Abstract
Background
Congenital microphthalmia and coloboma are severe developmental defects that are frequently associated with additional systemic anomalies and display a high level of genetic heterogeneity.
Methods
To identify the pathogenic variant in a patient with microphthalmia, coloboma, retinal dystrophy, microcephaly and other features, whole exome sequencing (WES) analysis of the patient and parental samples was undertaken. To further explore the identified variant/gene, expression and functional studies in zebrafish were performed.
Results
WES revealed a de novo variant, c.473_474delGA, p.(Arg158Lysfs*4), in EFTUD2 which encodes a component of the spliceosome complex. Dominant mutations in EFTUD2 cause Mandibulofacial Dysostosis, Guion-Almeida type (MFDGA) which does not involve microphthalmia, coloboma or retinal dystrophy; analysis of genes known to cause these ocular phenotypes identified several variants of unknown significance but no causal alleles in the affected patient. Zebrafish eftud2 demonstrated high sequence conservation with the human gene and broad embryonic expression. TALEN-mediated disruption was employed to generate a c.378_385 del, p.(Ser127Aspfs*23) truncation mutation in eftud2. Homozygous mutants displayed a reduced head size, small eye, curved body, and early embryonic lethality. Apoptosis assays demonstrated a striking increase in TUNEL-positive cells in the developing brain, eye, spinal cord and other tissues starting at 30 hours post fertilization.
Conclusion
This study reports a novel mutation in EFTUD2 in an MFDGA patient with unusual ocular features and the generation of a first animal model of eftud2 deficiency. The severe embryonic phenotype observed in eftud2 mutants indicates an important conserved role during development of diverse tissues in vertebrates.
Keywords: EFTUD2, coloboma, retinal dystrophy, microphthalmia, zebrafish
Introduction
Microphthalmia, anophthalmia and coloboma (MAC) represent a continuum of structural eye defects caused by disruption of the molecular program that directs ocular development. Microphthalmia or anophthalmia describe a small or an absent eye while coloboma is a segmental ocular defect which can affect multiple ocular structures and is typically caused by optic fissure closure defects (Morrison et al., 2002; Skalicky et al., 2013). Additional eye anomalies are often seen in patients with MAC and include cataract, glaucoma, anterior segment dysgenesis and posterior segment abnormalities (retinal detachment, hypoplasia of the optic nerve/disc, persistent fetal vasculature); retinal dystrophy is rarely reported (Huynh et al., 2013; Morrison et al., 2002). MAC phenotypes can be limited to the eye but are frequently associated with non-ocular features, including neurological, craniofacial, hearing, skeletal, cardiac, and urogenital abnormalities, in a variety of known and not yet recognized heritable syndromes (Hornby et al., 2000; Huynh et al., 2013; Morrison et al., 2002; Nakamura et al., 2011; Skalicky et al., 2013).
The causes of syndromic and isolated MAC conditions are highly heterogeneous (Bardakjian and Schneider, 2011; Williamson and FitzPatrick, 2014) with many cases remaining unexplained. Common causes of syndromic MAC include CHD7 in CHARGE syndrome (which includes coloboma) and SOX2 or OTX2 in anophthalmia/microphthalmia syndromes (Huynh et al., 2013; Skalicky et al., 2013; Williamson and FitzPatrick, 2014); other genes associated with microcephaly, craniofacial anomalies, and ocular coloboma include TFAP2A in Branchiooculofacial syndrome (Gestri et al., 2009), ZEB2 in Mowat-Wilson syndrome (Wenger et al., 2014), ACTB and ACTG1 in Baraitser-Winter cerebrofrontofacial syndrome (Riviere et al., 2012), and occasionally SALL1 in Townes-Brocks syndrome (Botzenhart et al., 2005) and PQBP1 in Renpenning syndrome (Kleefstra et al., 2004). For cases without causal variants in the most common MAC genes, whole exome sequencing (WES) represents an efficient method of screening numerous known ocular genes along with the entire exome to identify the causative variant (Deml et al., 2014; Need et al., 2012; Reis et al., 2013; Weh et al., 2014). The WES approach has demonstrated success in discovering novel causative genes in MAC conditions as well (Beleggia et al., 2015; Deml et al., 2015; Kelberman et al., 2014; Williamson et al., 2014; Zahrani et al., 2013). Interpretation of WES results is often challenging due to the large number of identified variants. Variants proposed as likely pathogenic can be further evaluated in animal models; this is particularly valuable when variation is observed in genes with an unexplored/unknown function (Beleggia et al., 2015; Deml et al., 2015; Kelberman et al., 2014; Manzini et al., 2012). The recent development of new genomic editing technologies based on engineered nucleases that allow for precise and rapid modification of vertebrate genomes has enabled more efficient generation of animal models for human disease-related genes in the laboratory (Sanjana et al., 2012).
In this manuscript we present the identification of a novel de novo EFTUD2 pathogenic variant in a patient with syndromic colobomatous microphthalmia using trio whole exome sequencing analysis. This study also describes generation of the first animal model for eftud2 deficiency, a zebrafish line with a frameshift mutation that is predicted to result in protein truncation, similar to the majority of pathogenic variants in the human EFTUD2 gene, and characterized by reduced head size, small eye, early embryonic lethality, and massive cell death.
Materials and Methods
Human study
The human study was approved by the Children’s Hospital of Wisconsin Institutional Review Board (protocol number CHW 03/56) with written informed consent obtained from each participant and/or their legal representative, as appropriate.
Whole exome sequencing of DNA samples from the proband and her parents was undertaken through Perkin Elmer, Inc (Branford, CT). Library capture was completed using the Agilent Sure Select v4+UTR capture kit (Santa Clara, CA) and 100 base pair paired end sequencing was performed using the Illumina HiSeq 2000. The obtained data were aligned using the Burrows-Wheeler Aligner (BWA) and variants were called using the Genome Analysis Toolkit (GATK v2.20) pipeline available through Perkin Elmer (Branford, CT). Exome data were analyzed using the SNP & Variation Suite (Golden Helix, Bozeman, MT); preliminary analysis of the proband’s data involved screening of 82 known MAC genes (Supplemental Table 1). Trio analysis was performed with the parental samples to identify de novo variants in the proband or recessive variants (homozygous or compound heterozygous) inherited from both parents. Variants of interest were prioritized based on their absence/rarity in the general population (as reported in publicly available databases dbSNP (http://www.ncbi.nlm.nih.gov/snp), NHLBI Exome Sequencing Project Exome Variant Server (EVS; http://evs.gs.washington.edu/EVS/), 1000 Genomes (http://www.1000genomes.org/data), and the Exome Aggregation Consortium (ExAC) Browser (http://exac.broadinstitute.org/), as well as our own data) and possible effect on protein function; for missense variants, functional profiling was performed using SIFT, Polyphen2, Mutation Taster, MutationAssessor, and FATHMM as well as nucleotide conservation scores GERP++ and PhyloP as accessed through dbNSFP (Liu et al., 2013). To rule out an alternative genetic cause for the proband’s retinal dystrophy, the data was also analyzed for variants in COH1 (Cohen syndrome), PRPF3, PRPF4, PRPF6, PRPF8, PRPF31 and SNRNP200 (other components of the U4/U6.U5 tri-snRNP of the major spliceosome), as well as 123 other genes included in the Casey Eye Institute Clinical Retinal Dystrophy Panel (http://www.ohsu.edu/xd/health/services/casey-eye/diagnostic-services/cei-diagnostics/upload/Retinal-Dystrophy-Panel-Gene-List.pdf).
To confirm the identified EFTUD2 changes and perform cosegregation analysis, DNA from the affected patient and unaffected parents was amplified using the following primers and conditions: EFTUD2-F - TTTCATTTCCTGAGGGATGC and EFTUD2-R – TCTATGCTCCCGAGAAAGTTG with an annealing temperature of 60°C (product size of 467 bp). PCR products were sequenced bidirectionally using Big Dye Terminator chemistry and ABI 3730XL sequencer (Life Technologies, Carlsbad, CA, USA). Sequences were reviewed manually and using Mutation Surveyor (SoftGenetics, State College, PA) and compared to the NM_004247.3 transcript.
