Abstract
Cell culture is an essential tool for drug discovery, tissue engineering, and stem cell research. Conventional tissue culture produces two-dimensional (2D) cell growth with gene expression, signaling, and morphology that can differ from those in vivo and thus compromise clinical relevancy1–5. Here we report a three-dimensional (3D) culture of cells based on magnetic levitation in the presence of hydrogels containing gold and magnetic iron oxide (MIO) nanoparticles plus filamentous bacteriophage. This methodology allows for control of cell mass geometry and guided, multicellular clustering of different cell types in co-culture through spatial variance of the magnetic field. Moreover, magnetic levitation of human glioblastoma cells demonstrates similar protein expression profiles to those observed in human tumor xenografts. Taken together, these results suggest levitated 3D culture with magnetized phage-based hydrogels more closely recapitulates in vivo protein expression and allows for long-term multi-cellular studies.
While protein-based gel environments or rotational/agitation-based bioreactors have been developed in attempts to allow three-dimensional (3D) cell culture1–5, broad practical application of such methods has not as yet been achieved2–4. Thus, a straightforward technology enabling 3D cell culture is still an unmet need3–4. To address this challenge, we introduce a “3D bio-assembler” that relies on magnetic forces and cell levitation6–8. The methodology is based on the cellular uptake and subsequent magnetic levitation of a bioinorganic hydrogel composed of bacteriophage (phage) plus magnetic iron oxide (MIO; Fe3O4, magnetite) and gold nanoparticles that self-assemble into hydrogels (Fig. 1a). Incorporation of MIO nanoparticles creates a new material that retains the biocompatibility of gold-phage hydrogels9, 10 while adding capabilities for the culture and magnetic manipulation of cells.
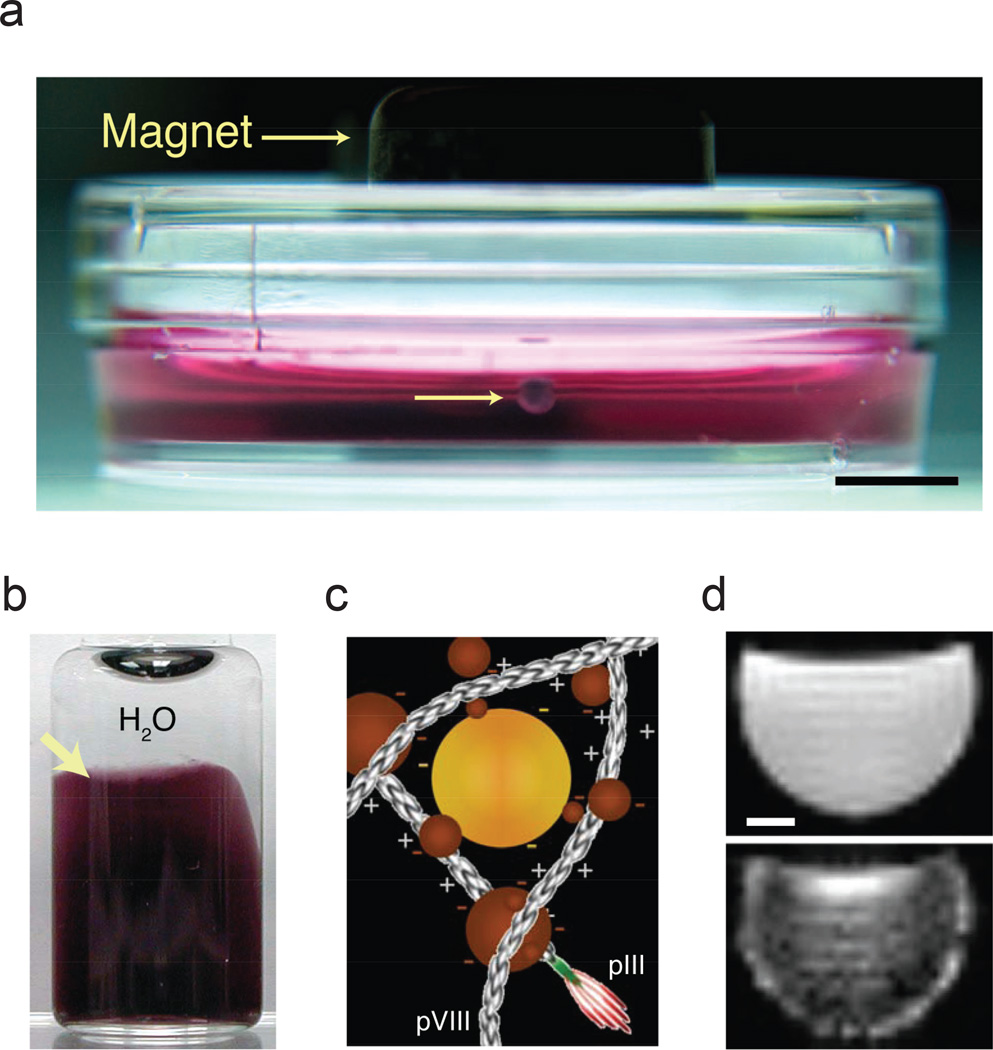
Figure 1. Magnetic iron oxide-containing hydrogels.
