Abstract
Many techniques have been proposed to segment organs from images, however the segmentation of diseased organs remains challenging and frequently requires lots of user interaction. The challenge consists of segmenting an organ while its appearance and its shape vary due to the presence of the disease in addition to individual variations. We propose a template registration technique that can be used to recover the complete segmentation of a diseased organ from a partial segmentation. The usual template registration method is modified in such a way that it is robust to missing parts. The proposed method is used to segment Mycobacterium tuberculosis infected lungs in CT images of experimentally infected mice. Using synthetic data, we evaluate and compare the performance of the proposed algorithm with the usual sum of squared difference cost function.
Index Terms: Registration, Template-based Segmentation, Diseased Organs
1. Introduction
More than 125 years after its discovery, tuberculosis (TB) is still surging, causing 9.2 million new cases and 1.7 million deaths each year [1]. New anti-TB drugs and vaccines are crucial to control TB. Their development requires a better understanding of the interactions between the host immune system and the bacteria. Unfortunately, traditional post-mortem tissue analysis techniques used in pre-clinical studies offer limited insight into host-bacteria interactions. On the other hand, longitudinal imaging studies provide a cost effective alternative to qualitatively follow the evolution of TB infection in the same experimentally infected animals while alive. The development of rigorous quantitative tools for rapid automated image analysis is needed to compare lesions across subjects and/or time points.
The first challenge for developing such tools consists of segmenting the lungs of the animal from Computerized Tomography (CT) images at all stages of infection. To do so, one not only needs to account for the evolution of the disease, but also for shape variations. These variations come from the position of the animal and various clinical factors. They are significant and cannot be easily controlled.
A common strategy for segmenting organs is to register a template image onto the target image which is to be segmented. The deformation that matches the template to the target is obtained by comparing intensity and/or image features. However, the presence of lesions in the diseased organ is an impediment to the success of such techniques. In some cases refinement procedure are used to correct the segmentation of the lungs [2]. More generally, robust matching algorithms have been proposed in which lesions are modeled as outliers and excluded from the matching cost. These techniques have been successfully applied in [3, 4] to detect multi-sclerosis lesions in brain MR images, as well as cancerous breast lesions in radiographic images. Another approach consists of modeling the lesion as an additional anatomical tissue, see e.g. [5], whose growth is assessed by template matching. One usually needs to place a seed in the template at the appropriate location to initialize the matching. This approach is particularly well suited to the analysis of cancerous lesions. However, because infectious diseases tend to affect an organ in multiple locations, the proper initialization of such method is problematic.
In this paper, we propose a new non-rigid registration method that allows one to match a template organ to a diseased organ, even in the presence of lesions. We use the proposed method to segment TB infected lungs. Using synthetic data, we show that we can recover almost 90% of the volume of the lungs, even when 50% of the lungs are infected. We prove that this performance could not be achieved with the usual sum of squared differences matching method.
2. Segmentation of Diseased Organs By Template Registration
2.1. Partial Segmentation
Obtaining a complete segmentation of a diseased organ is very challenging and time consuming. Instead, it is faster and much easier to obtain a partial segmentation. The registration method that we present relies on an initial partial segmentation of the diseased organ. We make the assumption that the segmentation contain only few errors, i.e. voxels that do not belong to the lungs. However, we tolerate that some parts of the diseased organ may be missing. In this work, the partial segmentation is provided by the user and is based on intensity thresholding and connected components. Most of the diseased parts of the organ are not included in this initial segmentation, see Figure 3(a).
Fig 3.
