Abstract
Schlafen 3 (Slfn3) mediates rodent enterocyte differentiation in vitro and in vivo, required for intestinal function. Little is known about Schlafen protein structure-function relationships. To define the Slfn3 domain that promotes differentiation, we studied villin and sucrase isomaltase (SI) promoter activity in Slfn3-null human Caco-2BBE cells transfected with full-length rat Slfn3 DNA or truncated constructs. Confocal microscopy and Western blots showed that Slfn3 is predominantly cytosolic. Villin promoter activity, increased by wild type Slfn3, was further enhanced by adding a nuclear exclusion sequence, suggesting that Slfn3 does not affect transcription by direct nuclear action. We therefore sought to dissect the region in Slfn3 stimulating promoter activity. Since examination of the Slfn3 N-terminal region revealed sequences similar to both an aminopeptidase (App) and a divergent P-loop resembling those in NTPases, we initially divided Slfn3 into an N-terminal domain containing the App and P-loop regions, and a C-terminal region. Only the N-terminal construct stimulated promoter activity. Further truncation indicated that both the App and the smaller P-loop constructs enhanced promoter activity similarly to the N-terminal sequence. Point mutations within the N-terminal region (R128L, altering a critical active site residue in the App domain, and L212D, conserved in Schlafens but variable in P-loop proteins) did not affect activity. These results show that Slfn3 acts in the cytosol to trigger a secondary signal cascade that elicits differentiation marker expression and narrow the active domain to the third of the Slfn3 sequence homologous to P-loop NTPases, a first step in understanding its mechanism of action.
Keywords: Schlafen 3, differentiation, divergent P-loop NTPases, functional annotation, structure-function relationship
1.1. Introduction
Schlafen (Slfn) proteins were first described in the mouse where they were shown to be differentially expressed during thymocyte maturation and T-cell activation [1]. Although most studies have examined the role of Slfn proteins in the immune response [2–8], some Slfns may influence development, cell differentiation or the control of tumorigenesis [9–14]. Our laboratory has focused on Slfn3 and rat intestinal epithelial cell differentiation, which is important in healthy intestinal function for absorption and barrier function, as well as in disease processes such as short gut syndrome, infectious enteritis, and inflammatory bowel disease [15–17]. We have previously demonstrated that Slfn3 promotes differentiation in rat intestinal epithelial cells (IEC) in vitro and in vivo [18–20]. In vitro, Slfn3 expression in IEC-6 cells is upregulated in response to disparate differentiation stimuli; Slfn3-specific siRNA lowers basal and blocks stimulated activity of dipeptidyl dipeptidase 4 (Dpp4), a canonical marker of enterocytic differentiation [18]. In vivo, mid-intestinal Slfn3 expression increases with gestational age; it decreases in the adult rat jejunal epithelium when it becomes atrophic because of loss of interaction with luminal contents [9, 20]. Furthermore, instillation of a Slfn3 adenovirus in the intestinal lumen enhances epithelial mRNA and protein levels of enterocytic differentiation markers. Reducing endogenous Slfn3 with small interfering RNA conversely reduces epithelial expression of these markers [20].
Slfn family members, widely expressed in mammals, are classified into three groups according to sequence homology and molecular size as short (Slfn 1, 2), intermediate (Slfn 3, 4, 12) or long (Slfn 5, 8–11, 14) [1, 21]. These broad classifications are indicative of how little is known about the molecular functions of Schlafens. Intermediate and long Slfns share a highly conserved “SWADL” (Ser-Trp-Ala-Asp-Leu) domain of unknown function, while long Slfns are characterized by a C-terminal extension that contains sequence motifs homologous to members of the superfamily I of DNA/RNA helicases [4, 22, 23]. All Slfns contain a “Slfn box” adjacent to a divergent AAA domain, and previous researchers have suggested that Slfns may have ATP/GTP binding functions similar to the classical AAA superfamily of proteins [4, 24]. Identification of these putative functional domains indicates that Slfns may play a role in protein degradation and folding, vesicle transport and transcription (AAA domain) or DNA/RNA metabolism (helicase domain) [25].
Despite identification of broad commonalities, the mechanisms by which Slfns act remain largely unknown. Subcellular localization often provides clues to protein function. The short and intermediate Slfns 1, 2 and 4 localize chiefly in the cytosol, while the longer Slfns 5, 9 and 11, some of which contain a nuclear localization sequence, are predominantly nuclear [23]. Although these observations suggest that individual Slfns may have specific roles, they do not explain most published results. We and others have shown that some of the predominantly cytoplasmic intermediate and short Slfns (Slfns 3, 4 and 12, and Slfns 1 and 2, respectively) promote differentiation and/or inhibit cell proliferation, often by affecting promoter activity or the abundance of cell cycle proteins [11, 12, 22]. Conversely, although some long Slfns co-localize with an active form of RNA polymerase II, to date, none of the long Slfns have been reported to have direct nuclear action. Indeed, Slfn11 inhibits HIV protein synthesis by binding to tRNA [26], a cytoplasmic action.
The current study focuses in on the molecular function of Slfn3 by defining its closest homologs and testing the activity of the corresponding Slfn3 domains. Mouse Slfn4 had been shown to be located in the cytosol but the distribution of the highly homologous mSlfn3 had not been tested [23]. Therefore, we first confirmed that endogenous rSlfn3 is predominantly cytosolic in rat IEC-6 intestinal epithelial cells. Then we transfected human Caco-2 cells that do not express Slfn3 with a rat EGFP-Slfn3 plasmid to demonstrate that while the Slfn3 protein remained predominantly cytosolic in these cells, it nevertheless enhanced villin promoter activity, which reflects downstream activity associated with colon differentiation. Because a small amount of Slfn3 immunoreactivity appeared within the nucleus, we added a nuclear exclusion sequence to the Slfn3 protein to target it to the cytoplasm. Slfn3 still stimulated villin promoter activity when it was excluded from the nucleus via the nuclear exclusion sequence. Since the 3-dimensional and coarser domain structure of Slfn3 is unknown, we utilized structural homology modeling techniques to predict functional regions in the rSlfn3 protein from its sequence, and then tested these functions by site-directed and truncation mutagenesis followed by villin reporter assay. Our results support that Slfn3 acts in the cytosol to initiate a signaling cascade that affects nuclear promoter activity and differentiation marker expression. Furthermore, our analysis narrowed the segment of rSlfn3 responsible for stimulating promoter activity to a 175 residue region in the N-terminal half, matching the most compact known folded structure of a P-loop domain. Demonstrating cytoplasmic activity and P-loop homology for Slfn3 provide the first steps in defining its mechanism of action.
1.2. Methods
1.2.1. Cell culture
The rat non-transformed intestinal epithelial IEC-6 and the human Caco-2BBE cell lines were obtained from ATCC (Manassas, VA) and maintained in Dulbecco’s Modified Essential Eagle’s Medium supplemented with 10% FBS as described by us previously [18–20]. We used Caco-2 cells transfected with the full-length pEGFP-C1/Slfn3 plasmid or truncated/mutated constructs for the confocal cellular localization and luciferase activity studies described below.
1.2.2. Western blot
IEC-6 cells were lysed at confluence and cytosolic and nuclear fractions prepared using a kit from BioVison, Inc (Milpitas, CA) according to the manufacturer’s directions. Protein concentration was determined via BCA (Thermo Fisher, Rockford, IL). The proteins were then separated by SDS-Page electrophoresis, transferred to nitrocellulose membranes and exposed to antibodies to Slfn3 (Santa Cruz Biotechnology, Santa Cruz, CA) and cytosolic (Rho-GDI; EMD Millipore, Billerica, MA) and nuclear (lamin B1; Abcam, Cambridge, MA) markers. Detection was accomplished on an Odyssey Infrared Imaging System using Li-Cor Biosciences antibodies (Lincoln, NB). Densitometry was performed on a Kodak 440CF Image Station (Rochester, NY).
1.2.3. Rat Slfn3 (rSlfn3) Truncation/Point Mutation Design
1.2.3.a. Region of Aminopeptidase Homology
The N-terminal region (residues 1–324) in rSlfn3 was found to match two protein structural folds with different functions. First identified was a putative leucyl M17 cytosolic aminopeptidase from Staphylococcus aureus, in Protein Data Bank (PDB) entry 3kzw chain A; this structure was determined by Dr. Boguslaw Nocek of the Midwest Center for Structural Genomics. The rSlfn3-aminopeptidase similarity was detected by performing protein BLAST [27] against all sequences in the PDB [28]. The alignment of residues 69–302 in rSlfn3 to the aminopeptidase (Supplementary Fig A) corresponds to its complete catalytic domain except for a short C-terminal beta-strand. The structure of the leucyl aminopeptidase domain matched by rSlfn3 is shown in Supplementary Fig. B, with its catalytic site indicated by the three carboxylate side chains at center. By homology with other aminopeptidases, this cleft positions the peptidyl substrate with its amino terminal peptide bond adjacent to the carboxylates.