For sequence comparison, the protein sequences encoded by human EFTUD2 (NM_004247.3) and zebrafish eftud2 (NM_200508.2) transcripts were aligned using the Kalign multiple sequence alignment tool (http://www.ebi.ac.uk/Tools/msa/kalign).
Expression studies
The animal protocol was approved by the Institutional Animal Care and Use Committee at the Medical College of Wisconsin (protocol number AUA00000351_AR_5).
Zebrafish (Danio rerio) maintenance and developmental staging were performed as previously described (Liu and Semina, 2012). The expression of eftud2 was studied in 18–72-hpf zebrafish embryos using transcript-specific antisense riboprobes and previously described protocols (Liu and Semina, 2012). For the eftud2 probe, a 715-bp fragment covering the sequence between nucleotides 233 and 947 (GenBank# NM_200508.2) was amplified using the following primers (F- ATGCTGATGAGGCGGATG; R- TTGAGCTCCACAATCAGACG) and cloned in pCRII-TOPO vector. For RT-PCR analysis, total RNA from whole embryos at different stages of development (~18-hpf (hours post fertilization) through 6-dpf (days post fertilization)) or dissected eyes (~24- and ~72-hpf) was isolated using the RNAqueous-Micro Total RNA Isolation Kit (Cat. #AM1931) (Life Technologies) and reverse transcribed using the SuperScript III Reverse Transcriptase system (Life Technologies, Carlsbad, CA, USA). The following primers were utilized to amplify a 425-bp product corresponding to the eftud2 transcript (to avoid amplification of genomic DNA, if present, the forward primer was designed to span exons 17–18 and the reverse- exons 21–22): eftud2-rt-F - TGAAGAGGCACAGATCTTCAG and eftud2-rt-R – TTTTCCTGTTCCATGTGATCT Amplification of the zebrafish gene for elongation factor 1 alpha (loading control) was performed with the following primers: ef1ax1-2F 5′-TCTCTCAATCTTGAAACTTATCAATCA-3′ and ef1ax3R 5′-AACACCCAGGCGTACTTGAA-3′ (PCR product equal 205 bp). The 1 Kb Plus DNA Ladder was utilized as the nucleic acid standard (Life Technologies, Carlsbad, CA, USA). The RT-PCR reactions to assess the levels of mutant and wild-type eftud2 transcripts were performed using 3 independent RNA samples, with each containing 3 homozygous mutant or wild-type eftud2 embryos from the same clutch. For semi-quantitative analysis, the PCR products were visualized by electrophoresis and quantitated by densitometry of the bands using Image J software (http://imagej.nih.gov/ij/) as previously described (Deml et al., 2015).
Generation and analysis of eftud2 mutant lines
TALENs were constructed following the previously published protocol (Sanjana et al., 2012). Briefly, eftud2 TALENs were designed to target the region surrounding the nucleotide corresponding to the mutation site in human patients (left TALEN – 5′-TGCTGATCTTATGGACAGC-3′, right TALEN – 5′-GTCCACACAGTGTGACATT-3′). Interruption of the eftud2 target sequence was predicted to affect the BclI restriction site (TGATCA) located between the left and right TALENs; thus BclI digestion of the 301-bp PCR-amplified genomic fragment generated with the following primers (eftud2-zf-T-F - GCCCATTATAAAACCTGTGAGG and eftud2-zf-T-R - AAACCCCCACATGATGCTC) was utilized to determine TALEN cutting efficiencies as well as to genotype mutant embryos/adult fish; undigested PCR products were sequenced for confirmation. Zebrafish embryos at the 1–4 cell stage were injected with TALEN RNA; initial analysis of injected larvae confirmed genome editing events and ~200 injected embryos were raised to adulthood to generate mosaic founders; these fish were then bred to produce embryos carrying germline eftud2 mutations that were raised to adulthood, genotyped as described above and bred to generate homozygous embryos.
Gross morphological and histological analysis of zebrafish embryos was performed as previously described (Liu and Semina, 2012). For apoptosis analysis, the whole mount TUNEL assay (that detects terminal deoxynucleotidyl transferase dUTP nick end labeling) protocol was followed (Eimon, 2014); for alcian blue staining, the earlier described technique was applied (Liu and Semina, 2012). For measurements, some previously developed metrics (McCollum et al., 2007) were utilized; lateral images of mutant (n=6) and wild-type (n=6) embryos at ~34 hpf were used to determine full body length (from the head top to the tail end), dorsoventral head height (from the yolk to the top of the head through the center of the lens) and ocular diameters (along the dorsoventral (DV) and nasotemporal (NT) axes, through the center of the lens) using ImageJ software. The averages and standard deviations were calculated for body length, head height, and ocular diameters as well as head height/body length and ocular diameter/body length ratios and compared between wild-type and mutant embryos; significant differences were determined based on p values.
Results
Identification of a pathogenic variant in EFTUD2 in a patient with syndromic microphthalmia and coloboma
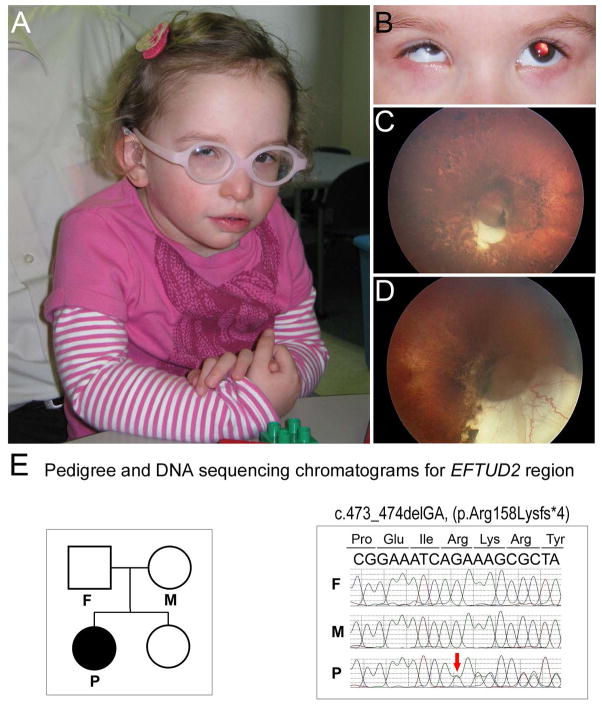
The proband is a 2.5-year-old Caucasian female with microphthalmia, microcornea, inferior optic nerve coloboma of the right eye, high myopia of the left eye, and bilateral pigmentary retinal dystrophy with optic pit/optic nerve atrophy and nystagmus. At dilated fundus exam at 2 years of age, the right eye showed a large optic pit and inferior coloboma, severely attenuated/absent retinal vasculature, and a mild peripheral pigmentary dystrophy; the left eye was noted to have a moderately sized optic pit with inferior cresent, lacunar ‘punched out’ defects with hyper- and hypo- pigmentation scattered in a ‘bird-shot’ fashion throughout the entire retina, attenuated vessels, and hypoplastic macula (Fig. 1). Electroretinogram (ERG) showed a 6 fold reduction in photopic response and no response from the scotopic in the right eye and diminished delayed response in photopic and more severe scotopic impairment in the left eye. Non-ocular anomalies include microcephaly (<3rd centile; at 25-months-old, OFC measured 44.5 cm, 50th centile for a 10-month-old), low-set crumpled ears, broad nasal bridge, upsplanting palpebral fissures, left epicanthic fold, micrognathia, triangular facies, mild facial asymmetry, conductive hearing loss, global developmental delay (functioning at a 15–18 month level at chronological age of 30 months) and a history of feeding difficulties and gastroesophageal reflux as an infant. Height and weight are normal at the 25th centile. The patient has had normal results from echocardiogram and brain magnetic resonance imaging, as well as chromosomal analysis and array-based comparative genomic hybridization. Because of the combination of craniofacial and ocular features, clinical genetic sequencing of CHD7 and SALL1 was undertaken and found to be normal for both genes. Additional clinical testing included Whole-Genome Oligonucleotide Array CGH (105K) that did not identify any copy number abnormalities of known clinical significance.
