherey Nagel), and first-strand cDNA synthesis was performed with the RevertAid™ First Strand cDNA synthesis Kit (Fermentas) according to the manufacturer's protocols. Real-time reverse transcriptase-polymerase chain reaction (RT-PCR) was performed in LightCycler 2.0 (Roche) using the FastStart DNA Master SYBR Green I Kit (Roche). The PCR conditions were 95°C for 10 min followed by 28 cycles of 95°C for 10 s, 59°C for 10 s, and 72°C for 20 s. In real-time PCR, the HPRT1 gene was used as a housekeeping gene and relative quantification of the products was carried out using the LightCycler Software 4.0. Primer sequences were as follows: for VANGL1 (179 bp product), forward 5′-ATCACCAACGGCATGACC-3′ and reverse 5′-AGGCTGAAGTCCAAGCAC-3′ and for HPRT1 (262 bp product), forward 5′-GTGGAGATGATCTCTCAACT-3′ and reverse 5′-ACATGATTCAAATCCCTGAAG-3′.
Construction of shRNA expression plasmid and transfection
The shRNA template for VANGL1 gene silencing was designed using a web-based tool (https://rnaidesigner.lifetechnologies.com/rnaiexpress/). The sequence of the shRNA template for VANGL1 gene was sense strand 5′-GATCCGCCACAACGAGTTGTATTA TTCAAGAGATAATACAACTCGTTGTGGCTTA-3′ and antisense strand 5′-AGCTT AAGCCACAACGAGTTGTATTATCTCTTGAATAATACAACTCGTTGTGGCG-3′. The oligonucleotides were annealed and inserted into pSilencer™ 4.1-CMV-Neo plasmid (Ambion) and maxipreparation of the plasmid was performed; then, the plasmid was controlled by the sequence analysis. To create stable clones of HEP-G2 cells carrying pSilencer 4.1-CMV Neo plasmid, HEP-G2 cells were transfected with the plasmid using Tfx™ 50 reagent (Promega). Selection of transfected HEP-G2 cells was performed with culturing the cells in complete DMEM supplemented with 1000 μg/mL Geneticin as a selection agent. HEP-G2 cells were incubated for 3 weeks and the cell culture medium was replaced every 48 h. At the end of the incubation period, colonies of the stably transfected cells were picked up by cloning disks (Sigma-Aldrich).
Western blot analysis
Proteins were subjected to SDS-PAGE under reducing conditions and then electrophoretically transferred to the nitrocellulose membrane. After blocking with 5% nonfat dried milk in the TBS-Tween 20 buffer at room temperature for 30 min, nitrocellulose membranes were sequentially blotted at 4°C for overnight with the anti-VANGL1 antibody (Sigma-Aldrich) and for 45 min at room temperature with horseradish peroxidase-conjugated anti-rabbit IG (Amersham). Protein bands were visualized with enhanced chemiluminescence.
Motility and invasion assays
Motility and invasion assays were performed in modified Boyden chambers with 8.0 μm pore size (BD Clontech). Motility chambers coated with the Matrigel matrix (BD Clontech) were used as invasion chambers. Cells were seeded into the upper reservoir of motility or invasion chambers and were cultured in the complete DMEM supplemented with 2% FBS for 48 h. The cells that migrated through the pores of the chambers were stained and then counted with an inverted microscope.
Statistical analyses
The GraphPad Prism 2.0 software (GraphPad Software, Inc.) was used for statistical analyses. The Mann–Whitney U test was performed to analyze the significance of the differences between the cells.
Results
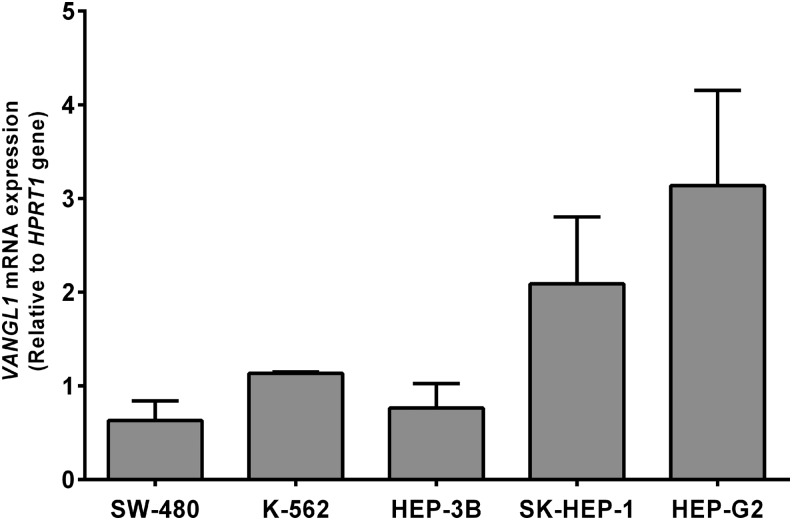
Relative quantification of VANGL1 gene expression in HEP-G2, HEP-3B, and SK-HEP-1 cell lines was determined by quantitative RT-PCR, while SW-480 and K-562 cell lines were used as controls. HEP-G2 cells were found to express VANGL1 more than HEP-3B and SK-HEP-1 cells (Fig. 1) and subsequent experiments were carried out with the HEP-G2 cell line.