1.2.3.b. Mutant Design to Test Aminopeptidase Homology
To test whether rSlfn3 activity localizes to the N-terminal aminopeptidase-like (App) domain, a construct was designed to truncate the rSlfn3 sequence 15 residues after the App domain match (Supplementary Fig A), as shown in Fig. 3A. This truncation position allowed for the inclusion of the sequence corresponding to the beta-strand at the terminus of the catalytic domain in 3kzw, followed by several residues, to allow for typical C-terminal structural flexibility without affecting the integrity of the App domain. Secondly, a site-directed mutation was made to the conserved Arg128 in rSlfn3, which conserves charge and potential metal-binding activity with the critical aminopeptidase active-site metal ligand, Lys259. The Arg128Leu mutation is conservative in side chain shape and bulk, while losing the ability to bind metal. This mutation tests conservation of the catalytic site between Leu aminopeptidases and rSlfn3.
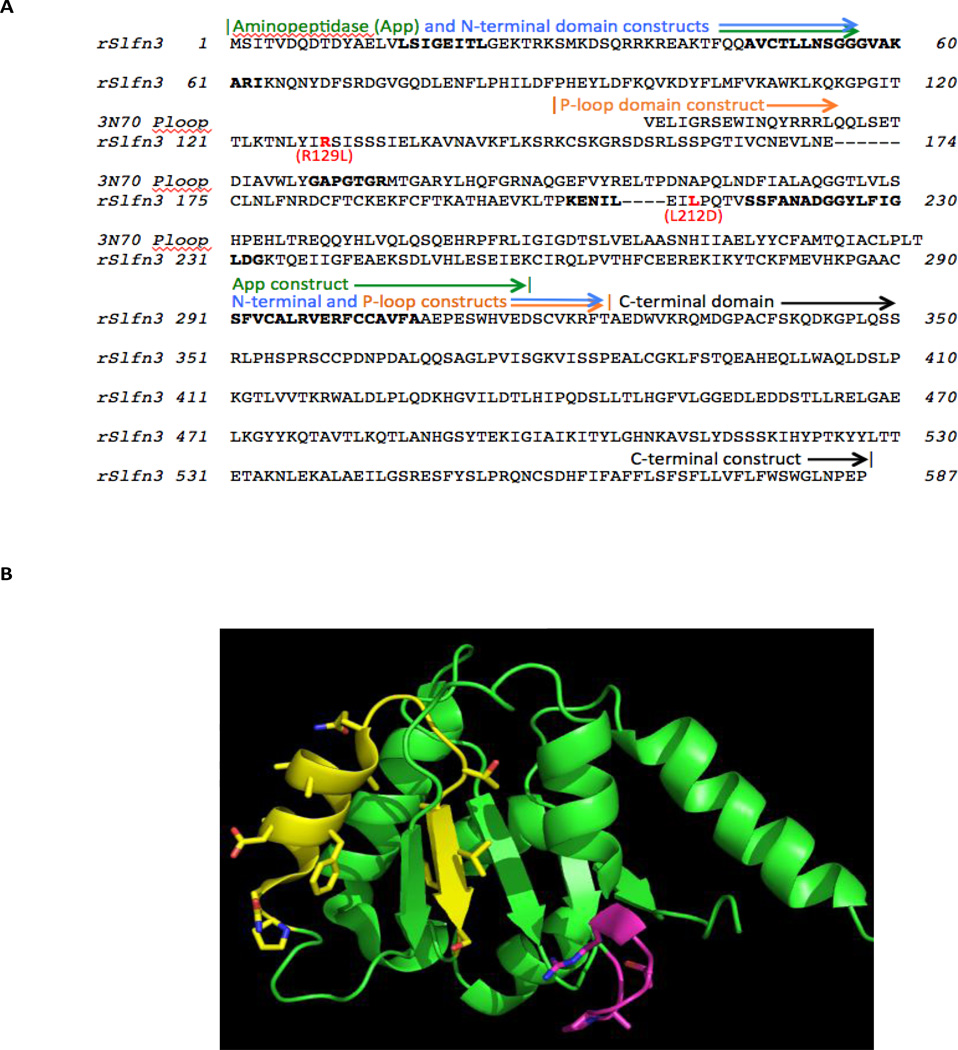
Figure 3.
BLAST-identified functional domain matches with rSlfn3, and corresponding constructs designed to test the correlation of Slfn3 function with maintenance of aminopeptidase-like (PDB entry 3kzw) versus AAA P-loop-like (PDB entry 3n70) function. (A) Sequence alignment, with boldfaced positions in the rSlfn3 sequence indicating the three most-conserved motifs in Schlafens (AVCTLLNSGGGVAKARI, SSFANADGGYLFIGLDG, and SFVCALRVERFCCAVFA). The position of the Walker A P-loop motif in the 3n70 sequence, GAPGTGR, appears in boldface, and two site-directed mutations in rSlfn3, Arg129Leu and Leu212Asp, are highlighted in red. (B) The main-chain fold of the P-loop domain crystal structure (PDB entry 3n70), corresponding to the BLAST Conserved Domain match in rSlfn3. The highest-homology region, PQLN…VLS, surrounding and including the second conserved Schlafen motif, is highlighted with side chains in yellow. The Walker A motif, which is not conserved in rSlfn3, consistent with its match to divergent AAA domains, is shown with side chains in magenta. The figure was rendered with PyMOL (Schrödinger LLC, NY).
1.2.3.c. AAA P-loop Domain Homology
When searching the larger non-redundant protein sequence database, BLAST identified a Conserved Domain match for rSlfn3 [29], corresponding to the AAA_4 or divergent AAA domain superfamily (Pfam04326). The expectation value for this sequence match, shown in Supplementary Fig. C, was 2.75 × 10−08, indicating a very low probability of occurring at random. The AAA domain match extends from residue 199 to 302 in rSlfn3 (sequence EVK…ERFC), beginning halfway into the putative App domain match with rSlfn3, and ending 19 residues after the App domain (Fig 3A). Another tool, InterProScan [30], also identified residues 203–305 in rSlfn3 as matching the AAA+ superfamily of ring-shaped P-loop NTPases, with a similar expectation value of 4.7 X e−08. The shortest stably folded example of a P-loop domain in the Protein Data Bank is entry 3N70. This 3-dimensional structure provides a context for assessing the P-loop region of homology with rSlfn3 and designing mutants (Fig 3B).
1.2.3.d. Mutations Testing Activity of the P-loop Region of rSfln3
A P-loop domain mutant was designed for rSlfn3 based on N-terminal and C-terminal truncation, as shown in Fig 3A, incorporating the region of highest conservation with divergent AAA P-loops. This construct focused on the correspondence with the P-loop crystal structure (PDB entry 3n70), since it folds stably without additional sequence. For the P-loop domain construct, several residues in rSlfn3 were included N- and C-terminal to the region of 3n70 alignment, to avoid domain destabilization from flexible termini. This construct removes 81 residues from the N-terminal region of rSlfn3 predicted as homologous to the aminopeptidase (starting at residue 69: DFS..FLKSR), and thus also effectively tests whether the P-loop rather than the aminopeptidase domain conserves villin promoter activity.
Finally, a site-directed mutation was made in the Leu212 position, which is highly conserved in Schlafens and immediately precedes the motif most highly conserved between Schlafens and P-loop domains (PQTVSS…). The Leu212Asp mutation is isosteric but greatly changes the chemical character of this residue. P-loop domains tolerate hydrophobic, polar and charged substitution in this position, so Leu212Asp would be expected to maintain activity if rSlfn3 acts as a P-loop, and to lose function if rSlfn3 conservation of Leu212 were required for an alternative function.
1.2.4. Plasmid Construction
We used the pEGFP-N1/schlafen3 construct described in Patel et al [10] as a template to generate our constructs inserted into the pEGFP-C1 expression vector from Clontech (Mountain View, Ca). The N-terminal, C-terminal, App and P-loop constructs were amplified with tailed PCR from the original pEGFP-C1/schlafen3 using specific primers for the BamHI and HindII cloning sites. For the NES signal, the leucine-rich amino acid stretch (ALLKKLVVLLLDE) that represents residues 32–44 of MEK1 was introduced in duplicate into the expression vector pEGFP-C1 (Clontech) between BamHI and XbalI sites; the EGFP-Slfn3 or N-terminal-EGFP-Slfn3 was then inserted so that the NES was located at the C-terminal end of each. The point mutations, 128 R to L and 212 L to D, were accomplished using the Quick Change II XL Site-Directed Mutagenesis kit from Agilent Technologies (Santa Clara, CA). All plasmids were purified via MiniPrep (QIAGEN, Valencia, CA)) prior to sequencing at the MSU Core Facility. After sequence confirmation, the plasmids were amplified and prepared for transient expression using the QIAGEN MaxiPrep kit.