Figure 1. Clinical images and EFTUD2 sequence of Patient 1.
A–D: Patient images. Please note craniofacial features of microcephaly, low-set crumpled ears, broad nasal bridge, upsplanting palpebral fissures, left epicanthic fold, mandibular and malar hypoplasia, triangular facies, and mild facial asymmetry as well as microphthalmia/microcornea (A–B). Retinal images demonstrate moderately sized optic pit with inferior cresent, lacunar ‘punched out’ defects with hyper- and hypo- pigmentation scattered in a ‘bird-shot’ fashion throughout the entire retina, attenuated vessels, and hypoplastic macula in the left eye (C) and large optic pit and inferior coloboma, severely attenuated/absent retinal vasculature, and a mild peripheral pigmentary dystrophy in the right eye (D). E. Pedigree (on the left) and DNA sequencing chromatograms (on the right). Individuals included in trio analysis are indicated on the pedigree; affected proband indicated with black symbol, P, proband, M, mother, F, father. Chromatogram shows the presence of the frameshift variant (red arrow) in the proband and its absence in both parents.
DNA samples from the affected patient and both unaffected parents were submitted for whole exome sequencing and produced a mean coverage of 62X ± 7 with 95.7% of bases having >10X coverage for the proband, 72.1% for paternal, and 90.7% for maternal samples. Initial analysis of the whole exome data for variants in 82 known genes associated with microphthalmia or coloboma (Deml et al., 2014) showed the genes were generally well-covered (Supplemental Table 1) and identified two relatively rare (0.5–0.8% allele frequency in European control populations) missense variants in LRP2, c.11092G>A, p.(Val3698Met) and c.9613A>G, p.(Asn3205Asp). Mutations in LRP2 are associated with Donnai-Barrow syndrome (DBS), a very rare condition characterized by agenesis of the corpus callosum, developmental delay, high myopia, proteinuria, and sensorineural hearing loss in >90% of published cases as well as congenital diaphragmatic hernia or omphalocele in about half of affected patients; coloboma and retinal dystrophy are occasionally reported (Pober et al., 2009). Characteristic facial features include hypertelorism, macrocephaly, down-slanting palpebral fissures, and flat nasal bridge with triangular tip. Typical LRP2 mutations result in protein truncation (nonsense, frameshift, and splicing variants); two cases with homozygous missense mutations have been reported with neither pathogenic allele being present in control populations (Pober et al., 2009; Schrauwen et al., 2014). Taken together, the absence of classic DBS features and the presence of anomalies not consistent with DBS (microcephaly, micrognathia, feeding disorder, upslanting palpebral fissures, and ear anomalies) in our patient, a higher than appropriate population frequency for both alleles and the presence of homozygous controls for each variant (6 homozygotes noted in ExAC) led us to consider the identified LRP2 variants non-pathogenic.
Analysis of the complete exome data for either de novo variants (under dominant inheritance model) or homozygous/compound heterozygous variants (under recessive model) identified three de novo and eight possible recessive alleles in 8 additional genes (Supplemental Table 2). Two of these genes, KMT2C (MLL3) and EFTUD2, have been previously studied in relation to human disorders: a heterozygous nonsense variant in KMT2C (MLL3) was reported in a patient with Kleefstra syndrome spectrum but determined to be of unknown significance (Kleefstra et al., 2012) and numerous dominant EFTUD2 alleles were reported in Mandibulofacial Dysostosis, Guion-Almeida type (MFDGA; MIM 610536). The MFDGA is characterized by microcephaly, midface and malar hypoplasia, micrognathia, microtia, dysplastic ears, preauricular skin tags, upslanting palpebral fissures, and significant developmental delay as well as choanal atresia/respiratory difficulties, conductive hearing loss, esophageal atresia/feeding difficulties, cleft palate, congenital heart defects, and radial ray anomalies in many cases (Table 1) (Gordon et al., 2012; Lehalle et al., 2014; Lines et al., 2012; Luquetti et al., 2013; Need et al., 2012; Voigt et al., 2013). The majority of dominant EFTUD2 pathogenic variants are truncating (nonsense/frameshift) or splicing alleles and most are de novo, similar to the c.473_474delGA, p.(Arg158Lysfs*4) variant identified in this study (Figs 1, 2). Additionally, upon re-examination, the proband’s features (including microcephaly, ear anomalies, mandibular/malar hypoplasia, upslanting palpebral fissures, facial asymmetry, hearing loss, feeding difficulties, and developmental delay) were found to be consistent with the previously reported cases of MFDGA (Table 1). Thus, the de novo variant c.473_474delGA, p.(Arg158Lysfs*4) in EFTUD2 was considered to be pathogenic in this patient. The variant was confirmed in the proband’s DNA and not found in samples from either unaffected parent by Sanger sequencing of PCR products generated with EFTUD2-specific primers (Fig. 1). The pathogenic variant affects exon 6 of 28 EFTUD2 exons and is predicted to result in nonsense-mediated decay (NMD) of the transcript or, if produced, a truncation of the encoded protein at 16% of its normal length.
Table 1.
Summary of features associated with pathogenic variants in EFTUD2 in previously reported cases of MFDGA and the affected patient in this study.
| Featuresa | Previously reported casesb | Frequency | This study |
|---|---|---|---|
| Craniofacial phenotype : | 60/60 | 100% | + |
| Zygomatic arch cleft | 7/7 | 100% | N/R |
| External ear anomalies | 57/58 | 98% | + |
| Mandibular hypoplasia | 54/58 | 93% | + |
| Malar hypoplasia | 42/46 | 91% | + |
| Hearing loss | 40/52 | 77% | + |
| Semicircular canal anomalies | 11/15 | 73% | N/R |
| Auditory canal stenosis | 30/48 | 63% | − |
| Facial asymmetry | 25/47 | 53% | + |
| Preauricular tags | 25/58 | 43% | − |
| Choanal atresia | 20/58 | 34% | − |
| Cleft palate | 19/58 | 33% | − |
| Eye anomalies | 9/30 | 30% | + |
| Psychomotor delay | 55/56 | 98% | + |
| Microcephaly | 53/60 | 88% | + |
| Brain MRI anomaly | 8/18 | 44% | − |
| Kidney anomalies | 6/14 | 43% | − |
| Congenital heart defect | 22/54 | 41% | − |
| Hand anomalies | 23/58 | 40% | − |
| Esophageal atresia | 22/59 | 37% | − |
| Epilepsy | 11/52 | 21% | − |
| Spine and thorax anomalies | 9/53 | 17% | − |
| Feet anomalies | 5/58 | 9% | − |
| Genitalia anomalies | 5/57 | 9% | − |
features are listed in order of decreasing frequency
as reported in Lehalle et al. 2014
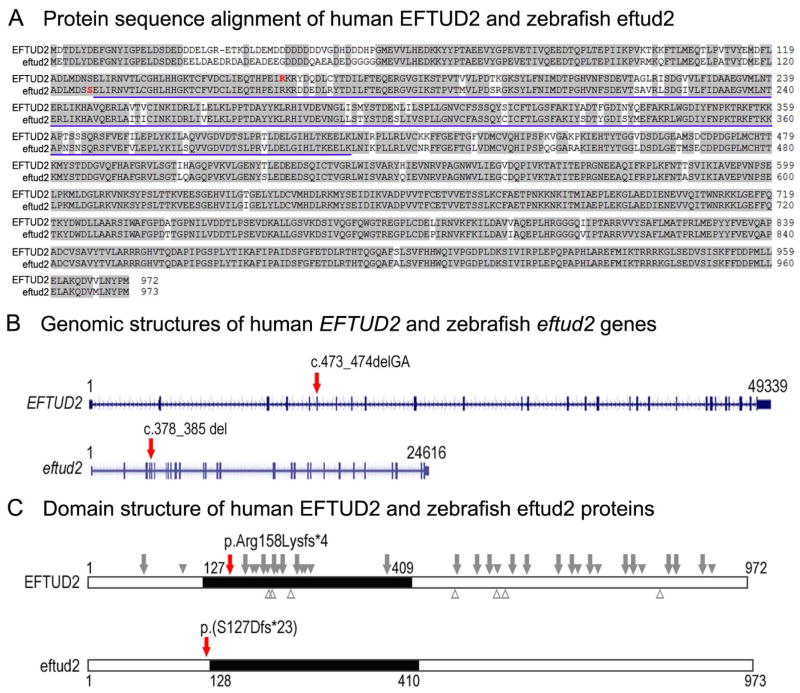
Figure 2. Comparison of human and zebrafish EFTUD2/eftud2 genomic and protein structure.