FIG. 1.
Relative quantification of VANGL1 expression with real-time polymerase chain reaction (PCR) in hepatocellular cancer cell lines.
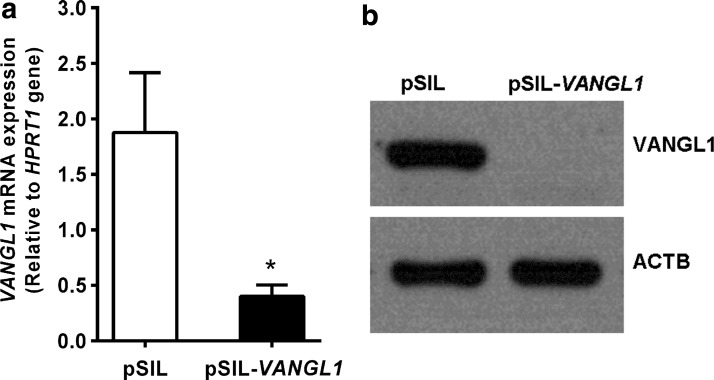
To silence the VANGL1 gene, HEP-G2 cells were transfected with the pSilencer 4.1 vector containing the template for shRNA expression. To detect the degree of gene expression at the mRNA level after shRNA transfection, relative quantification was performed by real-time PCR (Fig. 2a). Nearly 70% VANGL1 gene silencing at the mRNA level was achieved and western blot analysis confirmed the gene silencing at the protein level (Fig. 2b).
FIG. 2.
(a) VANGL1 expression of HEP-G2 cells transfected with pSilencer plasmid. pSIL-VANGL1 cells carry the shRNA template for VANGL1 and pSIL where the control cells transfected with the control pSilencer vector. Relative quantification of VANGL1 expression with real-time PCR in transfected cells showing the degree of gene silencing (p=0.0009). *Indicates the significance. (b) Western blot analysis of VANGL1 expression in the cells transfected with siRNA.
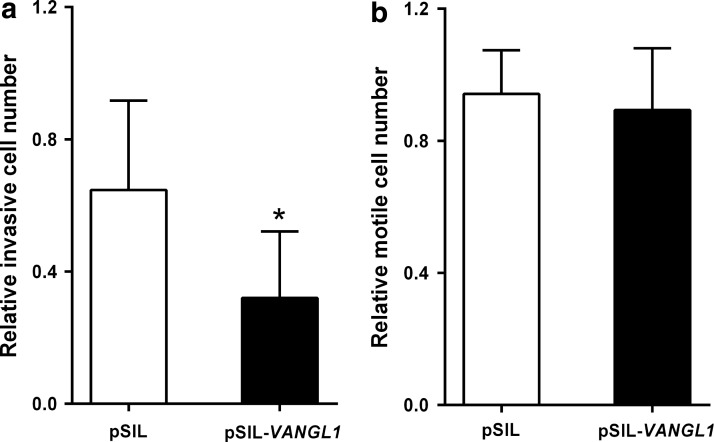
Motility and invasion assays to assess the invasion capacity of the cells after VANGL1 silencing were carried out with HEP-G2 cells with the silenced VANGL1 gene (pSIL-VANGL1) and a control colony transfected with the control plasmid (pSIL). When compared to the control cells, the cells with the silenced VANGL1 gene showed a more suppressed invasive capacity (Fig. 3a). Motility assay results showed that there is minimal difference between the control cells and the VANGL1 gene-silenced HEP-G2 cells (Fig. 3b).
FIG. 3.
The change in the number of (a) invasive (p=0.03) and (b) motile HEP-G2 cells after transfection with VANGL1 siRNA. *Indicates the significance. pSIL-VANGL1 cells carry the shRNA template for VANGL1 and pSIL where the control cells transfected with the control pSilencer vector.
Discussion
The effects of VANGL1 gene silencing on the cellular invasion and motility were investigated in this study. The shRNA application method of stable gene silencing was performed to inhibit the expression of the VANGL1 gene in the HEP-G2 hepatocellular carcinoma cell line. Invasion assays revealed a significantly reduced invasive capacity of the cells with the silenced VANGL1 gene, whereas motility assays showed a slight but insignificant difference compared to control cells. These results implicated that the VANGL1 protein was mostly involved in cell invasion rather than cell motility.