1.2.5. Fluorescence Confocal Microscopy
IEC-6 and Caco-2BBE cells (1.0 × 105) cultured on poly-D-lysine 4-well culture chambers were transfected with 2.0 µg of the relevant construct (pEGFP-Slfn3-C1, pEGFP-Slfn3-N1, pEGFP-Slfn3-N-terminus (1-1049)-C1 or pEGFP-Slfn3-NES-C1) using either Fugene (Promega, Madison, WI) or Lipofectamine (Invitrogen, Life Technologies, Grand Island, NY) according to the manufacturers’ directions. Forty eight hours after transfection, cells were fixed with formalin-free IHC Zinc fixative (PharmMingen, BD Biosciences, Sand Diego, CA). Endogenous Slfn3 in IEC-6 cells was detected using a primary Slfn3 antibody from Santa Cruz Biotechnology, and fluorescent second antibodies (FITC or Alexa 467 (Abcam)). Cells were incubated with 4’,6-diamidino-2-phenylindole (DAPI, Zyagen, San Diego, CA) to stain nuclei, and fluorescent images were captured using an Olympus FluoView FV1000 Confocal Laser Scanning Microscope and densitometry quantitated with Olympus Advanced Software (Olympus America, Inc, Center Valley, PA. Images were processed using Photoshop software (Adobe, San Jose, CA) without affecting the linearity of the images.
1.2.6. Luciferase reporter assay
Caco-2BBE cells (1.0 × 104) were seeded in 96-well (0.32 cm2) plates, and co-transfected the next day with 150 ng/well of the empty vector (pEGFP-C1, Clontech Laboratories, Inc., Mountain View, CA) or rat schlafen3 DNA constructs (pEGFP-Slfn3, pEGFP-Slfn3-NES, pEGFP-Slfn3-N terminus, pEGFP-Slfn3-C terminus, pEGFP-Sfn3-App, pEGFP-Slfn3-Ploop, pEGFP-Slfn3-Mutant 1(R128L) or pEGFP-Slfn3-Mutant 2 (L212D) with 50.0 ng/well of the human Villin (Vil1) or Sucrase Isomaltase (SI) LightSwitch promoter reporter goclone (Switchgear Genomics, Carlsbad, CA). Thus, cells in each well received a total of 150 ng of DNA with SLFN3 and Vil1 or SI promoter reporter constructs at a 3:1 ratio. The DNA was mixed with Opti-MEM medium (10 µl/well) for 5 min, and Fugene (Switchgear Genomics, Carlsbad, CA) was added (3.2 µl/1.0 µg DNA). In a separate experiment, Caco-2 cells were exposed to 0.1 ng/ml TGF-β or 5 mM sodium butyrate before addition of the Vil1 reporter. The mixture was incubated at room temperature for 30 minutes and added to the cells. Forty eight hours after transfection, the experiment was terminated by addition of luciferase as per the manufacturer’s recommendations. Briefly, 100ul/well (buffer + substrate) LightSwitch Luciferase assay reagent (Switchgear Genomics) was added and incubated for 30 minute at room temperature in the dark. IEC-6 cells were co-transfected similarly with rat villin or SI promoter reporters (Switchgear Genomics) but Lipofectamine (Invitrogen) was used as the transfection reagent and the experiment was terminated after 24 hours. Relative Luminescence (RLU) was measured using a VERITAS Microplate Luminometer (Turner Biosystems, Sunnyvale, CA).
1.2.7. Statistical Analysis
Densitometric results for cytosolic/nuclear protein expression were normalized to their respective controls and significance determined by Students’ T-test. Results of the luciferase reporter assays were normalized to the basal promoter activity, villin or SI, because of variations in the RLU readings between assays. Significance of the results, presented as X±SE, was ascertained by one-way ANOVA with Pairwise Multiple Comparisons (Holm-Sidak Method).
1.3. Results
1.3.1. Cytosolic Slfn3 enhances villin promoter activity
1.3.1.a. Cellular distribution of exogenously expressed Slfn3 mimics that of the endogenous molecule in IEC-6 cells
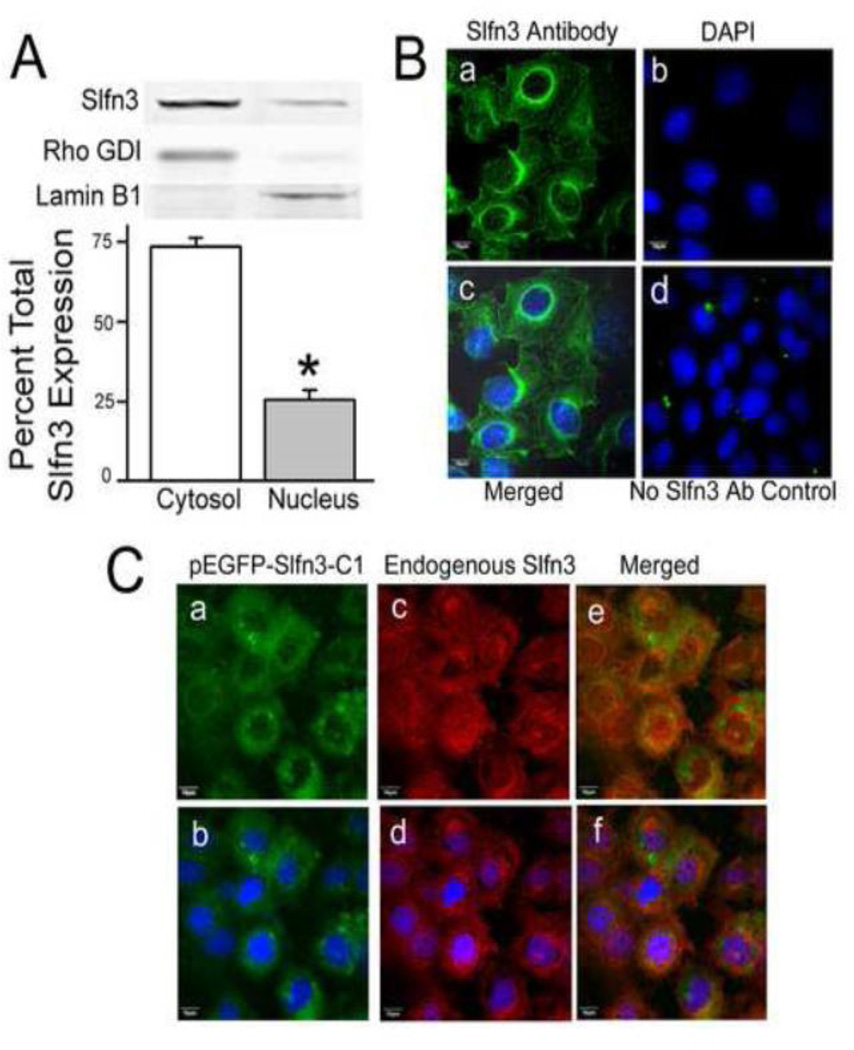
Since Caco-2 cells do not normally express Slfn3 and addition of the EGFP tag could conceivably alter cellular Slfn3 distribution, we first assessed endogenous Slfn3 protein distribution in rat intestinal epithelial IEC-6 cells by Western blot. In IEC-6 cells, Slfn3 was predominantly cytosolic (73.5 ± 2.9% of total), with much less Slfn3 in the nuclear fraction (25.6 ± 2.9%, n=8). Successful fractionation was validated by demonstrating enrichment of Rho-GDI in the cytoplasmic fraction and Lamin B1 in the nuclear fraction (Fig 1A). We confirmed this distribution by immunofluorescent staining, using antibody to Slfn3 and a FITC-tagged secondary antibody (Fig 1B). Visible nuclear staining (Fig 1B part a) clearly co-localized with the DAPI nuclear stain (Fig 1B part c). Fluorescence analysis of 30 cells indicated that 73 ± 11% of the total immunoreactive signal for Slfn3 signal is found in the cytosol while 27 ± 2% resides in the nucleus, nearly identical to the results obtained with cell fractionation.
Figure 1. Predominantly cytosolic and modest nuclear localization of exogenously expressed Slfn3 mimics that of the endogenous molecule in IEC-6 cells.