A. Protein sequence alignment of human EFTUD2 and zebrafish eftud2; amino acids identical between the homologs are highlighted with a light grey color, the amino acids representing the beginning of the frameshift in the human patient and zebrafish line are indicated in red font in the respective protein. The blue underline indicates the amino acids involved in the translational (tr)-type guanine nucleotide-binding (G) domain. B. Genomic structures of the human EFTUD2 and zebrafish eftud2 genes as indicated in the Genome Browser for the respective genes. The exons are shown as blue boxes with 5′ and 3′UTR shown as short and coding regions shown as tall blue boxes. The positions of the human and zebrafish variants are indicated with red arrows on the respective genes. C. Domain structure of human EFTUD2 and zebrafish eftud2 proteins. The black box shows the translational (tr)-type guanine nucleotide-binding (G) domain; the positions of the human and zebrafish variants that were identified/generated in this study are indicated with red arrows on the respective proteins; the approximate positions of the unique previously reported pathogenic variants are shown on the human protein with truncating variants indicated above as grey arrows (nonsense and frameshift) or solid grey arrowheads (splicing variants, indicated based on affected exon) above and missense variants indicated as empty grey arrowheads below.
Since neither MAC nor retinal dystrophy have been previously reported as part of the MFDGA spectrum, we decided to examine the possibility of multiple molecular diagnoses in the affected individual. The analysis of MAC genes, as presented above, did not detect any pathogenic or likely pathogenic variants. Similarly, analysis of genes associated with retinal dystrophy did not identify any disease-causing alleles that could explain the retinal features. Several heterozygous variants of uncertain significance were identified in genes associated with autosomal recessive retinitis pigmentosa (Supplemental Table 3); none could be clearly categorized as pathogenic (Richards et al., 2015) and none were paired with a second rare allele in the same gene (with good coverage of the genes of interest).
Zebrafish eftud2 expression studies
To investigate the role of EFTUD2/eftud2 in vertebrate development, we identified zebrafish eftud2 and studied its embryonic expression using in situ hybridization and RT-PCR. The zebrafish transcript corresponding to the eftud2 gene (NM_200508.2) was previously mapped to chromosome 3 and encodes a protein with ~91% identity to human EFTUD2 (Fig. 2). BLAST searches of the ZFIN database did not identify additional sequences with significant homology to the EFTUD2 gene, thus this transcript represents the only zebrafish ortholog of human EFTUD2. Similar to the human gene, zebrafish eftud2 consists of 28 exons that comprise ~24,616 kb, which is ~ ½ of the genomic size of human EFTUD2 (49,339 kb) (Fig. 2).
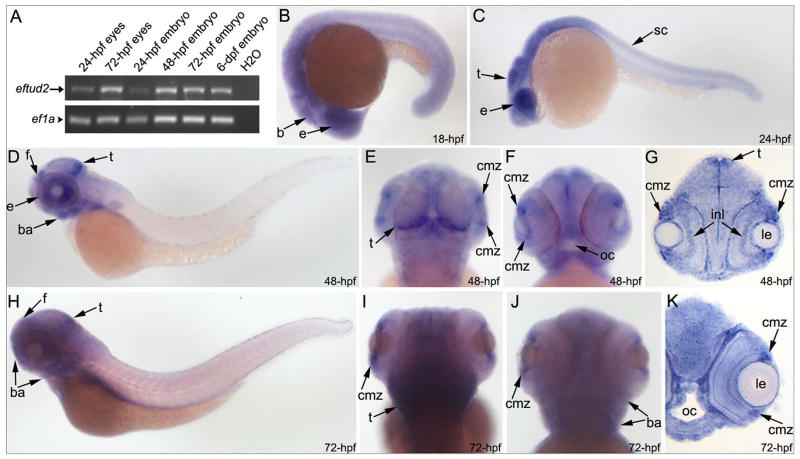
The eftud2 transcripts were detected by RT-PCR in all examined samples including 24- and 72-hpf embryonic eyes and 1–6-dpf whole embryos (Fig. 3A); in situ hybridization experiments confirmed broad eftud2 expression (Fig. 3). At 18- and 24-hpf, an elevated level of expression was observed in the head region (Fig. 3B–C). At 48–72-hpf, more robust staining for eftud2 transcripts was detected in the head, particularly in the developing forebrain and tectum, the ciliary marginal zone of the developing eye and the craniofacial region/branchial arches (Fig. 3D–K). This is consistent with recently published data reporting the presence of eftud2 transcripts in various samples, including the ovaries (indicating that this is a maternally provided transcript), 2-cell embryos, 1–6-dpf embryos as well as the adult head and body, with a possible enrichment in the head (Collins et al., 2012). The maternally supplied mRNAs were shown to be subject to early destabilization often triggered by fertilization and largely completed upon activation of the zygotic genome, which in zebrafish occurs during the 10th cell cycle or approximately 3 hours post fertilization (Aanes et al., 2013; Tadros and Lipshitz, 2009). Therefore, eftud2 transcripts detected at later stages of development are produced by zygotic transcription.
Figure 3. Embryonic expression of zebrafish eftud2.
RT-PCR and in situ hybridization expression data for eftud2 in zebrafish embryos. Please note the presence of eftud2 transcript in 24- and 72-hpf embryonic eyes as well as whole embryos as detected by RT-PCR (A). In situ hybridization revealed a broad expression throughout the embryo with a possible enrichment in the head and eye region. Whole mount images (B–F, H–J) and sections (G, K) are shown; developmental stage is indicated in the lower-right corner; b- brain, ba- branchial arches, cmz-ciliary marginal zone of the retina, e-eye, f- forebrain, inl- inner nuclear layer of the retina; le- lens, oc-oral cavity, sc- spinal cord, t-tectum.
Generation and analysis of eftud2 zebrafish mutants
To determine the effect of eftud2 deficiency on embryonic development, the eftud2 gene was disrupted in zebrafish embryos using TALEN genome editing technology. A mutant line was established for the c.378_385 del; p.(Ser127Aspfs*23) frameshift mutation (Fig. 2; Supplemental Fig. 1A) that is predicted to result in nonsense mediated decay of mRNA (NMD). Semi-quantitative RT-PCR analysis of eftud2 expression identified the presence of the mutant transcript in homozygous p.(Ser127Aspfs*23) embryos, with some reduction in comparison to the expression of normal transcript in wild-type embryos (Supplemental Fig. 1B), suggesting an incomplete NMD. The mutant transcript is predicted to produce a truncated protein (13% of its normal length) lacking the translational (tr)-type guanine nucleotide-binding (G) domain and thus to result in a complete loss-of-function, similar to human mutations.