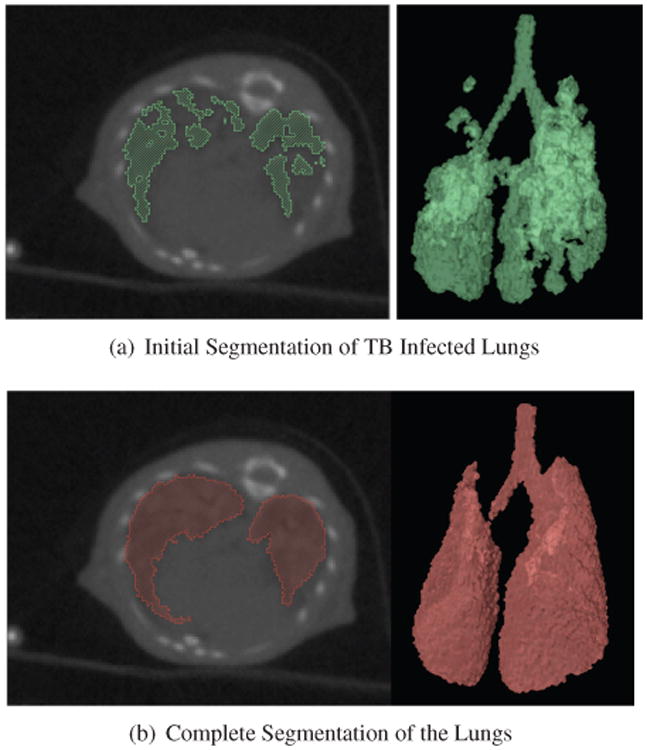
Top: Axial view of the CT image overlaid with the user-specified segmentation and 3D rendering of the partial segmentation, Bottom: Axial view of the CT image overlaid with the complete segmentation and 3D rendering of the complete lungs.
2.2. Template Registration with Missing Parts
Most of the proposed methods for template registration rely on an energy minimization formulation. The template, denoted by x0, is deformed by ϕ, which is a smooth deformation from ℝ3 to ℝ3. The energy function
| (1) |
is usually composed of two terms and a weighting factor, γ ∈ ℝ. The data term,
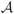 , measures the similarity between the deformed template, x0 ○ ϕ−1, and the target, x, while the regularization term, ℛ, penalizes for non-smooth deformations. In our case, x0 is the binary image corresponding to the segmentation of the lungs of an uninfected mouse, while x is the binary image resulting from the partial segmentation described previously.
, measures the similarity between the deformed template, x0 ○ ϕ−1, and the target, x, while the regularization term, ℛ, penalizes for non-smooth deformations. In our case, x0 is the binary image corresponding to the segmentation of the lungs of an uninfected mouse, while x is the binary image resulting from the partial segmentation described previously.
2.2.1. Modified Data Term
We use a simple statistical model for the segmentation problem in order to derive a likelihood function. This likelihood function is then used to infer a data term. We model the binary value x(s) at each voxel s as a random variable which follows a Bernoulli distribution. Indeed, if a voxel, u = ϕ−1(s), belongs to the template lungs (x0 ○ ϕ−1(s) = 1), then the corresponding voxel, s, in the target belongs to the lungs (x(s) = 1) with probability 1 − δ. δ accounts for the missing parts in the initial segmentation, and ranges between 0 and 0.5. If, on the contrary, a voxel, u = ϕ−1(s), does not belong to the template lungs (x0 ○ ϕ−1(s) = 0), then s belongs to the lungs with probability ε. Assuming that our initial segmentation contains few errors, we set ε to a small value, ∼ 10−4. In summary:
| (2) |
For simplicity, we assume that the collection of random variables, x(s), when s ranges across the set of voxels, are independent conditional on ϕ. With u=ϕ−1(s), the log-likelihood is
| (3) |
Keeping only the terms that depend on ϕ and using the fact that, since x0 and x are binary, and x(s) = x2(s), we obtain:
| (4) |
Henceforth, as suggested in [6], we use the negative log likelihood (4) as data term in the energy function (1):
| (5) |
The data term is a weighted sum of the usual sum of squared differences (SSD) and of a corrective term which is the volume of the deformed template. This corrective term allows for a certain amount of mismatch between the template and the target so that the missing parts may be recovered. In other words, there is a penalty for shrinking the template.
2.2.2. Regularization Term
The algorithm we use is a variant of the one described in [7], adapted to the proposed data attachment term. Letting L be a symmetric operator acting on vector fields (here, L = (Δ2 + αId)3 for some constant α), we define a Hilbert space V with norm , and generate diffeomorphisms ϕ according to the evolution equation:
| (6) |
with t ∈ [0; 1]. This equation is called the EPDiff equation [8, 9]. It has a unique solution specified by the initial conditions ϕ0 = identity and v0 = w, where w is a vector field. Denoting this solution by , the registration algorithm minimizes
| (7) |
Details on the minimization algorithm can be found in [7] when λseg = 0, the general case coming as a straightforward modification.