a, Human glioblastoma cells (lower arrow) treated with magnetic iron oxide (MIO)-containing hydrogel held at the air-medium interface by a magnet. The image was captured at 48 h of culture and depicts a ~1 mm spheroid. Scale bar, 5mm. b, Vial of a MIO-containing hydrogel (arrow) in water. c, Scheme of electrostatic interaction of nanoparticles (spheres) with phage (elongated structures). Gold (yellow sphere) and MIO (brown sphere) nanoparticles are depicted (not drawn to scale). d, MRI image (T2*-weighted) of purified hydrogel in solution: MIO-containing hydrogel (top panel), average T2* = 16 ms and MIO-free hydrogel control (bottom panel), average T2* = 46.2 ms. Scale bar, 2 mm.
The technology reported here provides an alternative to biodegradable porous scaffolds and protein matrices11–12. In general, biodegradable scaffolds may suffer from slow or delayed propagation of cells and establishment of cell-cell interactions. Existing commercial products, such as Matrigel11, are useful and not particularly expensive but their chemical composition is fixed (i.e., unchangeable). In contrast, our bio-assembler enables adaptable magnetic-based cell levitation and it may provide for improved 3D cell growth conditions in certain settings. The methodology is cost-effective because it does not require specific medium and it is compatible with standard 2D cell culture techniques.
Biological application of magnetic forces has long been studied13–18. For instance, Magnetic Resonance Imaging is a mainstay of clinical diagnostic radiology, relying on superconducting magnets and large magnetic fields. Magnets have also been used to levitate biological samples through the natural diamagnetism of organic materials6. Incorporation of MIO nanoparticles has further enabled manipulation of surface patterns7, 13, contrast-enhanced Magnetic Resonance Imaging14, cell sorting14, mechano-conditioning of cells14–16, studies of mechano-sensitive membrane properties17, and cellular micromanipulation18. However, while MIO nanoparticles can be modified to target proteins or coupled to cationic liposomes for delivery and concentration13, a combination of ligand peptide-mediated targeting and magnetic levitation towards 3D cell culture has not yet been systematically applied.
Our hydrogel uses M13-derived phage particles, displaying the ligand peptide CDCRGDCFC (termed RGD-4C) to target αv integrins19, 20, gold nanoparticles and MIO. Phage are being investigated for biotechnological and materials applications21 such as targeted gene delivery20, optical imaging microscopy9, and protein targeting19. Both phage and gold nanoparticles are potentially biocompatible and have even been approved by the United States Food and Drug Administration for use in several applications22. Under the conditions used, MIO nanoparticles are well-tolerated by mammalian cells, a result consistent with previous reports7, 13, 23. As such, magnetic-based levitation of cell-targeted hydrogels may become a useful biotechnology tool.
To first evaluate and next confirm the presence of MIO in the hydrogels, we employed several methodologies including elastic light scattering, Magnetic Resonance Imaging, and Inductively-coupled Plasma Atomic Emission Spectroscopy (Fig. 1b–d and Supplementary Figs. S1–S2). Having demonstrated the presence of MIO within the targeted hydrogel, we subsequently used murine C17.2 neural stem cells24 to evaluate the ability of the system to induce magnetic field-based cell levitation (Fig. 2). The admixture of neural stem cells and MIO-containing hydrogel co-rose to the air-medium interface but could not leave the medium, presumably due to surface tension (Fig. 2a–c). The observed cell levitation in the liquid medium confirms that the field from a permanent magnet is sufficient to overcome the gravitational force into a steady state, consistent with our theoretical estimates (Supplementary Fig. S3). While the liquid-air interface could also be thought of as a physical barrier for levitated cell culture, it lacks a solid material surface for cell attachment and growth. We observed that the magnetic field concentrated clusters of levitated cells in solution, triggering cell-cell interactions in a manner consistent with tissue engineering scaffolds3 designed to bestow cell growth advantage. In general, 3D cell cultures were axially symmetric and on a large-scale reflected the symmetry of the magnets used. But under magnification, we observed small-scale multicellular assembly features (Fig. 2d) with characteristic and reproducible branching morphogenesis25.
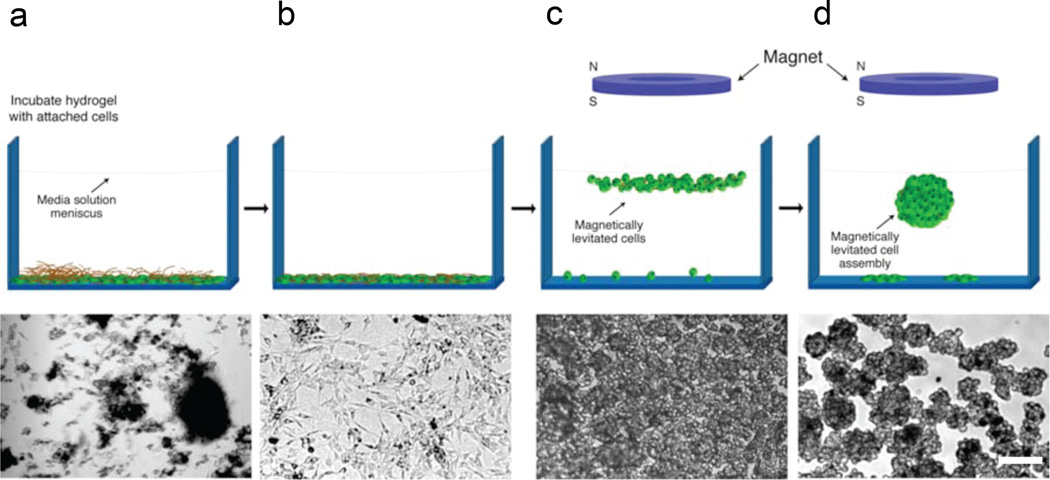
Figure 2. 3D cell culture with magnetic-based levitation.