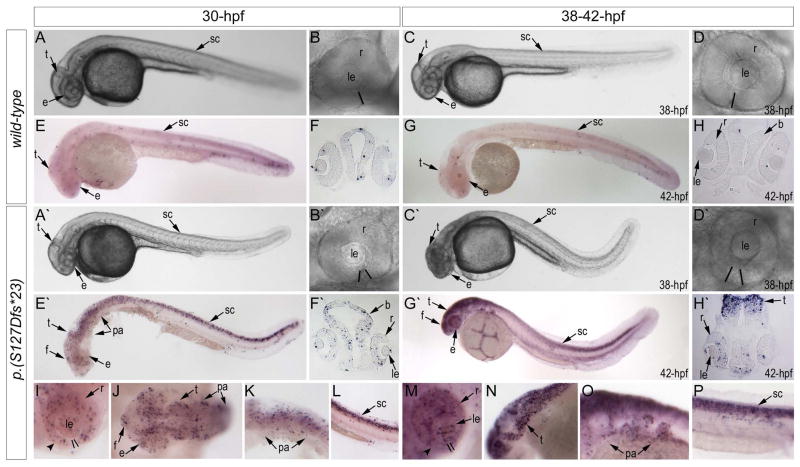
Initial crosses of heterozygous animals produced groups of abnormal embryos with a small head/eye and curved body (starting from 30-hpf) that died before 48-hpf (Fig. 4A′ and C′); genotyping analysis of a clutch of 50 embryos identified 11 wild-type homozygous and 29 heterozygous animals in the normal phenotypic group (40) while 10 out of 10 abnormal embryos were found to be homozygous for the eftud2 mutation. A disruption in optic fissure closure was observed in 47.9% (34/71) of 38–42-hpf mutant embryos with the optic fissure margins not coming into close contact with each other as seen in wild-type embryos (Fig. 4B′ and D′). Measurements of the head height and ocular diameters at 35-hpf identified a significant difference between mutant (n=6) and wild-type embryos (n=6) from the same clutch: the mutant embryos demonstrated a reduction in body length (96% of wild-type), head height (88% of wild-type), as well as dorsoventral (83% of wild-type) and nasotemporal (68% of wild-type) ocular diameters; after normalization of head/eye measurements to body length as suggested (Collery et al., 2014), the ratios for mutant embryos were found to be at ~92% of wild-type (head/body; p-value < 0.001), ~86% (for dorsoventral ocular diameter; p-value < 0.001) and ~71% (for nasotemporal ocular diameter; p-value < 0.001) (Supplemental Fig. 1C). Histological analysis in homozygous mutant embryos (n=8) indicated possible extensive cell death evidenced by the presence of cell corpses in the retina (which appear as intensely stained particles) that was not observed in wild-type embryos (n=6) (Supplemental Fig. 1D–G). Subsequent analysis of mutant embryos at 30-hpf (n=7) and 42-hpf (n=9) with TUNEL assay identified an excessive number of TUNEL-positive cells in mutants in comparison to wild-type (n=8 for 30-hpf; n=7 for 42-hpf) (Fig. 4). The increase in TUNEL staining was clearly observed at 30-hpf and became more intense by 42-hpf and included various embryonic tissues such as the developing head (especially the brain and retina), the region of the pharyngeal arches, and the spinal cord (Fig. 4I–P). Heterozygous embryos did not display any abnormal phenotype, based on gross morphological examination and alcian blue staining of 120-hpf embryos (n=19) and gross morphological examination of adult fish (n=5) (Supplemental Fig. 1H–K).
Figure 4. Phenotypic analysis of eftud2 mutants.
A–H. Morphology (A–D) and TUNEL staining (E–H) of wild-type embryos at 30 and 38–42-hpf (as indicated). A′–H′. Morphology (A′–D′) and TUNEL staining (E′–H′) of eftud2 mutant embryos at 30 and 38–42-hpf. Please note microcephaly and coloboma in mutant embryos. I–P. Enlarged whole mount images of TUNEL staining in eftud2 mutant embryos in representative embryonic regions. Please note increase in TUNEL positive cells throughout the embryo, particularly in the eye, branchial arches, tectum, and spinal cord. The black lines in B, B′, D, D′, I and M indicate the retinal edges and highlight the presence of coloboma in mutant embryos; arrowheads in I and M indicate thinning of the ventral portion of the retina in mutants; b- brain, e-eye, f- forebrain, le- lens, pa- pharyngeal arches, r-retina, sc- spinal cord, t-tectum.
Discussion
In this manuscript we report a novel mutation in human EFTUD2 and the generation of a zebrafish line with a similar eftud2 mutation and a severe embryonic phenotype. EFTUD2 encodes the U5-116kD GTPase, a core protein of the U5 snRNP and a homolog of Snu114p in S. cerevisiae which is essential for pre-mRNA splicing (Fabrizio et al., 1997). Snu114p is a critical part of the spliceosome and was shown to play a key role in spliceosome remodeling and function (Frazer et al., 2008). Overall, the spliceosome contains five small nuclear ribonucleoproteins (snRNPs), U1, U2, U4, U5, and U6, in combination with numerous other proteins (Singh and Cooper, 2012).
Lines et. al. (2012) identified twelve EFTUD2 de novo pathogenic variants in patients with mandibulofacial dysostosis type Guion-Almeida (MFDGA), also called mandibulofacial dysostosis with microcephaly. Since then, several additional manuscripts reported EFTUD2 pathogenic variants, typically de novo, in patients with craniofacial anomalies which overlap MFDGA; the majority of pathogenic variants are splicing (23/56; 41%) or nonsense/frameshift (20/56; 36%) with missense (9/56; 16%) and partial/whole gene deletions (4/56; 7%) also seen (Bernier et al., 2012; Gordon et al., 2012; Lehalle et al., 2014; Luquetti et al., 2013; Need et al., 2012; Voigt et al., 2013). Characteristic features noted in >75% of reported patients and also seen in our proband include external ear anomalies, mandibular and malar hypoplasia, hearing loss, psychomotor delay, and microcephaly (Table 1). There is also a wide spectrum of highly variable rare or unique features noted in less than a third of patients including spine, thorax, feet, and genital anomalies (Table 1;(Lehalle et al., 2014)).
Mandibulofacial dysostosis type Guion-Almeida (MFDGA) belongs to a broader group of facial dysostosis disorders which are primarily diagnosed based on anomalies in structures derived from the first and second pharyngeal arches (Trainor and Andrews, 2013). No animal models of eftud2 deficiency currently exist and thus the generation of an eftud2 zebrafish knockout line allows for the first insight into the conservation and developmental function of this factor. Our studies demonstrate that normal function of eftud2 is essential for normal embryonic development as eftud2-deficient zebrafish die before 2 days post fertilization. Prior to this premature death, the eftud2 homozygous mutants show reduced head/eye size and curved body accompanied by increased cell death in the developing head region as well as throughout the embryo. The phenotype observed in zebrafish appears to be more severe and follows a recessive pattern while pathogenic variants in human EFTUD2 are inherited in a dominant fashion. The lack of phenotype in heterozygous zebrafish suggests a less stringent requirement for full expression of eftud2 in zebrafish which may be due to variation in the expression of other splicing factors between humans and zebrafish and thus different compensation potentials. Variable gene dosage requirements between species have been described for numerous other genes (Zhang and Baldini, 2008).
As of yet, no consistent developmental ocular phenotype has been associated with pathogenic variants in EFTUD2, but eye anomalies have been reported in up to 28% of cases including myopia, hypermetropia, strabismus, astigmatism, epibulbar dermoids, epicanthic folds, lacrimal duct stenosis (Lehalle et al., 2014) and lower eyelid cleft (Lines et al., 2012). Interestingly, pathogenic variants in genes encoding other components of the U4/U6.U5 tri-snRNP of the major spliceosome including PRPF3, PRPF4, PRPF6, PRPF8, PRPF31 and SNRNP200 cause isolated retinitis pigmentosa (RP) (Chakarova et al., 2002; Chen et al., 2014; McKie et al., 2001; Tanackovic et al., 2011; Vithana et al., 2001; Zhao et al., 2006). It has been suggested that a decrease in tri-snRNP activity below a certain threshold may be responsible for RP (Linder et al., 2014); since EFTUD2 is also a component of the tri-snRNP, variants in EFTUD2 may also affect retinal development. Recently, novel missense variants in EFTUD2 were identified in two probands with autosomal dominant retinitis pigmentosa but the pathogenicity of these variants remains uncertain because no affected family members were available to confirm cosegregation with the ocular phenotype (Benaglio et al., 2014). Studies in zebrafish presented in this manuscript suggest that normal eftud2 function is necessary for normal development of head structures, including the developing eye. In homozygous mutant zebrafish embryos, the absolute and normalized (by body length) eye size was reduced and some embryos demonstrated optic fissure closure defects. In zebrafish embryos, the optic fissure becomes completely closed by 2 days post-fertilization, with most wild-type embryos demonstrating close contact between the fissure margins by 30–36 hpf (Weiss et al., 2012). This process appears to be interrupted in ~48% of mutant eftud2 embryos at 38–42 hpf; however, due to the premature death of eftud2 homozygous mutants accompanied by the massive cell death phenotype, further examination of ocular development was not possible.