(A) Slfn3 protein abundance is greatest in the cytosol in fractionated rat IEC-6. Bars represent densitometric analysis as a ratio of the total immunoreactivity (*p<0.05, n=8). A representative gel is shown above; Rho GDI and Lamin B were used as cytosolic and nuclear markers. (B) Confocal images confirm that endogenous immunoreactive Slfn3 is found in the nucleus but is predominantly localized to the cytosol: (a) Slfn3 primary antibody/FITC secondary antibody, (b) DAPI nuclear stain, (c) merged image, and (d) no primary antibody control with DAPI stain. Representative of 8 similar images. (C) (a) Exogenously expressed Slfn3 (pEGFP-Slfn3-C1) and (b) endogenous Slfn3 co-localize in IEC-6 cell (c). Cytosolic/nuclear expression is further delineated with nuclear DAPI staining (b, d, f). Representative of 6 similar.
We then determined whether exogenous p-EGFP-Slfn3 distribution parallels that of endogenous Slfn3 in IEC-6 cells. Since the distribution of each of our wild-type, full length constructs, pEGFP-Slfn3-C1 and pEGFP-Slfn3-N1, was similar regardless of the position of the GFP tag (Supplementary Fig D), we chose the N-terminal tagged pEGFP-Slfn3-C1 for all subsequent studies. Fig 1C illustrates the results of co-localization experiments. In the transfected cells, exogenous pEGFP-Slfn3 was most abundant in the cytosol but was also found in the nucleus (Fig 1C part a); immunoreactive endogenous Slfn3 showed a similar pattern (Fig 1C part c). The Slfn3 antibody reacts with both the endogenous and the exogenously expressed Slfn3 resulting in a stronger signal where they overlap in the transfected cells. Merging of the images (Fig 1C part e) and the addition of DAPI (Fig 1C parts b, d, and f) further support this co-localization.
1.3.1.b. Exogenously expressed Slfn3 enhances differentiation marker expression in Slfn3-null Caco-2 cells
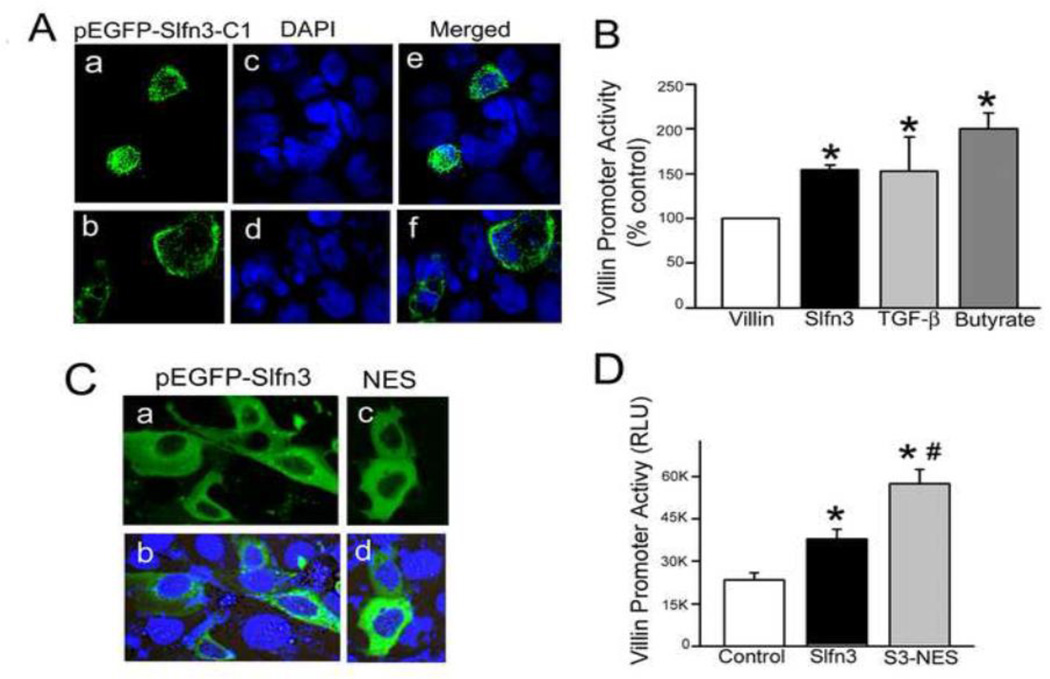
Cellular localization of the rat pEGFP-Slfn3-C1 product in human Caco-2 cells was similar to both endogenous and exogenous Slfn3 expression in rat IEC-6 cells: mostly cytosolic with a small amount of nuclear expression (Fig 2A). Two representative fields are shown because of low transfection efficiency (pEGFP (a, b); nuclear DAPI (c, d)). Merged images (e, f) highlight the localization of the expressed product. To verify that transfected Caco-2 cells are an appropriate model, we measured villin and SI promoter activity in both cell lines. Non-normalized RLU’s (Table 1) demonstrated several fold higher basal activity of the human villin promoter in Caco-2 than of the rat promoter in IEC-6 cells, but the response to exogenous expression of Slfn3 was of similar magnitude in each cell type. The differences in baseline promoter activity most likely reflect the nature of the reporter constructs themselves, which necessarily vary between rat IEC-6 cells and human Caco-2 cells, as well as differences in the transfection efficiency achieved in each cell line. To further validate the use of Caco-2 cells to study Slfn3 biology, we compared villin promoter activity in response to exogenous Slfn3 to the response of villin promoter activity to two known differentiation factors, TGF-β and sodium butyrate that we have previously studied in intestinal epithelial and other cells [31–36]. Responses to these disparate stimuli were relatively similar (Fig 2B), confirming that exogenous Slfn3 expression promotes a physiologically relevant response in the pEGFP-Slfn3-plasmid transfected Caco-2 cells. In a separate study, we infected Caco-2 cells with an adenovirus coding for Slfn3 and GFP or a control adenovirus coding only for GFP, each previously described, and confirmed that there was no difference in cell number between the two cell populations at 24 hours (data not shown), consistent with our previous observation in rat jejunal epithelium in vivo [20], and with the previous observation that knockdown of Slfn3 does not prevent the mitogenic effects of repetitive deformation or EGF in IEC-6 cells in vitro [18]. Although we did not study this further specifically here as beyond the scope of the current investigation, Caco-2 cells also respond to repetitive deformation and EGF similarly to IEC-6 cells [37].
Figure 2. Exogenously expressed Slfn3 stimulates villin promoter activity from a cytosolic location in Slfn3-null Caco-2 cells.
(A) Two representative confocal micrographs of pEGFP-Slfn3-C1 expressed in Caco-2 cells demonstrate strong cytosolic and minor nuclear expression (a, b). DAPI-stained nuclei reflect the number of non-transfected cells (c, d), Merged images (e, f) confirm that the pEGFP-Slfn3-C1 construct is expressed in Caco-2 as it is in IEC-6 cells. (B) Exogenous Slfn3 enhances villin promoter activity similarly to TGF-β and sodium butyrate (*p<0.05; n=3–6). (C) Cellular localization of GFP-tagged wild-type Slfn3 (pEGFP-Slfn3) transfected into Slfn3-null Caco-2 cells (a, b) and cytosolic localization with the addition of a nuclear exclusion sequence (pEGFP-Slfn3-NES; c, d) is shown in these representative confocal micrographs. (D) Villin promoter activity is greater in Caco-2 cells transfected with the Slfn3-NES construct (S3-NES) than with the full-length construct (Slfn3) (*p<0.05 vs control, #p<0.05 vs full length by one way ANOVA, n=4).
Table 1.
Villin and Sucrase Isomaltase (SI) promoter activity in IEC-6 vs. Caco-2 cells.
| IEC-6 Cells | Caco-2 cells | |
|---|---|---|
| Villin Promoter | ||
| Basal villin | 1115±93 | 15874±2394 |
| Slfn3 | 1944±56* | 28489±5001* |
| N-terminal Slfn3 | 2178±82** | 42180±9395** |
| SI Promoter | ||
| Basal SI | 1065±69 | 3060±198 |
| Slfn3 | 2022±75* | 4227±143* |
| N-terminal Slfn3 | 1844±66* | 4166±196* |
p<0.05 vs Basal
p<0.05 vs Slfn3
1.3.1.c. Slfn3 enhances villin promoter activity from a cytosolic location
To further determine whether cytosolic Slfn3 maintained full activity, we attached a nuclear exclusion sequence (NES) to our Slfn3 plasmid, monitored its localization by confocal microscopy and measured its effect on villin promoter activity in human Slfn3-null Caco-2 cells. As shown in Fig 2C, pEGFP-Slfn3 fluorescence was mostly confined to the cytosol, with slight nuclear immunofluorescence (a, b). No nuclear fluorescence could be discerned in the EGFP-Slfn3-NES cells (c, d). Villin promoter activity (Fig 2D) was significantly elevated in the Slfn3-transfected cells and greatest in the Slfn3-NES expressing cells (p<0.05 by one-way ANOVA; n=4), indicating that Slfn3 initiates its effects from its cytosolic location and not by direct nuclear action.