The top row shows the general cell levitation strategy and the bottom row shows the corresponding optical micrograph of neural stem cells at each stage. a, Hydrogel is dispersed over cells and the mixture is incubated. The dark blotches are fragments of hydrogel. b, Washing steps remove non-interacting hydrogel fragments. Fractions of phage, gold, and MIO nanoparticles enter cells or remain membrane-bound. c, Application of an external magnet causes cells to rise to the air-medium interface. Image shows culture 15 min after levitation. d, After 12 h of levitation, characteristic multicellular structures form (single structure is shown in the schematic). Scale bar, which applies to bottom row, 30 µm.
In order to assess cell growth within the bio-assembler, we visually and quantitatively monitored the formation rate, size, and viability of genetically-modified human glioblastoma cells over a period of 8 d by monitoring the fluorescence from stable protein expression of mCherry (Fig. 3a). Within 30 min of levitation, cells were brought together (Fig. 3a, 30 min); moreover, a cohesive multicellular assembly emerged by 24 h and a spheroid shape formed between 3–8 d, reaching a maximum diameter of ~1 mm. We obtained similar results--albeit with a lower initial yield--with shorter incubation of cells and MIO-containing hydrogels (Supplementary Fig. S4).
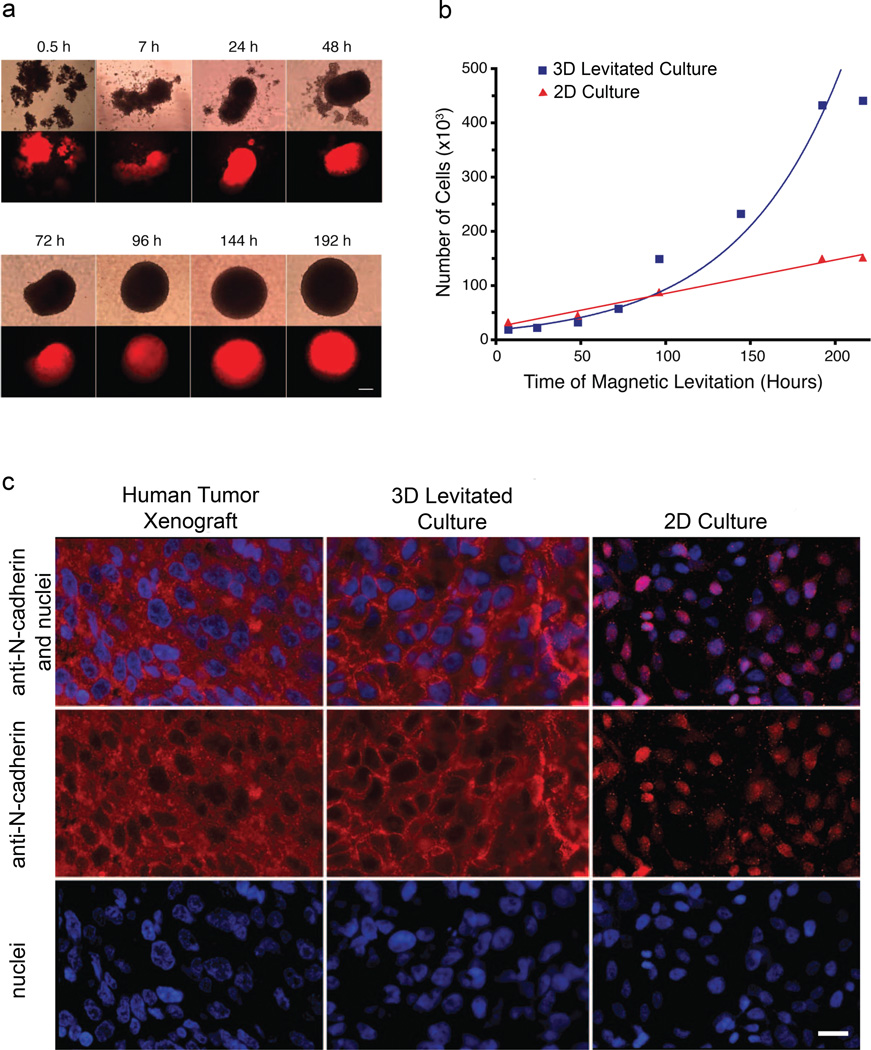
Figure 3. Comparison of 3D cell growth with standard 2D tissue culture.