Since the previously published EFTUD2- positive MFDGA cases seem to demonstrate normal eye size and retinal development, the ocular phenotype in the reported patient may be completely coincidental and resulting from an independent mutation in an unknown ocular gene or environmental factor. It is also possible that the causative mutation for the retinal phenotype is located within the mitochondrial genome, which was not sequenced in this patient. Alternatively, the ocular phenotype may be produced by an interaction between the pathogenic EFTUD2 allele and another, yet to be determined, factor(s); analysis of known A/M and retinal dystrophy genes identified several rare “variants of unknown significance” that may have contributed to the ocular phenotype and need to be further investigated. Additional screenings of EFTUD2 in patients with syndromic microphthalmia, ocular coloboma, and/or retinal dystrophy and careful ocular examination of patients with MFDGA will help to specify the role of EFTUD2, if any, in ocular disease. The eftud2 zebrafish mutant line will allow for further investigation of the mechanisms of MFDGA, evaluation of mutations identified in human patients, and testing of potential new therapeutic interventions.
Supplementary Material
Molecular and phenotypic examinations of eftud2 mutant embryos. A. DNA chromatogram for zebrafish mutant allele; red line indicates a deletion of 8-nt in mutant DNA. B. RT-PCR analysis of eftud2 transcripts. Top: image of gel electrophoresis of eftud2- specific (lanes 1–6) and ef1a-specific (lanes 7–12) RT-PCR products obtained using mutant (1–3 and 7–9) and wild-type (4–6 and 10–12) total RNA; red arrow indicates a 425-bp eftud2 product, black arrowhead points to 205-bp ef1a-specific band; M corresponds to molecular weight marker. Please note a reduction in eftud2 level in 3 independent RNA samples (lanes 1–3) extracted from mutant embryos in comparison to wild-type (lanes 4–6). Bottom: graph representing quantification of eftud2 expression using ImageJ in mutant and wild-type embryos. C. Measurements of body length, head height and ocular diameters in eftud2 homozygous mutant and wild-type embryos. DV- dorsoventral; NT- nasotemporal. D–G. Histological analysis of eftud2 homozygous fish. Sections at eye level of 38-hpf wild-type (C, D) and eftud2 mutants (E, F) are shown. c-cornea, le- lens, r-retina; arrowheads indicate intensely stained particles likely corresponding to dead cell corpses. H–K. Alcian blue staining in eftud2 120-hpf heterozygous fish showing normal craniofacial cartilage structure. ch- ceratohyal cartilage, ep- ethmoid plate, m- Meckel’s cartilage, pq- palatoquadrate cartilage components of upper and lower jaws.
Known human MAC genes and their WES coverage in the affected patient.
Summary of rare and novel variants identified via trio analysis of the affected family.
Summary of novel/rare alleles identified in retinal dystrophy genes.
Acknowledgments
Grant information: This work was supported by NIH grant EY015518 (EVS), and funds provided by the Children’s Research Institute, Children’s Hospital of Wisconsin, Clinical and Translational Science Award (CTSA) UL1RR031973, and T32 NIH grant EY014537.
The authors gratefully acknowledge the patient and her family for their participation in research studies.
Footnotes
The authors declare no competing financial interests.
Literature cited
- Aanes H, Ostrup O, Andersen IS, Moen LF, Mathavan S, Collas P, Alestrom P. Differential transcript isoform usage pre- and post-zygotic genome activation in zebrafish. BMC Genomics. 2013;14 doi: 10.1186/1471-2164-14-331. 331-2164-14-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardakjian TM, Schneider A. The genetics of anophthalmia and microphthalmia. Curr Opin Ophthalmol. 2011;22:309–313. doi: 10.1097/ICU.0b013e328349b004. [DOI] [PubMed] [Google Scholar]
- Beleggia F, Li Y, Fan J, Elcioglu NH, Toker E, Wieland T, Maumenee IH, Akarsu NA, Meitinger T, Strom T, Lang R, Wollnik B. CRIM1 haploinsufficiency causes defects in eye development in human and mouse. Hum Mol Genet. 2015 doi: 10.1093/hmg/ddu744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaglio P, San Jose PF, Avila-Fernandez A, Ascari G, Harper S, Manes G, Ayuso C, Hamel C, Berson EL, Rivolta C. Mutational screening of splicing factor genes in cases with autosomal dominant retinitis pigmentosa. Mol Vis. 2014;20:843–851. [PMC free article] [PubMed] [Google Scholar]
- Bernier FP, Caluseriu O, Ng S, Schwartzentruber J, Buckingham KJ, Innes AM, Jabs EW, Innis JW, Schuette JL, Gorski JL, Byers PH, Andelfinger G, Siu V, Lauzon J, Fernandez BA, McMillin M, Scott RH, Racher H, Majewski J, Nickerson DA, Shendure J, Bamshad MJ, Parboosingh JS FORGE Canada Consortium. Haploinsufficiency of SF3B4, a component of the pre-mRNA spliceosomal complex, causes Nager syndrome. Am J Hum Genet. 2012;90:925–933. doi: 10.1016/j.ajhg.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botzenhart EM, Green A, Ilyina H, Konig R, Lowry RB, Lo IF, Shohat M, Burke L, McGaughran J, Chafai R, Pierquin G, Michaelis RC, Whiteford ML, Simola KO, Rosler B, Kohlhase J. SALL1 mutation analysis in Townes-Brocks syndrome: twelve novel mutations and expansion of the phenotype. Hum Mutat. 2005;26:282. doi: 10.1002/humu.9362. [DOI] [PubMed] [Google Scholar]
- Chakarova CF, Hims MM, Bolz H, Abu-Safieh L, Patel RJ, Papaioannou MG, Inglehearn CF, Keen TJ, Willis C, Moore AT, Rosenberg T, Webster AR, Bird AC, Gal A, Hunt D, Vithana EN, Bhattacharya SS. Mutations in HPRP3, a third member of pre-mRNA splicing factor genes, implicated in autosomal dominant retinitis pigmentosa. Hum Mol Genet. 2002;11:87–92. doi: 10.1093/hmg/11.1.87. [DOI] [PubMed] [Google Scholar]
- Chen X, Liu Y, Sheng X, Tam PO, Zhao K, Chen X, Rong W, Liu Y, Liu X, Pan X, Chen LJ, Zhao Q, Vollrath D, Pang CP, Zhao C. PRPF4 mutations cause autosomal dominant retinitis pigmentosa. Hum Mol Genet. 2014;23:2926–2939. doi: 10.1093/hmg/ddu005. [DOI] [PubMed] [Google Scholar]
- Collery RF, Veth KN, Dubis AM, Carroll J, Link BA. Rapid, accurate, and non-invasive measurement of zebrafish axial length and other eye dimensions using SD-OCT allows longitudinal analysis of myopia and emmetropization. PLoS One. 2014;9(10):e110699. doi: 10.1371/journal.pone.0110699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JE, White S, Searle SM, Stemple DL. Incorporating RNA-seq data into the zebrafish Ensembl genebuild. Genome Res. 2012;22:2067–2078. doi: 10.1101/gr.137901.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deml B, Kariminejad A, Borujerdi RH, Muheisen S, Reis LM, Semina EV. Mutations in MAB21L2 result in ocular coloboma, microcornea and cataracts. PLoS Genet. 2015;11:e1005002. doi: 10.1371/journal.pgen.1005002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deml B, Reis LM, Maheshwari M, Griffis C, Bick D, Semina EV. Whole exome analysis identifies dominant COL4A1 mutations in patients with complex ocular phenotypes involving microphthalmia. Clin Genet. 2014;86:475–481. doi: 10.1111/cge.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eimon PM. Studying apoptosis in the Zebrafish. Methods Enzymol. 2014;544:395–431. doi: 10.1016/B978-0-12-417158-9.00016-9. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Laggerbauer B, Lauber J, Lane WS, Luhrmann R. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 1997;16:4092–4106. doi: 10.1093/emboj/16.13.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer LN, Nancollis V, O’Keefe RT. The role of Snu114p during pre-mRNA splicing. Biochem Soc Trans. 2008;36:551–553. doi: 10.1042/BST0360551. [DOI] [PubMed] [Google Scholar]