1.3.2. rSlfn3 N-terminal homology
1.3.2.a. Aminopeptidase
The aminopeptidase functional annotation is based on the complete preservation of the C-terminal catalytic domain between 3kzw and known leucyl aminopeptidases and their structures, such as Protein Data Bank entry 3kqx from Plasmodium falciparum (personal communication, Dr. Boguslaw Nocek, Argonne National Laboratory). This 293-residue match to the rSlfn3 sequence, with 23% sequence identity, is significant (only 1% chance of occurring at random) and close to the threshold that guarantees 3-dimensional structural homology (24.8% identity over more than 80 residues [38]). Based on this homology, the active site residues in 3kzw include Asp264, Asp341, and Glu343, of which the latter is conserved with Glu206 in the KENIL motif of rSlfn3 (Fig. 3A). The partial conservation of catalytic residues suggests that rSlfn3 could bind an N-terminal peptide similarly to the aminopeptidase while not sharing its peptide bond cleavage function.
1.3.2.b. Conserved Motifs in Schlafens and the P-loop Domain
According to ConSurf analysis [39] of hundreds of sequences with 30–80% sequence identity to rSlfn3, the three most highly conserved motifs in the N-terminal half of Schlafens begin at residue 47 in rSlfn3: AvCTLLNSGGGVAKARI, residue 217: SSFANAdGGYlFiGLDG, and residue 291: SFVCALrVerFCCaVFA, where the lower case letters represent moderately conserved positions. The region of significant match between rSlfn3 and the sequence motifs shared by divergent AAA P-loop proteins including 3n70 begins before the second motif, at rSlfn3 residue 199: EVK… The region of strongest P-loop match starts five residues prior to the second conserved Schlafen motif, at residue 212, and includes LPQ…FIG. The P-loop homology continues through the end of the third motif, where the similarity ends at …SCVK, residue 331 (Fig 3A). The ATP-binding Walker A motif GAPGTGR near the N-terminus of the P-loop domain (Fig 3B) is not conserved with rSlfn3, due to its match to divergent rather than canonical AAA P-loop domains. Together, these results suggest that this region in rSlfn3 folds like a P-loop domain but would not be expected to bind or hydrolyze ATP in the P-loop region.
1.3.2.c. The N-terminal segment of Slfn3 is sufficient for villin and SI expression
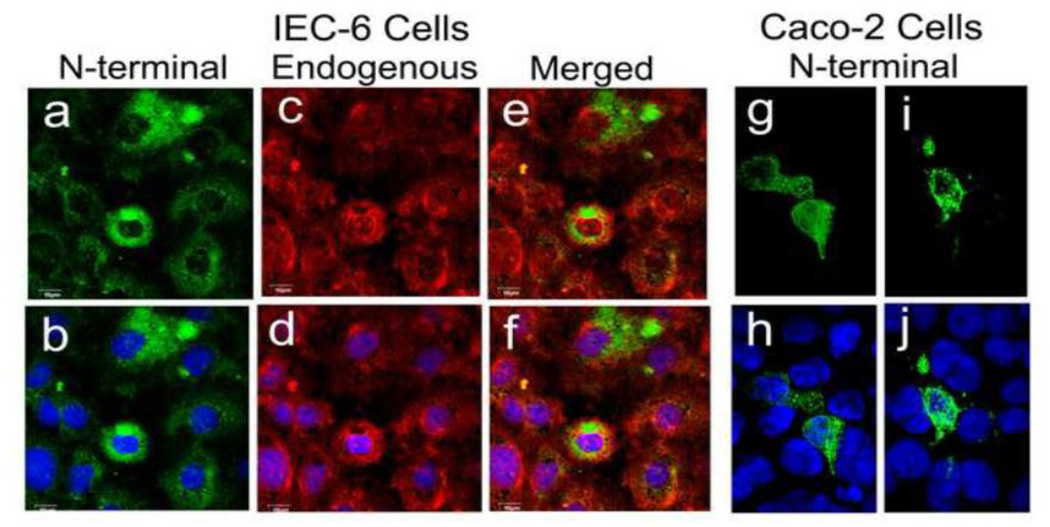
We therefore sought to determine whether the N-terminal segment of the Slfn3 molecule that exhibits greatest homology to both the App and P-loop structures promotes its differentiating effects, by designing the truncations and mutations described in section 1.2.4 and illustrated in Fig 3. In these studies, we again utilized human Caco-2 intestinal epithelial cells as a Slfn3-null cell. We initially ascertained that cellular distribution of the expressed N-terminal construct in the Caco-2 cells was the same as in the IEC-6 cells by confocal microscopy. Fluorescence was detected in both the nuclear and cytosolic compartments of each cell type, again with most of the expression in the cytosol (Fig 4). In the IEC-6 cells, the exogenously expressed N-terminal product (Fig 4 part a) co-localized with endogenously expressed Sfln3 (e). Two representative images of transfected Caco-2 cells also demonstrate predominant cytosolic staining (Fig 4 parts g, i) and minor nuclear expression (Fig 4 parts h, j).
Figure 4. Exogenously expressed Slfn3 N-terminal domain localizes predominantly to the cytoplasm in IEC-6 and Slfn3-null Caco-2 cells.
Confocal localization of N-terminal Slfn3 (a) closely matches that of the endogenously expressed Slfn3 (c) as shown in the merged image (e). Each image is shown with DAPI stain for clarity (b, d, f). Similar N-terminal localization in transfected Caco-2 cells (g, i) is confirmed by DAPI (h, i). IEC-6: representative of four similar; Caco-2: two representative images of four.
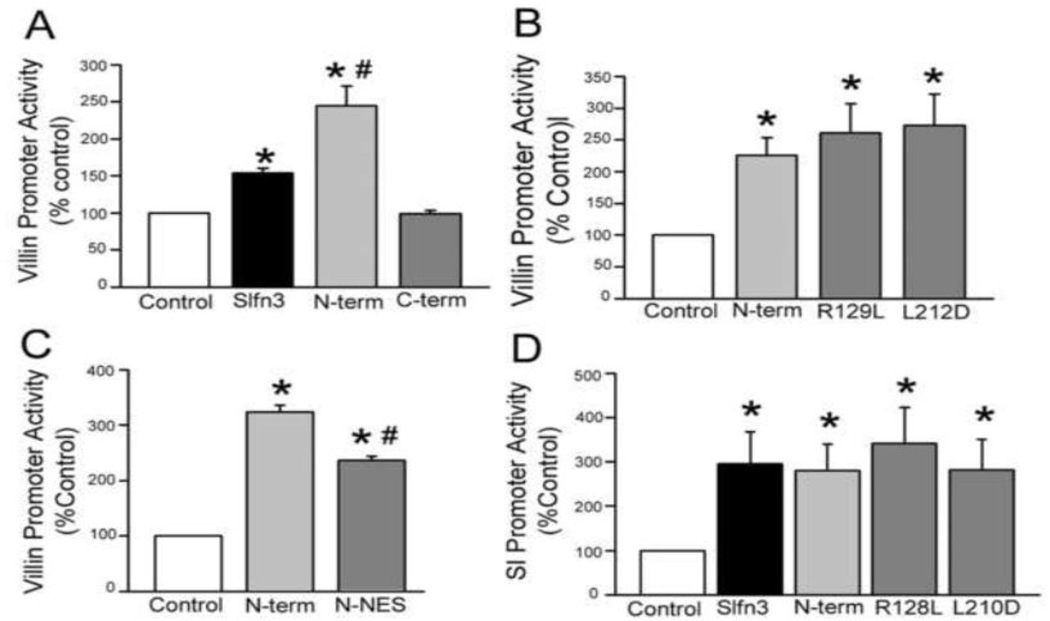
Villin promoter activity was significantly enhanced by co-transfection with the full Slfn3 construct and to an even greater extent by transfection with the N-terminal portion of the molecule (p<0.05 by one-way ANOVA; n=4). In contrast, luciferase activity in cells transfected with the C-terminal segment was not elevated above basal levels (Fig 5A). However, point mutation of an N-terminal domain residue required for App activity (Arg129 to Leu) and a second position conserved across Schlafen proteins but tolerant of mutation in other P-loop proteins (Leu212 to Asp) did not disrupt villin promoter activity (Fig 5B), supporting P-loop rather than App-like function. We then investigated whether cytosolic retention of the N-terminal construct was important to its activity. Villin expression was significantly elevated after co-transfection with the N-terminal-NES construct, albeit to a slightly lesser extent than that of the N-terminal construct alone (Fig 5C). We confirmed these results using a promoter for sucrase-isomaltase (SI), another marker of enterocytic differentiation. As in our observations with the villin promoter, increased SI promoter activity was observed after transfection of either wild type Slfn3 or the N-terminal segment, and the two point mutations had no effect (Fig 5D).