a, Phase contrast (top row) and fluorescence (mCherry; bottom row) images of levitated human glioblastoma cells monitored over 8 days. Cells coalesced within hours and formed spheroids by 24 h. Scale bar, 200 µm. b, Number of cells as a function of time for levitated cell culture (a; blue squares) and representative 2D culture (red triangles). Line fits indicate an exponential trend for levitated cells (blue line) and linear trend for surface attached (red line). c, Immunofluorescence detection of N-cadherin (red) and DAPI nuclear staining (blue) in a mouse xenograft, 3D magnetic levitation for 48 h, and 2D standard culture with human glioblastoma cells. Scale bar, 10 µm.
Furthermore, morphological analysis of the architectural integrity of 3D structures was performed with transmission and scanning electron microscopy. In “small” spheroids, we noted viable cells throughout the structure; in contrast, we observed central necrosis surrounded by a viable outer region in “large” spheroids (Supplementary Figs. S5–S7). While such features may actually be desirable in emulating conditions within the tumor microenvironment (such as ischemia, acidosis, or nutrient transport, among other parameters), the formation of a heterotypic vascular supply from exogenous angiogenic endothelial cells is an active area of research and development. Together, future use of serial co-cultures of endothelium-derived cells and different ligand peptides or varying magnetic fields can also be envisioned.
The intense red fluorescence from mCherry protein expression confirmed cell viability within the 3D assembly, and cultures could be maintained for at least 12 weeks at which timepoint the experiment was terminated (Supplementary Fig. S8). We compared the growth rate of magnetically levitated cells with that of cells in standard 2D cultures (Fig. 3b). In contrast to the indicated exponential trend for the growth of levitated cells, cell culture in 2D showed a linear growth pattern, a feature of surface attached cultures26. Because of the volume accessible during 3D growth, a large assembly can be accomplished without the de-attachment/re-plating cycles (“passage”) required in standard tissue culture.
To explore the biological attributes afforded by magnetic levitation in our system, we cultured human glioblastoma cells and observed not only morphological but also molecular similarity to orthotopic human tumor xenografts from immunodeficient mice (Fig. 3c). We evaluated the marker N-cadherin, a transmembrane protein mediating cell-cell contact through homotypic cell adhesion interactions27. N-cadherin expression in 3D cultures suggests that magnetic levitation may recapitulate at least some in vivo-like traits. Indeed, 2D cultures show N-cadherin scattered in the cytoplasm and nucleus but absent from the membrane, while 3D-levitated cells express N-cadherin in the membrane, cytoplasm, and cell junctions (akin to the protein expression pattern observed in tumor xenografts). Our results are qualitatively consistent with those recently reported by Ofek et al.27 in which cartilage grown in vitro also yielded differential N-cadherin expression patterns in 3D- relative to 2D-culture. In control experiments, we observed no alterations in N-cadherin expression under conditions including: (i) MIO-containing hydrogels with no magnetic field, (ii) MIO-containing hydrogels with a magnetic field but cultured in 2D, and (iii) cells alone under a magnetic field (Supplementary Fig. S9). Thus, magnetically induced 3D tissue culture in vitro may become a complementary and cheaper surrogate for the labor- and cost-intensive generation and maintenance of human brain tumor xenografts in immunodeficient mice.