- Gestri G, Osborne RJ, Wyatt AW, Gerrelli D, Gribble S, Stewart H, Fryer A, Bunyan DJ, Prescott K, Collin JR, Fitzgerald T, Robinson D, Carter NP, Wilson SW, Ragge NK. Reduced TFAP2A function causes variable optic fissure closure and retinal defects and sensitizes eye development to mutations in other morphogenetic regulators. Hum Genet. 2009 doi: 10.1007/s00439-009-0730-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon CT, Petit F, Oufadem M, Decaestecker C, Jourdain AS, Andrieux J, Malan V, Alessandri JL, Baujat G, Baumann C, Boute-Benejean O, Caumes R, Delobel B, Dieterich K, Gaillard D, Gonzales M, Lacombe D, Escande F, Manouvrier-Hanu S, Marlin S, Mathieu-Dramard M, Mehta SG, Simonic I, Munnich A, Vekemans M, Porchet N, de Pontual L, Sarnacki S, Attie-Bitach T, Lyonnet S, Holder-Espinasse M, Amiel J. EFTUD2 haploinsufficiency leads to syndromic oesophageal atresia. J Med Genet. 2012;49:737–746. doi: 10.1136/jmedgenet-2012-101173. [DOI] [PubMed] [Google Scholar]
- Hornby SJ, Adolph S, Gilbert CE, Dandona L, Foster A. Visual acuity in children with coloboma: clinical features and a new phenotypic classification system. Ophthalmology. 2000;107:511–520. doi: 10.1016/s0161-6420(99)00140-2. [DOI] [PubMed] [Google Scholar]
- Huynh N, Blain D, Glaser T, Doss EL, Zein WM, Lang DM, Baker EH, Hill S, Brewer CC, Kopp JB, Bardakjian TM, Maumenee IH, Bateman BJ, Brooks BP. Systemic diagnostic testing in patients with apparently isolated uveal coloboma. Am J Ophthalmol. 2013;156:1159–1168. e4. doi: 10.1016/j.ajo.2013.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelberman D, Islam L, Lakowski J, Bacchelli C, Chanudet E, Lescai F, Patel A, Stupka E, Buck A, Wolf S, Beales PL, Jacques TS, Bitner-Glindzicz M, Liasis A, Lehmann OJ, Kohlhase J, Nischal KK, Sowden JC. Mutation of SALL2 causes recessive ocular coloboma in humans and mice. Hum Mol Genet. 2014;23:2511–2526. doi: 10.1093/hmg/ddt643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleefstra T, Franken CE, Arens YH, Ramakers GJ, Yntema HG, Sistermans EA, Hulsmans CF, Nillesen WN, van Bokhoven H, de Vries BB, Hamel BC. Genotype-phenotype studies in three families with mutations in the polyglutamine-binding protein 1 gene (PQBP1) Clin Genet. 2004;66:318–326. doi: 10.1111/j.1399-0004.2004.00308.x. [DOI] [PubMed] [Google Scholar]
- Lehalle D, Gordon CT, Oufadem M, Goudefroye G, Boutaud L, Alessandri JL, Baena N, Baujat G, Baumann C, Boute-Benejean O, Caumes R, Decaestecker C, Gaillard D, Goldenberg A, Gonzales M, Holder-Espinasse M, Jacquemont ML, Lacombe D, Manouvrier-Hanu S, Marlin S, Mathieu-Dramard M, Morin G, Pasquier L, Petit F, Rio M, Smigiel R, Thauvin-Robinet C, Vasiljevic A, Verloes A, Malan V, Munnich A, de Pontual L, Vekemans M, Lyonnet S, Attie-Bitach T, Amiel J. Delineation of EFTUD2 haploinsufficiency-related phenotypes through a series of 36 patients. Hum Mutat. 2014;35:478–485. doi: 10.1002/humu.22517. [DOI] [PubMed] [Google Scholar]
- Linder B, Hirmer A, Gal A, Ruther K, Bolz HJ, Winkler C, Laggerbauer B, Fischer U. Identification of a PRPF4 loss-of-function variant that abrogates U4/U6.U5 tri-snRNP integration and is associated with retinitis pigmentosa. PLoS One. 2014;9:e111754. doi: 10.1371/journal.pone.0111754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lines MA, Huang L, Schwartzentruber J, Douglas SL, Lynch DC, Beaulieu C, Guion-Almeida ML, Zechi-Ceide RM, Gener B, Gillessen-Kaesbach G, Nava C, Baujat G, Horn D, Kini U, Caliebe A, Alanay Y, Utine GE, Lev D, Kohlhase J, Grix AW, Lohmann DR, Hehr U, Bohm D, Majewski J, Bulman DE, Wieczorek D, Boycott KM FORGE Canada Consortium. Haploinsufficiency of a spliceosomal GTPase encoded by EFTUD2 causes mandibulofacial dysostosis with microcephaly. Am J Hum Genet. 2012;90:369–377. doi: 10.1016/j.ajhg.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013;34:E2393–402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Semina EV. pitx2 Deficiency results in abnormal ocular and craniofacial development in zebrafish. PLoS One. 2012;7:e30896. doi: 10.1371/journal.pone.0030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luquetti DV, Hing AV, Rieder MJ, Nickerson DA, Turner EH, Smith J, Park S, Cunningham ML. “Mandibulofacial dysostosis with microcephaly” caused by EFTUD2 mutations: expanding the phenotype. Am J Med Genet A. 2013;161A:108–113. doi: 10.1002/ajmg.a.35696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzini MC, Tambunan DE, Hill RS, Yu TW, Maynard TM, Heinzen EL, Shianna KV, Stevens CR, Partlow JN, Barry BJ, Rodriguez J, Gupta VA, Al-Qudah AK, Eyaid WM, Friedman JM, Salih MA, Clark R, Moroni I, Mora M, Beggs AH, Gabriel SB, Walsh CA. Exome sequencing and functional validation in zebrafish identify GTDC2 mutations as a cause of Walker-Warburg syndrome. Am J Hum Genet. 2012;91:541–547. doi: 10.1016/j.ajhg.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum CW, Amin SR, Pauerstein P, Lane ME. A zebrafish LMO4 ortholog limits the size of the forebrain and eyes through negative regulation of six3b and rx3. Dev Biol. 2007;309:373–85. doi: 10.1016/j.ydbio.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKie AB, McHale JC, Keen TJ, Tarttelin EE, Goliath R, van Lith-Verhoeven JJ, Greenberg J, Ramesar RS, Hoyng CB, Cremers FP, Mackey DA, Bhattacharya SS, Bird AC, Markham AF, Inglehearn CF. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13) Hum Mol Genet. 2001;10:1555–1562. doi: 10.1093/hmg/10.15.1555. [DOI] [PubMed] [Google Scholar]
- Morrison D, FitzPatrick D, Hanson I, Williamson K, van Heyningen V, Fleck B, Jones I, Chalmers J, Campbell H. National study of microphthalmia, anophthalmia, and coloboma (MAC) in Scotland: investigation of genetic aetiology. J Med Genet. 2002;39:16–22. doi: 10.1136/jmg.39.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura KM, Diehl NN, Mohney BG. Incidence, ocular findings, and systemic associations of ocular coloboma: a population-based study. Arch Ophthalmol. 2011;129:69–74. doi: 10.1001/archophthalmol.2010.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need AC, Shashi V, Hitomi Y, Schoch K, Shianna KV, McDonald MT, Meisler MH, Goldstein DB. Clinical application of exome sequencing in undiagnosed genetic conditions. J Med Genet. 2012;49:353–361. doi: 10.1136/jmedgenet-2012-100819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pober BR, Longoni M, Noonan KM. A review of Donnai-Barrow and facio-oculo-acoustico-renal (DB/FOAR) syndrome: clinical features and differential diagnosis. Birth Defects Res A Clin Mol Teratol. 