Figure 5. Villin and sucrase isomaltase (SI) promoter stimulatory activity resides in the N-terminal portion of Slfn3.
(A) Co-transfection of the Slfn3 N-terminal domain (N-term) has a greater effect on villin promoter activity than the full Slfn3 construct (*p<0.05 vs control, #p<0.05 vs the full-length construct); the C-terminal region (C-term) has no effect. (B) Point mutation of N-terminal domain residues (A129L and L212D) does not reduce villin promoter activity when compared to the N-terminal (N-term) construct (*p<0.05 vs control). (C) Cytosolic retention of the N-terminal construct (Slfn3-NES) enhances villin promoter activity but to a lesser degree than the N-terminal construct itself (*p<0.05 vs control, #p<0.05 vs the N-terminal construct). (D) SI promoter activity is increased by co-transfection of the full-length (Slfn3) and N-terminal (N-term) Slfn3 constructs and is unchanged by the point mutations (*p<0.05 vs control). (One way ANOVA and n=4 for each of the above).
1.3.4 The P-loop segment, totally contained within the N-terminal and App (aminopeptidase-like) domains of Slfn3, stimulates both villin and SI expression
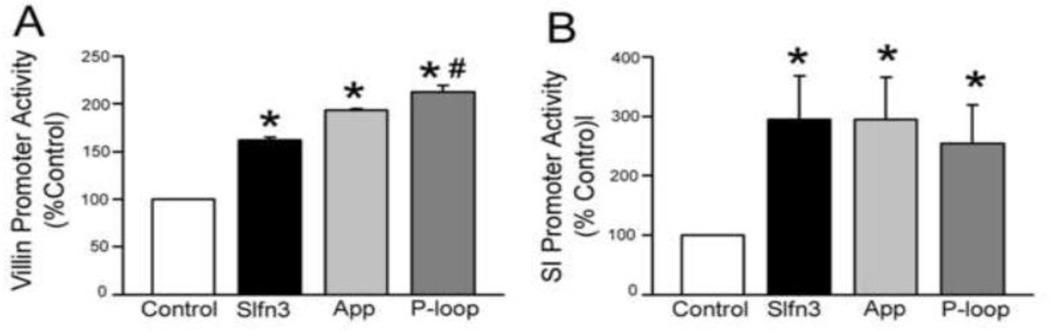
Analysis of the N terminal half of Slfn3 identified two potential functions for this region based on homology to an aminopeptidase, and in a more compact region, to a divergent AAA P-loop domain (Fig 3A). We therefore further truncated the N-terminal domain to create two smaller proteins corresponding to each of these motifs. Transfection with either the App segment or the smaller P-loop domain each significantly increased villin promoter activity (Fig 6A). Analysis by ANOVA revealed that the P-loop segment exhibited significantly greater activity than even the full-length Slfn3 construct (p<0.05, n=3). SI promoter activity was also significantly enhanced by each construct, but all appeared to be equally effective (Fig 6B). The results suggest that the P-loop domain is responsible for the villin and SI promoter stimulatory effect of the larger N-terminal and App domains.
Figure 6. The P-loop, the smallest segment within the N-terminal domain, increases both villin and SI promoter expression.
(A)Transfection with either the aminopeptidase (App) or the smaller P-loop domain each enhances villin promoter activity. The P-loop effect is greater than that of the full-length Slfn3 construct (*p<0.05 vs control, #p<0.05 vs full-length Slfn3 by one way ANOVA, n=3). (B) SI promoter activity is equally increased by the full-length Slfn3, App and P-loop constructs (*p<0.05 vs control, n=3).
1.4. Discussion
The data presented above indicate that, although Slfn3 resides mostly in the cytoplasm, it engenders nuclear effects including villin and SI promoter transcriptional activation. This suggests that Slfn3 acts via one or more intermediate proteins, initiating a signaling cascade in the cytosol that ultimately terminates in the nucleus. It has been suggested that Slfns may act within the nucleus as DNA/RNA helicases as they possess a highly conserved sequence (CDD:COG2865) with similarity to putative transcriptional regulators [4]. Indeed, others have demonstrated that Slfn1, a short Slfn also found in the cytosol, is transported to the nucleus after binding to the Hsp40 chaperone protein DnaJB6 where it precipitates cell cycle arrest [40]. However, our results with the EGFP-Slfn3-NES construct argue against a direct action for Slfn3 within the nucleus.
Localization of the domains responsible for specific protein activities can be obtained by manipulating either large sections of the protein by truncation [41] or by point mutation of residues likely to be critical for function. For example, the N-terminal region of p97/VCP, a ubiquitous AAA ATPase, was shown to be required for ATPase activity, while the C-terminal domain proved to be necessary for oligomer stability [42]. The only such study done in Slfns reported that a Slfn1 mutant lacking 27 N-terminal amino acids abrogated its growth inhibitory properties when overexpressed in NIH 3T3 cells [4]. The same authors described the gene organization of the long Slfns and ascribed putative DNA/RNA helicase function to the extended C-terminal domain, lacking in short and intermediate Slfns, on the basis of recurrent motifs with structural homology to known Superfamily I helicases [4]. Slfns have been shown to have regions of homology with AAA family ATPases [4, 22, 23], and here we show this is the case for an N-terminal region in rSlfn3. The AAA superfamily includes a broad range of functions: DNA and RNA helicases (some of which are cytosolic in action), DNA clamp loaders, DNA replication initiators, protein remodelers including proteasomal subunits, and Mg2+ chelatases [43]. Some act as molecular switches and others are processive machines. AAA and AAA+ domains both contain an N-terminal P-loop or Rossmann fold associated with ATP binding and hydrolysis, plus a C-terminal alpha-helical domain. AAA+ family members also share a conserved “second region of homology” (SRH) in the C-terminal domain [44]; this SRH was not matched in rSlfn3.
AAA ATPases belong to the larger P-loop NTPase family characterized by ATP binding domains that contain Walker A and Walker B motifs that bind ATP/GTP [45]. The divergent AAA superfamily members are overall similar to those with the canonical P-loop fold, while showing sequence divergence in the Walker ATP binding motifs including the P-loop itself. AAA ATPases form oligomeric assemblies with a number of factors and exert their effects through energy-dependent conformational changes [24]. Although rSlfn3 does not contain canonical Walker motifs and Slfns have not been shown to hydrolyze ATP, structural homology led us to focus on the N-terminal segment of rSlfn3 as a possible binding site for protein partners. Further search revealed a match between rSlfn3 and the P-loop domain of PDB entry 3n70. Full maintenance of downstream promoter activity upon critical mutations removing half of the potential aminopeptidase domain and one of its critical active-site residues supports that aminopeptidase activity is unlikely for rSlfn3. This is consistent with no observation of enzymatic function for Schlafens in the literature. Since the smaller P-loop construct was fully active, the N-terminal sequence upstream of the P-loop sequence is unlikely to be involved in rSlfn3 stimulation of promoter activity. Co-transfection with a construct containing the 175-residue 3n70 P-loop segment was as effective as the entire Slfn3 construct in stimulating both villin and SI promoter activity.
While villin promoter activity in response to transfection with the N-terminal construct was significantly greater than that to the full-length Slfn3, no such difference was seen with the SI promoter. These results were replicated in the IEC-6 cells where SI activation also appeared similar between the N-terminal and full-length constructs (Table 1), suggesting that the effect is not cell-line specific. Although they contain some common response elements, organization of the upstream regulatory region of villin and sucrase-isomaltase differs, reflecting the complex control of their expression [46–49]. For example, nutrient availability plays a greater role in SI expression [50]. Thus, it is possible that the downstream signal generated by the full-length and N-terminal Slfn sequences may bind to different response elements in each promoter sequence. More specifically, the C-terminal region of Slfn3 may have an inhibitory effect on the villin promoter that is absent in the sucrase-isomaltase promoter.