Transient siRNA silencing of the VANGL1 gene in various cancer cell lines was reported to regulate cellular invasion and motility. In gastric carcinoma, colorectal cancer, and oral cancer cell lines, fibronectin-stimulated cellular invasion of gelatin was shown to be decreased, and motility of the cells was found to be reduced with the scratch wound assay following transient siRNA silencing of the VANGL1 gene (Ryu et al., 2010; Lee et al., 2011; Yoon et al., 2013). Although our invasion assay results were similar, our motility assay results were not similar to these reports and there were differences in the assay methods used to analyze cellular motility and invasion. In our assays, invasion was measured with Matrigel matrix-coated transwell chambers and there was no stimulating agent to induce cell invasion. While the Matrigel matrix invasion assay has a superior capacity to analyze cellular invasion since it contains numerous molecules, including laminin, collagen IV, enactin, and fibronectin, only the gelatin matrix has collagen molecules (Kleinman and Martin, 2005). Associated with the multimolecular composition of the Matrigel matrix, cellular response to invade the Matrigel matrix is more complex than the gelatin matrix (Brinckerhoff et al., 2000). Therefore, we preferred to use the Matrigel matrix assay method to analyze basal invasion capacity of cells without stimulation.
In the previous studies, motility assays were performed with the scratch wound assay, but in our experiments, transwell chambers were used to investigate cell motility (Ryu et al., 2010; Lee et al., 2011; Yoon et al., 2013). Since the cytoskeletal movement of the cell has a similar pattern in both transwell migration and invasion experiments, we preferred to use the transwell migration assay instead of the wound closure assay (Albini et al., 1987; Mierke, 2013). In one study, the effect of VANGL1 silencing on cellular migration was analyzed with the transwell assay as in our experiments, and their results showed that motility of the breast cancer cells reduced with VANGL1 silencing (Anastas et al., 2012). Our findings are not concordant with this report, and we suppose that VANGL1 silencing may be resulting with a different effect on cellular motility depending on the cell type. In addition, the influence of the VANGL1 protein on cellular motility might be more pronounced under specific conditions, which are stimulating the cellular motility response. It has been shown that VANGL1 silencing reduced the ITF-stimulated migration of intestinal epithelial cells, but did not affect the basal motility of these cells (Kalabis et al., 2006). Moreover, the discrepancy of our motility assay results with the previous studies may be related with the siRNA silencing method whether transient or stable.
The mechanism of the inhibition of cell invasion with transient VANGL1 silencing was reported to be associated with the regulation of the extracellular matrix. Results showed that the silencing of the VANGL1 gene inhibits the transcription of metalloproteinase enzymes, which are degrading the extracellular matrix proteins with their proteolytic enzyme activity. The association of Vangl1 protein with cell invasion and motility might also be related by its effect on the cell adhesion. A number of molecules, which are located downstream of the VANGL1 on the Wnt-PCP pathway, have a role in the regulation of both cell to cell and cell to extracellular matrix adhesion. Particularly, the activity of small GTPases, like Rac and RhoA proteins, contributes to the inactivation of cadherin-mediated cell to cell adhesion in human cancer (Evers et al., 2000; Fort and Théveneau, 2014). Moreover, Rac and RhoA proteins are involved in cell motility and the invasion processes with their role in the formation of lamellipodia, membrane ruffles, focal contacts, and cytoskeletal changes (Hall, 1998). Both the loss of intercellular adhesion and the stimulation of migration are well-known characteristic features of invasive cancers. Recently, studies with breast cancer and HCC tissues showed that cancer cells have a higher expression level of the VANGL1 gene compared to normal cells (Cho et al., 2011; Anastas et al., 2012). Furthermore, both in colon adenocarcinomas and gastric tumors, metastatic cancer tissues were found to have an elevated VANGL1 protein expression (Kho et al., 2009; Ryu et al., 2010). These data are implying that the increased levels of the VANGL1 protein might be associated with the invasion and metastatic progression of cancer cells.
Taken together, we propose that VANGL1 is associated to the natural invasion capacity of HCC cells rather than motility, and further studies need to be performed to understand both basal and stimulated cellular invasion related with the VANGL1 protein in distinct types of cancer.
Acknowledgments
The authors gratefully thank Prof. Dr. Mehmet Ozturk for the cell lines. This study was supported by the Scientific Research Funds of Dokuz Eylul University (Grant No: 04.KB.SAG.094).
Author Disclosure Statement
No competing financial interests exist.
