SUMMARY
We propose an automatic structure recovery method for additive models, based on a backfitting algorithm coupled with local polynomial smoothing, in conjunction with a new kernel-based variable selection strategy. Our method produces estimates of the set of noise predictors, the sets of predictors that contribute polynomially at different degrees up to a specified degree M, and the set of predictors that contribute beyond polynomially of degree M. We prove consistency of the proposed method, and describe an extension to partially linear models. Finite-sample performance of the method is illustrated via Monte Carlo studies and a real-data example.
Keywords: Backfitting, Bandwidth estimation, Kernel, Local polynomial, Measurement-error model selection likelihood, Model selection, Profiling, Smoothing, Variable selection
1. Introduction
Because of recent developments in data acquisition and storage, statisticians often encounter datasets with large numbers of observations or predictors. The demand for analysing such data has led to the current heightened interest in variable selection. For parametric models, classical methods include backward, forward and stepwise selection. More recently, many approaches to variable selection have been developed that use regularization via penalty functions. Examples include the lasso (Tibshirani, 1996), smoothly clipped absolute deviation (Fan & Li, 2001), the adaptive lasso (Zou, 2006; Zhang & Lu, 2007), and the L0 penalty (Shen et al., 2013). Fan & Lv (2010) give a selective overview of variable selection methods.
Variable selection for nonparametric modelling has advanced at a slower pace than for parametric modelling, and has been studied primarily in the context of additive models, which are an important extension of multivariate linear regression. An additive model presupposes that each predictor contributes a possibly nonlinear effect, and that the effects of multiple predictors are additive. Such models were proposed by Friedman & Stuetzle (1981) and serve as surrogates for fully nonparametric models. Most nonparametric variable selection methods studied so far deal with additive models; see Ravikumar et al. (2009), Huang et al. (2010), Fan et al. (2011), and references therein. An exception is Stefanski et al. (2014), which considers variable selection in a fully nonparametric classification model. Additionally, variable selection and structure recovery for the varying coefficient model, a popular extension of the additive model, have been studied: Xia et al. (2004) considered structure recovery towards semi-varying coefficient modelling; Fan et al. (2014) studied a new variable screening method; and for the longitudinal setting Cheng et al. (2014) investigated variable screening and selection, as well as structure recovery.
Partially linear models were proposed by Engle et al. (1986). Combining the advantages of linearity and additivity, these models assume that some covariates have nonlinear additive effects while others contribute linearly. Estimation for a partially linear model requires knowing which covariates have linear effects and which have nonlinear effects, information that is usually not available a priori. Recently, Zhang et al. (2011) and Huang et al. (2012) proposed methods to identify covariates that have linear effects and ones that have nonlinear effects.
We consider an additive model for a scalar response Y and predictors X = (X1, … , XD)T,
| (1) |
under the identifiability conditions E{md (Xd)} = 0 (d = 1 , … , D). Denote a random sample from model (1) by {(Yi, Xi): i = 1, … , n}, where Xi =(Xi1, … , XiD)T. The goal is to estimate α and md (·) (d = 1, … , D).
We a method for estimating md(·) that first distinguishes between important predictors and predictors that are unimportant, i.e., those Xd for which md(·) = 0. Next, motivated by Zhang et al. (2011) and Huang et al. (2012), the method identifies predictors that have linear effects from the estimated set of important predictors. Then, the method identifies the predictors that have quadratic effects, and so on. This process continues and results in estimates of sets of predictors that have polynomial effects at different degrees, up to some degree M, and the set of predictors for which the corresponding md(·) are not polynomial of any degree up to M.
At the core of our structure recovery method is a new nonparametric kernel-based variable selection method derived from the measurement-error model selection likelihood approach of Stefanski et al. (2014). They studied the relationship between lasso estimation and measurement error attenuation in linear models, and used that connection to develop a general approach to variable selection in nonparametric models.
2. Backfitting algorithm
Backfitting coupled with smoothing is commonly used for fitting model (1). Here we use univariate local polynomial smoothing, with denoting the univariate local polynomial smoother with kernel function K (·), bandwidth h and degree p.
Local polynomial smoothing (Fan & Gijbels, 1996) is a well-studied nonparametric smoothing technique. To estimate the regression function g(t) = E(Z…T = t) from an independent and identically distributed random sample {(Ti, Zi) : i = 1, … , n}, local polynomial regression uses Taylor series approximations and weighted least squares. The local polynomial smoothing estimate of g(t0) based on smoother is given by , the optimizer of
See Fan & Gijbels (1996) for a detailed account of local polynomial modelling.
Using univariate local polynomial smoothing with kernel K(·), bandwidth hd and degree pd to estimate md(·) in the additive model (1), the backfitting algorithm consists of the following steps.
Step 1
Initialize by setting and for d = 1, … , D.
Step 2
For d = 1, … , D:
(a) apply the local polynomial smoother to and set the estimated function to be the updated estimate for md (·);
(b) if necessary, apply centring by updating with .
Step 3
Repeat Step 2 until the changes in all (d = 1, … , D) between successive iterations are less than a specified tolerance.
Denote the estimates at convergence by (d = 1, … , D) and , where h = (h1, … , hD)T and p = (p1, … , pD)T are the vectors of smoothing bandwidths and local polynomial degrees, respectively. For simplicity we use the same kernel K (·) for each component, and thus K (·) is omitted from the notation .
Backfitting works well for problems of moderate dimension D. However, even though backfitting entails only univariate smoothing, its performance deteriorates as D gets larger. Consequently, variable selection is important for the additive model.