Rodents are known to express two intermediate Slfns, Slfn3 and Slfn4 [1, 21]. Although highly homologous, mouse Slfn3 and Slfn4 appear to be differentially regulated in hematopoietic cells [4–6], suggesting that they may play different roles in the differentiation and maturation of these cells. In the rat, there is an extension with an alternate start site at the 5’ end of the Slfn4 coding sequence and a small alignment gap that was used in the design of the present Slfn3 construct but the two sequences appear to yield a nearly identical product. Whether Slfn4 differs in function in the rat remains unknown. Indeed, there is little information on how intermediate and other Slfns carry out their actions. Our findings that a specific N-terminal domain is required for Slfn3 stimulation of differentiation in intestinal epithelial cells may have broader implications. If a similar mode of action applies to other Slfns, it may shed light on Slfn modulation of leukocyte maturation and of growth and differentiation in other tissues.
In summary, although the 3-D structure of Slfn3 has not yet been determined, structural and functional homology predictions can be used to design constructs to test stable structural and functional domains, with the eventual goal of identifying binding partners that transduce specific Slfn-mediated signals. The data presented above point to the P-loop domain as able to stimulate villin and SI expression, and thus a promising candidate to examine further as a partner in molecular signaling interactions, an important step towards understanding Slfn mechanisms of action.
Supplementary Material
Highlights.
Schlafen 3 is shown to localize predominantly to the cytosol
Schlafen 3 elicits rat intestinal epithelial cell differentiation marker expression
Schlafen 3 expression also stimulates rat intestinal epithelial cell differentiation in vivo
The N-terminal sequence shares homology with the P-loop superfamily of NTPases
A rSlfn3 construct containing only the P-loop domain maintains promoter activity
Acknowledgements
We wish to thank Dr. Boguslaw Nocek, at the Midwest Center for Structural Genomics, Argonne National Laboratory, for his helpful discussion of the structure of the App domain, Lisi Yuan for technical assistance and Venkat Katkoori for help with immunohistochemistry.
This work was supported by an NIH/PHS grant (1R01DK096137) to Marc D. Basson.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Schwarz DA, Katayama CD, Hedrick SM. Schlafen, a new family of growth regulatory genes that affect thymocyte development. Immunity. 1998;9(5):657–668. doi: 10.1016/s1074-7613(00)80663-9. PMID: 9846487. [DOI] [PubMed] [Google Scholar]
- 2.Berger M, Krebs P, Crozat K, Li X, Croker BA, Siggs OM, Popkin D, Du X, Lawson BR, Theofilopoulos AN, Xia Y, Khovananth K, Moresco EM, Satoh T, Takeuchi O, Akira S, Beutler B. An Slfn2 mutation causes lymphoid and myeloid immunodeficiency due to loss of immune cell quiescence. Nat Immunol. 2010;11(4):335–343. doi: 10.1038/ni.1847. PMID: 20190759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Condamine T, Le Luduec JB, Chiffoleau E, Beriou G, Louvet C, Heslan M, Tilly G, Cuturi MC. Characterization of Schlafen-3 expression in effector and regulatory T cells. J Leukoc Biol. 2010;87(3):451–456. doi: 10.1189/jlb.0609410. PMID: 15351786. [DOI] [PubMed] [Google Scholar]
- 4.Geserick P, Kaiser F, Klemm U, Kaufmann SH, Zerrahn J. Modulation of T cell development and activation by novel members of the Schlafen (slfn) gene family harbouring an RNA helicase-like motif. Int Immunol. 2004;16(10):1535–1548. doi: 10.1093/intimm/dxh155. PMID: 15351786. [DOI] [PubMed] [Google Scholar]
- 5.van Zuylen WJ, Garceau V, Idris A, Schroder K, Irvine KM, Lattin JE, Ovchinnikov DA, Perkins AC, Cook AD, Hamilton JA, Hertzog PJ, Stacey KJ, Kellie S, Hume DA, Sweet MJ. Macrophage activation and differentiation signals regulate schlafen-4 gene expression: evidence for Schlafen-4 as a modulator of myelopoiesis. PLoS One. 2011;6(1):e15723. doi: 10.1371/journal.pone.0015723. PMID: 21249125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mavrommatis E, Fish EN, Platanias LC. The schlafen family of proteins and their regulation by interferons. J Interferon Cytokine Res. 2013;33(4):206–210. doi: 10.1089/jir.2012.0133. PMID: 23570387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee NK, Choi HK, Yoo HJ, Shin J, Lee SY. RANKL-induced schlafen2 is a positive regulator of osteoclastogenesis. Cell Signal. 2008;20(12):2302–2308. doi: 10.1016/j.cellsig.2008.08.019. PMID: 18796328. [DOI] [PubMed] [Google Scholar]
- 8.Sohn WJ, Kim D, Lee KW, Kim MS, Kwon S, Lee Y, Kim DS, Kwon HJ. Novel transcriptional regulation of the schlafen-2 gene in macrophages in response to TLR-triggered stimulation. Mol Immunol. 2007;44(13):3273–3282. doi: 10.1016/j.molimm.2007.03.001. PMID: 17434208. [DOI] [PubMed] [Google Scholar]
- 9.Walsh MF, Hermann R, Sun K, Basson MD. Schlafen 3 changes during rat intestinal maturation. Am J Surg. 2012;204(5):598–601. doi: 10.1016/j.amjsurg.2012.07.004. PMID: 22906252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel BB, Yu Y, Du J, Rishi AK, Sarkar FH, Tarca AL, Wali A, Majumdar AP. Schlafen 3, a novel gene, regulates colonic mucosal growth during aging. Am J Physiol Gastrointest Liver Physiol. 2009;296(4):G955–G962. doi: 10.1152/ajpgi.90726.2008. PMID: 19228883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel VB, Yu Y, Das JK, Patel BB, Majumdar AP. Schlafen-3: a novel regulator of intestinal differentiation. Biochem Biophys Res Commun. 2009;388(4):752–756. doi: 10.1016/j.bbrc.2009.08.094. PMID: 19703412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao L, Neumann B, Murphy K, Silke J, Gonda TJ. Lack of reproducible growth inhibition by Schlafen1 and Schlafen2 in vitro. Blood Cells Mol Dis. 2008;41(2):188–193. doi: 10.1016/j.bcmd.2008.03.006. PMID: 18479948. [DOI] [PubMed] [Google Scholar]
- 13.Zoppoli G, Regairaz M, Leo E, Reinhold WC, Varma S, Ballestrero A, Doroshow JH, Pommier Y. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc Natl Acad Sci U S A. 2012;109(37):15030–15035. doi: 10.1073/pnas.1205943109. PMID: 22927417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mavrommatis E, Arslan AD, Sassano A, Hua Y, Kroczynska B, Platanias LC. Expression and regulatory effects of murine Schlafen (Slfn) genes in malignant melanoma and renal cell carcinoma. J Biol Chem. 2013;288(46):33006–33015. doi: 10.1074/jbc.M113.460741. PMID: 24089532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Sadi R, Ye D, Said HM, Ma TY. Cellular and molecular mechanism of interleukin-1beta modulation of Caco-2 intestinal epithelial tight junction barrier. J Cell Mol Med. 2011;15(4):970–982. doi: 10.1111/j.1582-4934.2010.01065.x. PMID: 20406328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paunovic B, Deng X, Khomenko T, Ahluwalia A, Tolstanova G, Tarnawski A, Szabo S, Sandor Z. Molecular mechanisms of basic fibroblast growth factor effect on healing of ulcerative colitis in rats. J Pharmacol Exp Ther. 2011;339(2):430–437. doi: 10.1124/jpet.111.183665. PMID: 21841041. [DOI] [PubMed] [Google Scholar]