References
- Albini A, Iwamoto Y, Kleinman HK, et al. (1987) A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res 47:3239–3245 [PubMed] [Google Scholar]
- An SM, Ding QP, Li LS. (2013) Stem cell signaling as a target for novel drug discovery: recent progress in the WNT and Hedgehog pathways. Acta Pharmacol Sin 34:777–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastas JN, Biechele TL, Robitaille M, et al. (2012) A protein complex of SCRIB, NOS1AP and VANGL1 regulates cell polarity and migration, and is associated with breast cancer progression. Oncogene 31:3696–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinckerhoff CE, Rutter JL, Benbow U. (2000) Interstitial collagenases as markers of tumor progression. Clin Cancer Res 6:4823–4830 [PubMed] [Google Scholar]
- Cho SB, Park YL, Park SJ, et al. (2011) KITENIN is associated with activation of AP-1 target genes via MAPK cascades signaling in human hepatocellular carcinoma progression. Oncol Res 19:115–123 [DOI] [PubMed] [Google Scholar]
- Evers EE, Zondag GCM, Malliri A, et al. (2000) Rho family proteins in cell adhesion and cell migration. Eur J Cancer 36:1269–1274 [DOI] [PubMed] [Google Scholar]
- Fort P, Théveneau E. (2014) PleiotRHOpic: Rho pathways are essential for all stages of neural crest development. Small GTPases 5:e27975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Xiao G, Hu J. (2014) Regulation of Wnt/β-catenin signaling by posttranslational modifications. Cell Biosci 4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Orte E, Sáenz-Narciso B, Moreno S, et al. (2013) Multiple functions of the noncanonical Wnt pathway. Trends Genet 29:545–553 [DOI] [PubMed] [Google Scholar]
- Hall A. (1998) RhoGTPases and the actin cytoskeleton. Science 279:509–514 [DOI] [PubMed] [Google Scholar]
- Ikenoue T, Ijichi H, Kato N, et al. (2002) Analysis of the beta-catenin/T cell factor signaling pathway in 36 gastrointestinal and liver cancer cells. Jpn J Cancer Res 93:1213–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A, Darken RS, Wilson PA, et al. (2003) Prickle and strabismus form a functional complex to generate a correct axis during planar cell polarity signaling. EMBO J 22:4409–4420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalabis J, Rosenberg I, Podolsky DK. (2006) Vangl1 protein acts as a downstream effector of intestinal trefoil factor (ITF)/TFF3 signaling and regulates wound healing of intestinal epithelium. J Biol Chem 281:6434–6441 [DOI] [PubMed] [Google Scholar]
- Kho DH, Bae JA, Lee JH, et al. (2009) KITENIN recruits Dishevelled/PKC delta to form a functional complex and controls the migration and invasiveness of colorectal cancer cells. Gut 58:509–519 [DOI] [PubMed] [Google Scholar]
- Kleinman HK, Martin GR. (2005) Matrigel: basement membrane matrix with biological activity. Semin Cancer Biol 15:378–386 [DOI] [PubMed] [Google Scholar]
- Lee S, Song YA, Park YL, et al. (2011) Expression of KITENIN in human colorectal cancer and its relation to tumor behavior and progression. Pathol Int 61:210–220 [DOI] [PubMed] [Google Scholar]
- Llovet JM, Burroughs A, Bruix J. (2003) Hepatocellular carcinoma. Lancet 362:1907–1917 [DOI] [PubMed] [Google Scholar]
- Mierke CT. (2013) Physical break-down of the classical view on cancer cell invasion and metastasis. Eur J Cell Biol 92:89–104 [DOI] [PubMed] [Google Scholar]
- Moeini A, Cornellà H, Villanueva A. (2012) Emerging signaling pathways in hepatocellular carcinoma. Liver Cancer 1:83–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pez F, Lopez A, Kim M, et al. (2013) Wnt signaling and hepatocarcinogenesis: molecular targets for the development of innovative anticancer drugs. J Hepatol 59:1107–1117 [DOI] [PubMed] [Google Scholar]
- Rosenbluh J, Wang X, Hahn WC. (2014) Genomic insights into WNT/β-catenin signaling. Trends Pharmacol Sci 35:103–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso SB, Inestrosa NC. (2013) WNT signaling in neuronal maturation and synaptogenesis. Front Cell Neurosci 7:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu HS, Park YL, Park SJ, et al. (2010) KITENIN is associated with tumor progression in human gastric cancer. Anticancer Res 30:3479–3486 [PubMed] [Google Scholar]
- Takahashi-Yanaga F, Kahn M. (2010) Targeting Wnt signaling: can we safely eradicate cancer stem cells? Clin Cancer Res 16:3153–3162 [DOI] [PubMed] [Google Scholar]
- Yoon TM, Kim SA, Lee JK, et al. (2013) Expression of KITENIN and its association with tumor progression in oral squamous cell carcinoma. Auris Nasus Larynx 40:222–226 [DOI] [PubMed] [Google Scholar]
- Walck-Shannon E, Hardin J. (2014) Cell intercalation from top to bottom. Nat Rev Mol Cell Biol 15:34–48 [DOI] [PMC free article] [PubMed] [Google Scholar]