2009;85:76–81. doi: 10.1002/bdra.20534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis LM, Tyler RC, Muheisen S, Raggio V, Salviati L, Han DP, Costakos D, Yonath H, Hall S, Power P, Semina EV. Whole exome sequencing in dominant cataract identifies a new causative factor, CRYBA2, and a variety of novel alleles in known genes. Hum Genet. 2013;132:761–770. doi: 10.1007/s00439-013-1289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015 Mar 5; doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere JB, van Bon BW, Hoischen A, Kholmanskikh SS, O’Roak BJ, Gilissen C, Gijsen S, Sullivan CT, Christian SL, Abdul-Rahman OA, Atkin JF, Chassaing N, Drouin-Garraud V, Fry AE, Fryns JP, Gripp KW, Kempers M, Kleefstra T, Mancini GM, Nowaczyk MJ, van Ravenswaaij-Arts CM, Roscioli T, Marble M, Rosenfeld JA, Siu VM, de Vries BB, Shendure J, Verloes A, Veltman JA, Brunner HG, Ross ME, Pilz DT, Dobyns WB. De novo mutations in the actin genes ACTB and ACTG1 cause Baraitser-Winter syndrome. Nat Genet. 2012;44:440–4. S1–2. doi: 10.1038/ng.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen I, Sommen M, Claes C, Pinner J, Flaherty M, Collins F, Van Camp G. Broadening the phenotype of LRP2 mutations: a new mutation in LRP2 causes a predominantly ocular phenotype suggestive of Stickler syndrome. Clin Genet. 2014;86:282–286. doi: 10.1111/cge.12265. [DOI] [PubMed] [Google Scholar]
- Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med. 2012;18:472–482. doi: 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalicky SE, White AJ, Grigg JR, Martin F, Smith J, Jones M, Donaldson C, Smith JE, Flaherty M, Jamieson RV. Microphthalmia, anophthalmia, and coloboma and associated ocular and systemic features: understanding the spectrum. JAMA Ophthalmol. 2013;131:1517–1524. doi: 10.1001/jamaophthalmol.2013.5305. [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Tanackovic G, Ransijn A, Ayuso C, Harper S, Berson EL, Rivolta C. A missense mutation in PRPF6 causes impairment of pre-mRNA splicing and autosomal-dominant retinitis pigmentosa. Am J Hum Genet. 2011;88:643–649. doi: 10.1016/j.ajhg.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor PA, Andrews BT. Facial dysostoses: Etiology, pathogenesis and management. Am J Med Genet C Semin Med Genet. 2013;163C:283–294. doi: 10.1002/ajmg.c.31375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vithana EN, Abu-Safieh L, Allen MJ, Carey A, Papaioannou M, Chakarova C, Al-Maghtheh M, Ebenezer ND, Willis C, Moore AT, Bird AC, Hunt DM, Bhattacharya SS. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11) Mol Cell. 2001;8:375–381. doi: 10.1016/s1097-2765(01)00305-7. [DOI] [PubMed] [Google Scholar]
- Voigt C, Megarbane A, Neveling K, Czeschik JC, Albrecht B, Callewaert B, von Deimling F, Hehr A, Falkenberg Smeland M, Konig R, Kuechler A, Marcelis C, Puiu M, Reardon W, Riise Stensland HM, Schweiger B, Steehouwer M, Teller C, Martin M, Rahmann S, Hehr U, Brunner HG, Ludecke HJ, Wieczorek D. Oto-facial syndrome and esophageal atresia, intellectual disability and zygomatic anomalies - expanding the phenotypes associated with EFTUD2 mutations. Orphanet J Rare Dis. 2013;8 doi: 10.1186/1750-1172-8-110. 110-1172-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weh E, Reis LM, Happ HC, Levin AV, Wheeler PG, David KL, Carney E, Angle B, Hauser N, Semina EV. Whole exome sequence analysis of Peters anomaly. Hum Genet. 2014;133:1497–1511. doi: 10.1007/s00439-014-1481-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss O, Kaufman R, Michaeli N, Inbal A. Abnormal vasculature interferes with optic fissure closure in lmo2 mutant zebrafish embryos. Dev Biol. 2012;369:191–198. doi: 10.1016/j.ydbio.2012.06.029. [DOI] [PubMed] [Google Scholar]
- Wenger TL, Harr M, Ricciardi S, Bhoj E, Santani A, Adam MP, Barnett SS, Ganetzky R, McDonald-McGinn DM, Battaglia D, Bigoni S, Selicorni A, Sorge G, Monica MD, Mari F, Andreucci E, Romano S, Cocchi G, Savasta S, Malbora B, Marangi G, Garavelli L, Zollino M, Zackai EH. CHARGE-like presentation, craniosynostosis and mild Mowat-Wilson Syndrome diagnosed by recognition of the distinctive facial gestalt in a cohort of 28 new cases. Am J Med Genet A. 2014;164A:2557–2566. doi: 10.1002/ajmg.a.36696. [DOI] [PubMed] [Google Scholar]
- Williamson KA, FitzPatrick DR. The genetic architecture of microphthalmia, anophthalmia and coloboma. Eur J Med Genet. 2014 doi: 10.1016/j.ejmg.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Williamson KA, Rainger J, Floyd JA, Ansari M, Meynert A, Aldridge KV, Rainger JK, Anderson CA, Moore AT, Hurles ME, Clarke A, van Heyningen V, Verloes A, Taylor MS, Wilkie AO, Fitzpatrick DR UK10K Consortium. Heterozygous loss-of-function mutations in YAP1 cause both isolated and syndromic optic fissure closure defects. Am J Hum Genet. 2014;94:295–302. doi: 10.1016/j.ajhg.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahrani F, Aldahmesh MA, Alshammari MJ, Al-Hazzaa SA, Alkuraya FS. Mutations in c12orf57 cause a syndromic form of colobomatous microphthalmia. Am J Hum Genet. 2013;92:387–391. doi: 10.1016/j.ajhg.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Baldini A. In vivo response to high-resolution variation of Tbx1 mRNA dosage. Hum Mol Genet. 2008;17:150–157. doi: 10.1093/hmg/ddm291. [DOI] [PubMed] [Google Scholar]
- Zhao C, Lu S, Zhou X, Zhang X, Zhao K, Larsson C. A novel locus (RP33) for autosomal dominant retinitis pigmentosa mapping to chromosomal region 2cen-q12.1. Hum Genet. 2006;119:617–623. doi: 10.1007/s00439-006-0168-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Molecular and phenotypic examinations of eftud2 mutant embryos. A. DNA chromatogram for zebrafish mutant allele; red line indicates a deletion of 8-nt in mutant DNA. B. RT-PCR analysis of eftud2 transcripts. Top: image of gel electrophoresis of eftud2- specific (lanes 1–6) and ef1a-specific (lanes 7–12) RT-PCR products obtained using mutant (1–3 and 7–9) and wild-type (4–6 and 10–12) total RNA; red arrow indicates a 425-bp eftud2 product, black arrowhead points to 205-bp ef1a-specific band; M corresponds to molecular weight marker. Please note a reduction in eftud2 level in 3 independent RNA samples (lanes 1–3) extracted from mutant embryos in comparison to wild-type (lanes 4–6). Bottom: graph representing quantification of eftud2 expression using ImageJ in mutant and wild-type embryos. C. Measurements of body length, head height and ocular diameters in eftud2 homozygous mutant and wild-type embryos. DV- dorsoventral; NT- nasotemporal. D–G. Histological analysis of eftud2 homozygous fish. Sections at eye level of 38-hpf wild-type (C, D) and eftud2 mutants (E, F) are shown. c-cornea, le- lens, r-retina; arrowheads indicate intensely stained particles likely corresponding to dead cell corpses. H–K. Alcian blue staining in eftud2 120-hpf heterozygous fish showing normal craniofacial cartilage structure. ch- ceratohyal cartilage, ep- ethmoid plate, m- Meckel’s cartilage, pq- palatoquadrate cartilage components of upper and lower jaws.
Known human MAC genes and their WES coverage in the affected patient.
Summary of rare and novel variants identified via trio analysis of the affected family.
Summary of novel/rare alleles identified in retinal dystrophy genes.