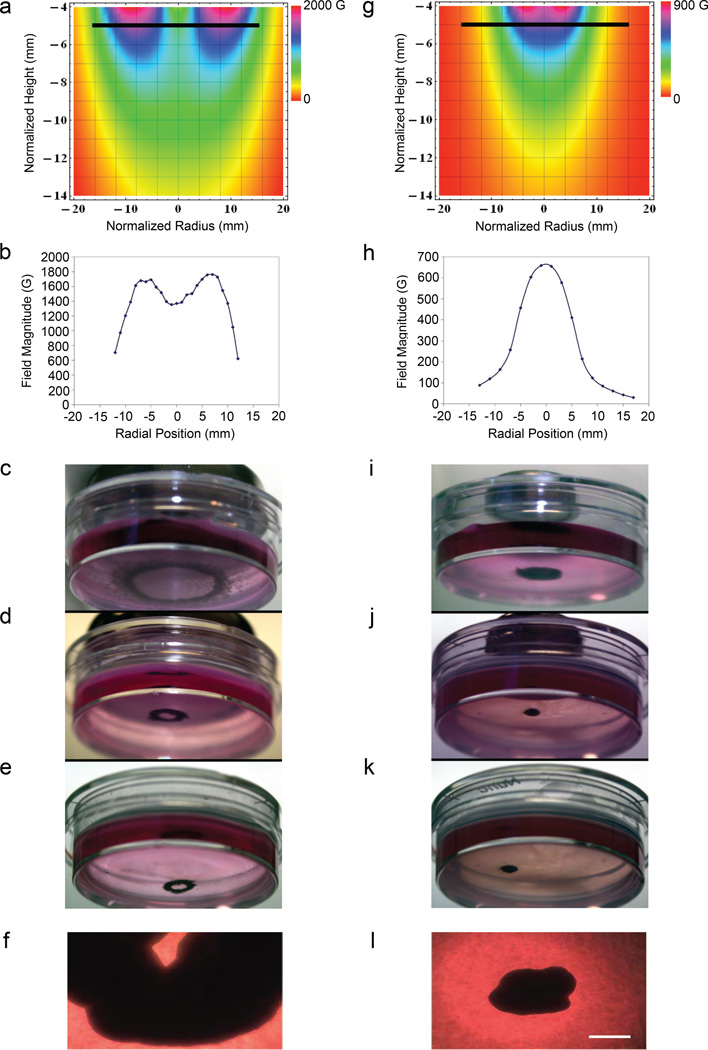
To evaluate the ability of magnetic fields to provide a means to engineer cell culture shape, we levitated tumor cells with different ring-shaped magnets, assessed the magnetic field strength, and serially recorded culture morphology (Fig. 4). The resulting structures reflected the properties of the estimated and Hall probe measured magnetic field. For the large radius magnet (Fig. 4a–f), the field at the height of the culture had a central minimum, which led to a ring-shaped cell pattern at the onset of levitation produced by the force pulling cells towards the field maximum (Fig. 4a, b); this shape persisted as cells assembled (Fig. 4c–f) and formed a cohesive structure with enough integrity to remain intact when the magnet was removed (Fig. 4e–f). While ischemia can cause central necrosis of spheroids cultured for long periods, the shape generated at the early time point was attributable to the magnetic field profile at the liquid-air interface. The small magnet produced a central maximum of the field at the height of the culture and a compact cellular assembly (Fig. 4g–l). In this initial work, the volume of medium and the magnetic fields were held constant and volumetric variation did not adversely affect 3D culture geometry.
Figure 4. Shape control of magnetically levitated culture.
3D culture derived from a magnet with a 12 mm (a–f) and 6 mm (g–l) outer radius. a, g, Magnetic field profiles were obtained by direct integration of the Biot-Savart law using Mathematica (Wolfram Research Inc.; Champaign, IL) and the surface current K⃗ = M⃗ × n̂ (M⃗, magnetization and n̂, unit vector normal to magnet surface). b, h, Hall probe measurements along a diameter perpendicular to the symmetry axis at the air-medium interface. Lines are spline fits. c–e, i–k, Human glioblastoma cells at the onset of levitation (c, i), 30 h (d, j), and following magnet removal at 30 h (e, k). f, l, Spheroid images after growth for one week. Scale bar, 400 µm.
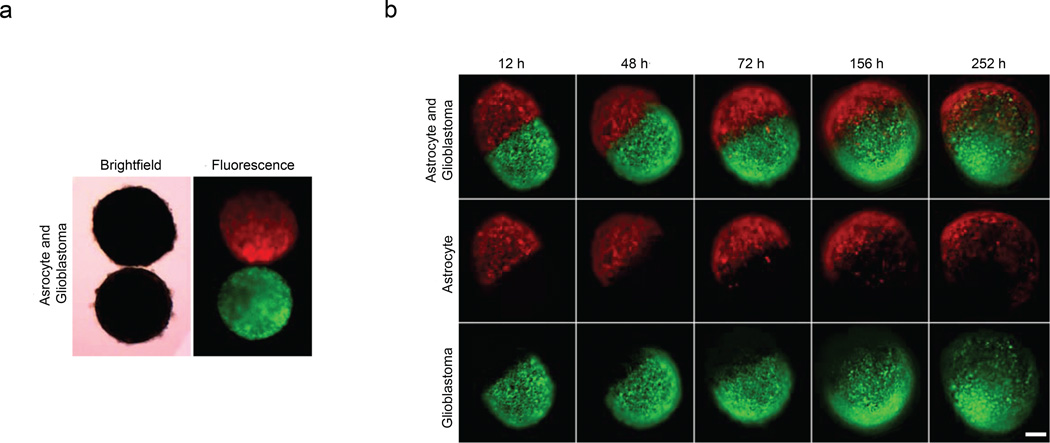
Finally, magnetic field manipulation may provide precise temporal and/or spatial control of distinct cell populations in an environment conducive to visualization or molecular imaging in situ. We demonstrate the feasibility of confrontation assays with co-culture of human glioblastoma cells (GFP-transfected; green cells) and normal human astrocytes (mCherry-transfected; red cells). Initially, a clear interface separating the cell structures was evident. By 12 h, the populations began to fuse and lose their individual spherical shapes. After 3 d, the populations coalesced into a single spheroid with the human glioblastoma cells invading the structure composed of normal human astrocytes. Glioblastoma is the most common, invasive, and lethal type of astrocytic brain tumor28, and these results illustrate the potential of this methodology for analysis of brain tumor invasiveness of normal brain in confrontation culture assays, which have been correlated with clinical outcome29.