2.3. Choice of the Model Parameters
The matching algorithm depends on the choice of 3 parameters: ε, δ, and γ. As previously mentioned, δ and ε respectively represents the proportion of missing parts and the segmentation error in the incomplete target volume. Assuming that all the mice used in a study have comparable lung volume, we roughly estimate the proportion of missing data by computing the ratio of the volume of the incomplete target volume over the volume of the template. We determine the value of γ when ε = δ, i.e. when λseg = 0, which corresponds to the case with no missing parts. In that case, the proposed algorithm boils down to the classical SSD registration algorithm. We experimentally determine that λ = λ0 = 0.3 gives satisfying matching results in the case of complete volume. Given ε and δ,
| (8) |
3. Experiments
3.1. Image Data and Generation of Synthetic Data
In order to follow the evolution of M. tuberculosis infection, a group of genetically identical mice is experimentally infected and imaged at different time points. 14 days after initial infection, the mice are treated with a daily dose of antibiotics. 3D CT images are acquired after 14, 28, 56 and 84 days of treatment at an isotropic resolution of 0.17 mm. A group of uninfected mice is also imaged at each time point.
Two complete lungs are manually segmented from the CT scans of two uninfected mice. Since the lungs appear at a lower intensity than the surrounding tissue, the segmentation is easily obtained by intensity thresholding. We model the TB lesions by randomly creating spherical holes in one of the lung volumes. At each voxel, we sample a Bernoulli distribution with low success probability and assign a sphere center in case of success. All the voxels located at less than a given distance r from a sphere center are excluded from the lung segmentation. Depending on the strain of the mouse, the size of the lesions may vary. We therefore generate two types of synthetic data with either large (r = 5) or small (r = 3) lesions, see Figure 1. The amount of missing data is controlled and varies between 0 and 50%. In order to recover the complete lung volume, the template is first registered onto the target with an affine registration. It is used as a starting point for the proposed algorithm. The segmentation is given by the deformed template registered onto the target.
Fig 1.
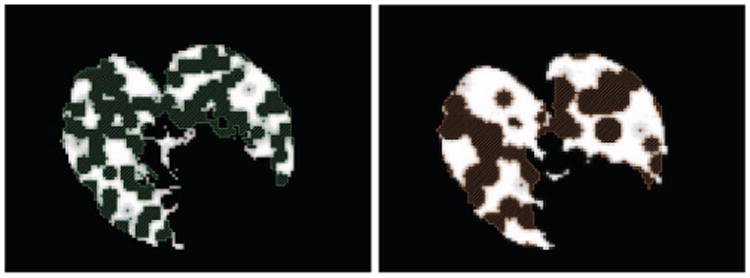
Axial view of the lungs with synthetic lesions and the recovered complete lung volume. Left: Small lesions Right: Large lesions.
3.2. Results
We use the Dice coefficient to assess the performance of the segmentation algorithm. We denote by DT the deformed template and T the complete target, i.e. the complete lung volume. The target is partitioned into the lesions, denoted by L, and TL the incomplete segmentation of the diseased lungs. We denote by | · | the volume, and by ⨪ the complement. We write the segmentation error:
| (9) |
These 3 terms respectively correspond to the deformed template overgrowth, the missed lesions, and the missed target.