- 17.Xiao L, Rao JN, Zou T, Liu L, Cao S, Martindale JL, Su W, Chung HK, Gorospe M, Wang JY. miR-29b represses intestinal mucosal growth by inhibiting translation of cyclin-dependent kinase 2. Mol Biol Cell. 2013;24(19):3038–3046. doi: 10.1091/mbc.E13-05-0287. PMID: 23904268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan L, Yu Y, Sanders MA, Majumdar AP, Basson MD. Schlafen 3 induction by cyclic strain regulates intestinal epithelial differentiation. Am J Physiol Gastrointest Liver Physiol. 2010;298(6):G994–G1003. doi: 10.1152/ajpgi.00517.2009. PMID: 20209602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovalenko PL, Basson MD. The correlation between the expression of differentiation markers in rat small intestinal mucosa and the transcript levels of schlafen 3. JAMA Surg. 2013;148(11):1013–1019. doi: 10.1001/jamasurg.2013.3572. PMID: 24005468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovalenko PL, Yuan L, Sun K, Kunovska L, Seregin S, Amalfitano A, Basson MD. Regulation of epithelial differentiation in rat intestine by intraluminal delivery of an adenoviral vector or silencing RNA coding for Schlafen 3. PLoS One. 2013;8(11):e79745. doi: 10.1371/journal.pone.0079745. PMID: 24244554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bustos O, Naik S, Ayers G, Casola C, Perez-Lamigueiro MA, Chippindale PT, Pritham EJ, de la Casa-Esperon E. Evolution of the Schlafen genes, a gene family associated with embryonic lethality, meiotic drive, immune processes and orthopoxvirus virulence. Gene. 2009;447(1):1–11. doi: 10.1016/j.gene.2009.07.006. PMID: 19619625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brady G, Boggan L, Bowie A, O'Neill LA. Schlafen-1 causes a cell cycle arrest by inhibiting induction of cyclin D1. J Biol Chem. 2005;280(35):30723–30734. doi: 10.1074/jbc.M500435200. PMID: 15946944. [DOI] [PubMed] [Google Scholar]
- 23.Neumann B, Zhao L, Murphy K, Gonda TJ. Subcellular localization of the Schlafen protein family. Biochem Biophys Res Commun. 2008;370(1):62–66. doi: 10.1016/j.bbrc.2008.03.032. PMID: 18355440. [DOI] [PubMed] [Google Scholar]
- 24.Frickey T, Lupas AN. Phylogenetic analysis of AAA proteins. J Struct Biol. 2004;146(1–2):2–10. doi: 10.1016/j.jsb.2003.11.020. PMID: 15037233. [DOI] [PubMed] [Google Scholar]
- 25.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317(1):41–72. doi: 10.1006/jmbi.2001.5378. PMID: 11916378. [DOI] [PubMed] [Google Scholar]
- 26.Li M, Kao E, Gao X, Sandig H, Limmer K, Pavon-Eternod M, Jones TE, Landry S, Pan T, Weitzman MD, David M. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature. 2012;491(7422):125–128. doi: 10.1038/nature11433. PMID: 23000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. PMID: 2231712. [DOI] [PubMed] [Google Scholar]
- 28.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. PMID: 10592235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39(Database issue):D225–D229. doi: 10.1093/nar/gkq1189. PMID: 21109532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zdobnov EM, Apweiler R. InterProScan--an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17(9):847–848. doi: 10.1093/bioinformatics/17.9.847. PMID: 11590104. [DOI] [PubMed] [Google Scholar]
- 31.Basson MD, Emenaker NJ, Hong F. Differential modulation of human (Caco02) colon cancer cell line phenotype by short chain fatty acids. Exp Biol Med. 1998;217:476–483. doi: 10.3181/00379727-217-44261. PMID: 9521097. [DOI] [PubMed] [Google Scholar]
- 32.Emenaker NJ, Basson MD. Short-chain fatty acids inhibit human (SW1116) colon cancer cell invasion by reducing urokinase plasminogen activator activity and stimulating TIMP-1 and TIMP-2 activities, rather than via MMP modulation. J Surg Res. 1998;76:41–46. doi: 10.1006/jsre.1998.5279. PMID: 9695737. [DOI] [PubMed] [Google Scholar]
- 33.Basson MD, Lin YW, Ham AM, Emenaker NJ, Shenoy SG, Rothberg BEG. Identification and comparative analysis of human colonocyte short-chain fatty acid response genes. J. Gastrointest. Surg. 2000;4:501–512. doi: 10.1016/s1091-255x(00)80093-1. PMID: 11077326. [DOI] [PubMed] [Google Scholar]
- 34.Walsh MF, Ampasala DR, Hatfield J, Vander Heide R, Suer S, Rishi AK, Basson MD. Transforming growth factor-beta stimulates intestinal epithelial focal adhesion kinase synthesis via Smad- and p38-dependent mechanisms. Am J Pathol 2008. 173:385–399. doi: 10.2353/ajpath.2008.070729. PMID: 18583311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walsh MF, Ampasala DR, Rishi AK, Basson MD. TGF-beta1 modulates focal adhesion kinase expression in rat intestinal epithelial IEC-6 cells via stimulatory and inhibitory Smad binding elements. Biochim Biophus Acta. 2009;1789:88–98. doi: 10.1016/j.bbagrm.2008.11.002. PMID: 19059368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Powell RJ, Bhargava J, Basson MD, Sumpio SE. Coculture conditions alter endothelial modulation of Tgf-β1 activation and smooth muscle growth morphology. Am J Physiol. 1998;274:H642–H649. doi: 10.1152/ajpheart.1998.274.2.H642. PMID: 9486269. [DOI] [PubMed] [Google Scholar]
- 37.Han O, Li GD, Sumpio BE, Basson MD. Strain induces Caco-2 intestinal epithelial proliferation and differentiation via PKC and tyrosine kinase signals. Am J Physiol. 1998;275:G534–G541. doi: 10.1152/ajpgi.1998.275.3.G534. PMID: 9724266. [DOI] [PubMed] [Google Scholar]
- 38.Sander C, Schneider R. Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins. 1991;9(1):56–68. doi: 10.1002/prot.340090107. PMID: 2017436. [DOI] [PubMed] [Google Scholar]
- 39.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38(Web Server issue):W529–W533. doi: 10.1093/nar/gkq399. PMID: 20478830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Yang Z, Cao Y, Zhang S, Li H, Huang Y, Ding YQ, Liu X. The Hsp40 family chaperone protein DnaJB6 enhances Schlafen1 nuclear localization which is critical for promotion of cell-cycle arrest in T-cells. Biochem J. 2008;413(2):239–50. doi: 10.1042/BJ20071510. PMID: 18373498. [DOI] [PubMed] [Google Scholar]
- 41.Eaves-Pyles T, Bu HF, Tan XD, Cong Y, Patel J, Davey RA, Strasser JE. Luminal-applied flagellin is internalized by polarized intestinal epithelial cells and elicits immune responses via the TLR5 dependent mechanism. PLoS One. 2011;6(9):e24869. doi: 10.1371/journal.pone.0024869. PMID: 21949773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niwa H, Ewens CA, Tsang C, Yeung HO, Zhang X, Freemont PS. The role of the N-domain in the ATPase activity of the mammalian AAA ATPase p97/VCP. J Biol Chem. 287(11):8561–8570. doi: 10.1074/jbc.M111.302778. 201. PMID: 22270372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. PMID: 16689629. [DOI] [PubMed] [Google Scholar]
- 44.Karata K, Inagawa T, Wilkinson AJ, Tatsuta T, Ogura T. Dissecting the role of a conserved motif (the second region of homology) in the AAA family of ATPases. Site-directed mutagenesis of the ATP-dependent protease FtsH. J Biol Chem. 1999;274(37):26225–26232. doi: 10.1074/jbc.274.37.26225. PMID: 10473576. [DOI] [PubMed] [Google Scholar]
- 45.Pathak E, Atri N, Mishra R. Role of highly central residues of P-loop and it’s flanking region in preserving the archetypal conformation of Walker A motif of diverse P-loop NTPases. Bioinformation. 2013;9(1):23–28. doi: 10.6026/97320630009023. PMID: 23390340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robine S, Sahuquillo-Merino C, Louvard D, Pringault E. Regulatory sequences on the human villin gene trigger the expression of a reporter gene in differentiating HT29 intestinal cell line. J Biol Chem. 1993;268:11426–11432. PMID: 8496191. [PubMed] [Google Scholar]
- 47.Yamamichi N, Inada K, Furukawa C, Sakurai K, Tando T, Ishizka A, et al. Cdx2 and the Brm-type SW/SNF complex cooperatively regulate villin expression in gastrointestinal cells. Exp Cell Res. 2009;315:1779–1789. doi: 10.1016/j.yexcr.2009.01.006. PMID: 19371634. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki T, Mochizuki K, Goda T. Histone H3 modifications and Cdx-2 binding to the sucrase-isomaltase (SI) gene is involved in induction of the gene in the transition from the crypt to the villus in the small intestine of rats. Biochem Biophys Res Comm. 2008;369:788–793. doi: 10.1016/j.bbrc.2008.02.101. PMID: 18313392. [DOI] [PubMed] [Google Scholar]
- 49.Boudreau F, Rings EH, van Wering HM, Kim RK, Swain GP, Krasinski SD, et al. Hepatocyte nuclear factor-1alpha, GATA-4, and caudal related homeodomain protein Cdx2 interact functionally to modulate intestinal gene transcription. Implication for the developmental regulation of the sucrase-isomaltase gene. J Biol Chem. 2002;277:31909–31917. doi: 10.1074/jbc.M204622200. PMID: 12060663. [DOI] [PubMed] [Google Scholar]
- 50.Inoue S, Mochizuki K, Goda T. Jejunal induction of SI and SGLT1 genes in rats by high-starch/low fat diet is associated with histone acetylation and binding of GCN5 on the genes. J Nutr Sci Vitaminol (Tokyo) 2011;57:162–169. doi: 10.3177/jnsv.57.162. PMID: 21697636. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.