In conclusion, we have described a bio-assembler based on magnetic levitation with gold-phage-MIO as an enabling technology for 3D cell culture. We demonstrated control of culture shape, and the ability to bring cultures together for controlled interaction in a confrontation assay with in situ monitoring. The magnetic levitation methodology reported here does not require specific medium, engineered scaffolds, matrices, or molded gels. This simple, flexible, and effective method may be suitable for a range of applications in biotechnology, drug discovery, stem cell research, or regenerative medicine. Indeed, a potential long-term goal is the possibility of accomplishing the “engineering” of normal tissues or complex organs12. Side-by-side comparison to various other 3D culture methods (including a panel of porous and biodegradable scaffolds) will ultimately determine the value of this technology in different experimental settings or applications.
Methods
Hydrogel self-assembly
Hydrogels were generated as described9, 10 except for the inclusion of MIO nanoparticles. MIO-containing hydrogels were prepared by mixing the gold nanoparticle solution (Optical absorbance 530 nm = 1.2–1.5 units) with MIO nanopowder (magnetite[Fe3O4], polydisperse particle size < 50 nm; stabilized with a surfactant of polyvinyl pyrrolidone; Sigma-Aldrich) to a concentration of 0.3 mg/ml. Magnetite nanoparticles were not placed in an oxidizing environment (i.e., chemical or heat) to avoid oxidation into another state such as maghemite [γ-Fe2O3]. Phage dilutions was prepared with 109 transducing units (TU)/µl in picopure water. Finally, the phage solution and the gold nanoparticle plus iron oxide solution were mixed with equal volumes and allowed to stand overnight at 4°C for hydrogel formation.
Magnets
Ring-shaped, neodymium rare earth magnets from Gauss Boys (Portland, OR; part numbers MR32 and MR16) have outer (inner) radii of 12 (2.8), and 6 (1.7) mm and thicknesses of 5 mm and 3 mm. Permanent magnetization is parallel to the symmetry axis.
Cell lines
Bosc, normal human astrocytes, and human glioblastoma (LN-229 or U-251MG) cells were cultured in Dulbecco modified Eagle’s medium containing 10% fetal bovine serum (FBS). Transfections and retroviral infections were performed as described, and cells were selected and maintained with blasticidin (mCherry) or puromycin (eGFP) selection30. C17.2 murine neural stem cells were cultured in high-glucose Dulbecco modified Eagle’s medium containing 10% FBS supplemented with sodium pyruvate 2 mM, glutamine, penicillin, and streptomycin24.
Magnetic levitation of cell culture in MIO-containing hydrogels
For levitated cell culture, surface attached cells (grown to ~80% confluence) were treated with 1 µl of hydrogel per 1 cm2 of surface area available for cell culture and incubated overnight. Treated cells were de-attached by PBS containing trypsin and EDTA and placed into a tissue culture Petri dish9, 10. A cover top with an attached neodymium magnet was immediately put in place. Trypan-blue excluding cells were de-attached and counted. Half of the sample (~3×104 cells) was transferred to seed a 2D surface-attached culture and the other half was seeded for a 3D levitated culture.
Orthotopic glioma xenograft models
Human glioblastoma cells (2×106 cells per 10 µl of PBS; LN229 vector-GFP containing cells) were orthotopically implanted into the brains of immunodeficient (SCID) mice by direct intracranial injection at the right frontal lobe. Tumor-bearing mice were closely monitored and killed upon any neurological signs of intracranial tumor burden. Tumor-containing brains were surgically collected and paraformaldehyde (PFA)-fixed for pathological analysis.
Immunofluorescence detection of N-cadherin
Human glioblastoma cells used for 2D cell culture were prepared by first plating 105 cells on poly-d-lysine coated glass slide and allowing cells to attach overnight. Cells were subsequently washed with PBS and fixed with PBS containing 4% PFA for 30 min, and washed again. Next, cells were permeabilized with PBS containing 0.1% triton X-100 for 5 min, washed with PBS, and treated with Image-iT FX signal enhancer (Invitrogen) for 30 min. Blocking was performed with PBS containing 0.2% gelatin and 10% donkey serum (PBS-gel) for 30 min. 3D cell structures from levitated cultures or brain xenografts were prepared with standard histopathology procedures. Briefly, 3D structures were collected, washed twice in PBS, and fixed in PBS containing 4% PFA for 30 min, then washed again. 3D structures were subsequently embedded in paraffin blocks and sectioned. Samples were treated in xylene, hydrated through a series of ethanols (100%, 95%, 70%) and water. Samples were steamed in antigen retrieval solution (Dako) for 20 min, rinsed in water, and transferred to PBS. Blocking was done with PBS containing 10% donkey serum for 30 min.
Microscope slides of the samples were incubated with primary antibody (anti-N-Cadherin, Zymed) overnight in blocking buffer at 4°C. Cells were washed with PBS then treated with secondary antibody Alexa 488, 555, or 568 (Molecular Probes/Invitrogen) in PBS-gel for 1 h at room temperature (RT). Cells were washed with PBS-gel then treated with DAPI in PBS-gel for 5 min, washed, and mounted onto slides. Images (Z-stacks) were taken on a Zeiss deconvolution microscope and de-convolved with the Axiovert software.