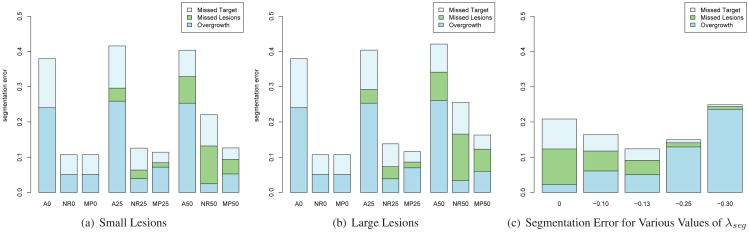
Figures 2(a) and 2(b) present the segmentation error between the ground truth and the recovered segmentation in the simulation experiments. We compare the 3 components of the error for different types of lesions and different amount of missing data, after affine registration and after classical non-rigid registration (i.e. λseg=0) and with the proposed algorithm. Recall that λseg controls the tolerance to missing parts, and that when λseg = 0, the data term boils down to the classical sum of squared differences. Even when there is no missing data, affine registration is not enough to capture the shape variation, while non-rigid registration reduces the segmentation error to ∼ 10%. Even with 50% of lesions, the proposed algorithm recovers almost 90% of the complete volume, against 80% only with usual SSD matching. Figure 2(c) illustrates the performance of the proposed algorithm at segmenting the complete lung volume depending on the parameter λseg. We compare the segmentation results for 8 volumes with 50% of small lesions, when λseg = 0 or 0.1274 and show that adding the corrective term significantly improves the segmentation (paired Wilcoxon test p-value=0.007). Note that at the optimal value of λseg, the 3 components of the segmentation error are balanced.
Fig 2.
Left and Middle Segmentation error for different amounts and types of lesions after template registration. A25, for example, corresponds to 25%; of lesions and template matching by Affine registration. NR stands for Non-Rigid registration, MP stands for non-rigid matching with Missing Parts. Right: Residual segmentation error in the case of MP50 with small lesions for different values of λseg.
3.3. Segmentation of M. Tuberculosis Infected Lungs
We apply the proposed segmentation method to M. tuberculosis infected lungs, using a user-specified partial segmentation of the diseased lungs obtained by intensity thresholding. Because infected parts have higher intensity, they are usually missing in the partial segmentation. Figure 3(a) presents an incomplete segmentation of the lungs. We use the lungs of an uninfected mouse as template. The matching is initialized by the affine registration of the template onto the incomplete target. Figure 3(b) illustrates an example of segmented lungs and shows that most of the missing parts have been recovered.
4. Conclusion
We have shown that the proposed method can be used to obtain a segmentation of diseased organs at various stages of infection and outperforms classical SSD registration. This technique is generic and therefore is applicable to other organs, pathologies and/or species.
Acknowledgments
This work was funded by the Bill and Melinda Gates foundation TB Drug Accelerator grant 48793, the Johns Hopkins Fund for Medical Discovery and the Potts Memorial Foundation.
References
- 1.Global tuberculosis control - surveillance, planning, financing. WHO; 2008. [Google Scholar]
- 2.Sluimer I, Prokop M, van Ginneken B. Toward automated segmentation of the pathological lung in CT. IEEE T Med Imaging. 2005;24(8):1025–1038. doi: 10.1109/TMI.2005.851757. [DOI] [PubMed] [Google Scholar]
- 3.Van Leemput K, Maes F, Vandermeulen D, Colchester A, Suetens P. Automated segmentation of multiple sclerosis lesions by model outlier detection. IEEE T Med Imaging. 2001 Aug;20(8):677–688. doi: 10.1109/42.938237. [DOI] [PubMed] [Google Scholar]
- 4.Hachama M, Desolneux A, Richard F. A probabilistic approach for simultaneous mammogram registration and abnormality detection. IWDM. 2006:205–212. [Google Scholar]
- 5.Cuadra MB, Pollo C, Bardera A, Cuisenaire O, Villemure JG, Thiran JP. Atlas-based segmentation of pathological MR brain images using a model of lesion growth. IEEE Trans Med Imaging. 2004 Oct;23(10):1301–1314. doi: 10.1109/TMI.2004.834618. [DOI] [PubMed] [Google Scholar]
- 6.Vidal C, Jedynak B. Learning to match: Deriving optimal template-matching algorithms from probabilistic image models. Int J Comput Vision. 2009 [Google Scholar]
- 7.Younes L. Jacobi fields in groups of diffeomorphisms and applications. Quart Appl Math. 2007;65:113–134. [Google Scholar]
- 8.Holm DD, Marsden JE, Ratiu TS. The Euler– Poincaré equations and semidirect products with applications to continuum theories. Adv in Math. 1998;137:1–81. [Google Scholar]
- 9.Miller MI, Trouvé A, Younes L. Geodesic shooting for computational anatomy. J Math Image and Vision. 2005 doi: 10.1007/s10851-005-3624-0. [DOI] [PMC free article] [PubMed] [Google Scholar]