Magnetic resonance measurements
MRI was acquired with a 4.7 T, 40 cm Bruker Biospec instrument. The MRI images were derived from a multi-gradient echo sequence with variance in T2*-weighting (500 ms repetition time, 16 echo times ranging from 1.5 ms to 39 ms, 64 × 64 image matrix, 32 mm × 32 mm field-of-view). The characteristic time constant for exponential transverse signal decay (T2*) was calculated from the average signal level of a region-of-interest in the center of each phantom as a function of echo time. The image contrast between the MIO-containing hydrogel and the MIO-free control results from the reduction in T2* relaxation constant in the presence of iron oxide nanoparticles.
Supplementary Material
Figure 5. Confrontation assay of magnetically levitated multicellular spheroids.
a, Brightfield and fluorescence images of human glioblastoma cells (green; GFP-expressing cells) and normal human astrocytes (red; mCherry-labeled) cultured separately and then magnetically guided together (time=0). b, Confrontation between human glioblastoma cells and normal astrocytes monitored for 10.5 d. Invasion of the spheroid composed of normal human astrocytes by human glioblastoma cells serves as a standard assay of glioma invasiveness29. Scale bar, 200 µm.
Acknowledgments
We thank Ricardo R. Brentani, Neal R. Pellis, and E. Helene Sage for helpful discussions and Kenneth Dunner Jr. for technical assistance. G.R.S. was supported by the Odyssey Scholar Program of the University of Texas M. D. Anderson Cancer Center and by the Breast Cancer Research Program of the U.S. Department of the Defense (DOD). D.J.S. received support from the National Science Foundation. T.C.K. received support from the David and Lucille Packard Foundation. W.A. and R.P. received support from the Gillson-Longenbaugh Foundation, the Marcus Foundation, AngelWorks, DOD, NIH, and NCI.
Footnotes
Author contributions
G.R.S., J.R.M., R.M.R., D.J.S., C.S.L, J.M., T.C.K., W.A., R.P. conceived and designed the experiments. G.R.S., J.R.M., T.C.K., D.J.S., C.S.L., J.S.A., J.M. performed the experiments. G.R.S., M.G.O., D.J.S., C.S.L., L.F.B., J.S.A., J.A.B., T.C.K., W.A., R.P. analyzed the data. R.M.R., M.M.G., J.A.B., J.G.G., T.C.K., W.A., R.P. contributed materials/analysis tools. G.R.S., R.M.R., M.G.O., L.F.B., J.A.B. J.G.G., T.C.K., W.A., R.P. co-wrote the paper.
The authors declare competing financial interests: details accompany the paper at www.nature.com/naturenanotechnology.
Supplementary information accompanies this paper at www.nature.com/naturenanotechnology.
Reprints and permission information is available online at http://npg.nature.com/reprintsandpermissions/.
Competing Interests Statement
The University of Texas M. D. Anderson Cancer Center (UTMDACC) and Rice University (RU), along with their researchers, have filled patents on the technology and other intellectual property reported here. If licensing or commercialization occurs, the researchers will be entitled to standard royalties. G.R.S., R.M.R., C.S.L., and T.C.K. have equity in Nano3D Biosciences, Inc. UTMDACC and RU manage the terms of these arrangements in accordance to their established institutional conflict-of-interest policies.
Contributor Information
T. C. Killian, Email: killian@rice.edu.
Wadih Arap, Email: warap@mdanderson.org.
Renata Pasqualini, Email: rpasqual@mdanderson.org.
References
- 1.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 2.Abbott A. Biology's new dimension. Nature. 2003;424:870–872. doi: 10.1038/424870a. [DOI] [PubMed] [Google Scholar]
- 3.Pampaloni F, Reynaud EG, Stelzer EH. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 2007;8:839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 4.Griffith LG, Swartz MA. Capturing complex 3D tissue physiology in vitro. Nat. Rev. Mol. Cell Biol. 2006;7:211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 5.Atala A. Engineering tissues, organs and cells. J. Tissue Eng. Regen. Med. 2007;1:83–96. doi: 10.1002/term.18. [DOI] [PubMed] [Google Scholar]
- 6.Coleman CB, et al. Diamagnetic levitation changes growth, cell cycle, and gene expression of Saccharomyces cerevisiae. Biotechnol. Bioeng. 2007;98:854–863. doi: 10.1002/bit.21526. [DOI] [PubMed] [Google Scholar]
- 7.Dobson J. Remote control of cellular behaviour with magnetic nanoparticles. Nat. Nanotechnol. 2008;3:139–143. doi: 10.1038/nnano.2008.39. [DOI] [PubMed] [Google Scholar]
- 8.Ito A, Shinkai M, Honda H, Kobayashi T. Medical application of functionalized magnetic nanoparticles. J. Biosci. Bioeng. 2005;100:1–11. doi: 10.1263/jbb.100.1. [DOI] [PubMed] [Google Scholar]
- 9.Souza GR, et al. Networks of Gold Nanoparticles and Bacteriophage as Biological Sensors and Cell Targeting Agents. Proc. Natl. Acad. Sci. USA. 2006;103:1215–1220. doi: 10.1073/pnas.0509739103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Souza GR, et al. Bottom-up assembly of hydrogels from bacteriophage and Au nanoparticles: the effect of cis- and trans-acting factors. PLoS ONE. 2008;3:e2242. doi: 10.1371/journal.pone.0002242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc. Natl. Acad. Sci. USA. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mikos AG, et al. Engineering complex tissues. Tissue Eng. 2006;12:3307–3339. doi: 10.1089/ten.2006.12.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito A, Ino K, Kobayashi T, Honda H. The effect of RGD peptide-conjugated magnetite cationic liposomes on cell growth and cell sheet harvesting. Biomaterials. 2005;26:6185–6193. doi: 10.1016/j.biomaterials.2005.03.039. [DOI] [PubMed] [Google Scholar]
- 14.Pankhurst Q, Connolly J, Jones SK, Dobson J. Applications of magnetic nanoparticles in biomedicine. J. Phys. D: Appl. Phys. 2003;36:R167–R181. [Google Scholar]
- 15.Alsberg E, Feinstein E, Joy MP, Prentiss M, Ingber DE. Magnetically-guided self-assembly of fibrin matrices with ordered nano-scale structure for tissue engineering. Tissue Eng. 2006;12:3247–3256. doi: 10.1089/ten.2006.12.3247. [DOI] [PubMed] [Google Scholar]
- 16.Dobson J, Cartmell SH, Keramane A, El Haj AJ. Principles and design of a novel magnetic force mechanical conditioning bioreactor for tissue engineering, stem cell conditioning, and dynamic in vitro screening. IEEE Trans. Nanobioscience. 2006;5:173–177. doi: 10.1109/tnb.2006.880823. [DOI] [PubMed] [Google Scholar]
- 17.Meyer CJ, et al. Mechanical control of cyclic AMP signaling and gene transcription through integrins. Nat. Cell Biol. 2000;2:666–668. doi: 10.1038/35023621. [DOI] [PubMed] [Google Scholar]
- 18.Matthews BD, La Van DA, Overby DR, Karavitis J, Ingber DE. Electromagnetic needles with submicron pole tip radii for nanomanipulation for biomolecules and living cells. Appl. Phys. Lett. 2004;85:2968–2970. [Google Scholar]
- 19.Arap W, Pasqualini R, Ruoslahti E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science. 1998;279:377–380. doi: 10.1126/science.279.5349.377. [DOI] [PubMed] [Google Scholar]
- 20.Hajitou A, et al. A hybrid vector for ligand-directed tumor targeting and molecular imaging. Cell. 2006;125:385–398. doi: 10.1016/j.cell.2006.02.042. [DOI] [PubMed] [Google Scholar]
- 21.Nam KT, et al. Virus-enabled synthesis and assembly of nanowires for lithium ion battery electrodes. Science. 2006;312:885–888. doi: 10.1126/science.1122716. [DOI] [PubMed] [Google Scholar]
- 22.Langer R, Tirrell DA. Designing materials for biology and medicine. Nature. 2004;428:487–492. doi: 10.1038/nature02388. [DOI] [PubMed] [Google Scholar]
- 23.Hautot D, et al. Preliminary observation of elevated levels of nanocrystalling iron oxide in the basal ganglia of neuroferritinopathy patients. Biochim. Biophys. Acta. 2007;1772:21–25. doi: 10.1016/j.bbadis.2006.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snyder EY, et al. Multipotent neural cell lines can engraft & participate in development of mouse cerebellum. Cell. 1992;68:33–51. doi: 10.1016/0092-8674(92)90204-p. [DOI] [PubMed] [Google Scholar]
- 25.Wan X, Li Z, Lubkin SR. Mechanics of mesenchymal contribution to clefting force in branching morphogenesis. Biomech. Model. Mechanobiol. 2008;7:417–426. doi: 10.1007/s10237-007-0105-y. [DOI] [PubMed] [Google Scholar]
- 26.Csikasz-Nagy A, Battogtokh D, Chen KC, Novak B, Tyson JJ. Analysis of a generic model of eukaryotic cell-cycle regulation. Biophys. J. 2006;90:4361–4379. doi: 10.1529/biophysj.106.081240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ofek G, et al. Matrix development in self-assembly of articular cartilage. PLoS ONE. 2008;3:e2795. doi: 10.1371/journal.pone.0002795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhowmick DA, Zhuang Z, Wait SD, Weil RJ. A functional polymorphism in the EGF gene is found with increased frequency in glioblastoma multiforme patients and is associated with more aggressive disease. Cancer Res. 2004;64:1220–1223. doi: 10.1158/0008-5472.can-03-3137. [DOI] [PubMed] [Google Scholar]
- 29.Chicoine MR, Silbergeld DL. Mitogens as motogens. J. Neurooncol. 1997;35:249–257. doi: 10.1023/a:1005808315821. [DOI] [PubMed] [Google Scholar]
- 30.Georgescu MM, Kirsch KH, Akagi T, Shishido T, Hanafusa H. The tumor-suppressor activity of PTEN is regulated by its carboxyl-terminal region. Proc. Natl. Acad. Sci. USA. 1999;96:10182–10187. doi: 10.1073/pnas.96.18.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.