3. Variable selection via backfitting local constants
3·1. Variable selection
In Step 2(a) of the backfitting algorithm, any smoothing method can be applied. In this section we consider the local constant smoother, i.e., the local polynomial smoother of degree 0, and propose a nonparametric variable selection method for the additive model (1).
With pd =0 in Step 2(a) of the backfitting algorithm, we update the estimate of md (xd) by the minimizer of
| (2) |
Note that K{hd−1(Xid – xd )} = K(0) for all i when hd = ∞. In this case, md (·) is globally approximated by a0. The corresponding optimizer of (2) is given by , which equals zero because and centring is applied in Step 2(b) of the backfitting algorithm.
Thus, when pd = 0, the backfitting algorithm leads to the constant zero function when hd = ∞. Consequently, the jth predictor is excluded from the model when hd = ∞ and is included only when hd < ∞. The equivalence, hd < ∞ if and only if the jth predictor is included, is key to the approach described in Stefanski et al. (2014), and we exploit it repeatedly in our method. We assume in this section that the degree of local polynomial smoothing for every function component is 0, i.e., pd 0 for d = 1, … , D. In this way, the smoothing bandwidth relates directly to the importance of each predictor, with small hd corresponding to important predictors.
As in Stefanski et al. (2014), we reparameterize hd as λd = 1/hd, so that large λd will correspond to important predictors and λd = 0 to unimportant predictors. Predictor smoothing is now determined by the inverse bandwidths λd. Stefanski et al. (2014) show that variable selection can be obtained by minimizing a loss function with respect to the λd, subject to a bound on the total amount of smoothing as determined by the sum of the λd or, equivalently, a bound on the harmonic mean of the bandwidths.
A suitable loss function for backfitting is the sum of squared errors
where in the second two arguments of , λ−1 = (1/λ1, … , 1/λD)T and 0D denotes the D × 1 zero vector. We shall also write 1s for the s × 1 vector of ones.
In the absence of constraints, the minimum of sse with respect to λ is 0, and corresponds to overfitted models. As in Stefanski et al. (2014), appropriate regularization is achieved by solving the constrained optimization problem
| (3) |
Solving (3) distinguishes important predictors, , from those that are unimportant, .
3·2. Modified coordinate descent algorithm
The constrained optimization problem (3) is not convex because of the complicated dependence on λ through the backfitting algorithm and the univariate local polynomial smoothing. We have had success using a modified coordinate descent algorithm.
Coordinate descent for high-dimensional lasso regression (Fu, 1998; Daubechies et al., 2004) cycles through variables one at a time, solving simple marginal univariate optimization problems at each step, and thus is computationally efficient. It has been studied extensively, for example by Friedman et al. (2007) and Wu & Lange (2008), among others. However, standard coordinate descent cannot be applied directly to (3).
In our modified coordinate descent algorithm, we initialize with equal smoothing, i.e., we set λd = τ0/D for all d = 1, … , D. We then cycle through all components and make univariate updates. Current solutions are denoted by , where the superscript c means current. Suppose that we are updating the d’th component. Let , a vector whose elements are all zero except for the d’th, which is 1, and set . The components of , and sum to unity. Thus, for any γ ∈ [0, τ0], the components of , sum to τ0 and satisfy the first constraint in (3). We then update the set of current solutions to the components of the vector , where is the optimizer of
subject to 0 ≤ γ ≤ τ0. Cycling through d’ = 1, … , D completes one iteration of the algorithm. Iterations continue until the change in solutions between successive iterations becomes small enough.
3·3. Tuning
The tuning parameter τ0 controls the total amount of smoothing, and can be selected by standard methods such as crossvalidation, aic or bic. Denote the optimizer of (3) by . If an independent tuning dataset is available, then τ0 can be selected by minimizing the sum of squared prediction errors of the estimator over the tuning set. For methods such as aic and bic, the degrees of freedom is needed. The local constant smoothing estimator is a linear smoother, and we couple the backfitting algorithm to marginal local constant smoothing. For each function component, the trace of the corresponding smoothing matrix minus 1 can be used as the degrees of freedom. The trace is reduced by 1 to account for the centring applied in Step 2(b) of the backfitting algorithm.
4. Higher-degree local polynomial regression
The approach in § 3 extends readily to higher-degree local polynomial regression. Suppose that we use local degree p* polynomial regression in Step 2(a) of the backfitting algorithm. With pd = p* in Step 2(a), we update the estimate of md (xd) by the minimizer of
As remarked previously, K{hd−1(Xid – xd)} = K(0) for all i when hd = ∞. Consequently, is a global approximation of md (xd). For this case, in Step 2(a) of the backfitting algorithm the estimate is updated by , where are the optimizers of .
Thus, when pd = p*, the backfitting algorithm yields a polynomial of degree p* as the estimate of md(·) when hd = ∞. The interpretation is that the dth predictor makes a polynomial contribution of degree up to p*. Based on this, we derive a method for detecting predictors that contribute polynomially up to degree p* in the additive model.
Now we use backfitting with univariate local polynomial smoothing of degree p* for every function component, again parameterizing via inverse bandwidths λd = 1/hd. As in the previous section, the general approach of Stefanski et al. (2014) leads to solving
| (4) |
where τp* plays the same role as τ0 in (3), and in the last two arguments of , λ−1 = (1/λ1, … , 1/λD)T and p*1D is the product of p* and 1D. In this case, an optimizer means that the dth predictor contributes polynomially at a degree no greater than p*.
The modified coordinate descent algorithm and the tuning procedures discussed in § § 3·2 and 3·3 extend naturally to (4).
5. Automatic structure recovery
The procedures in § § 3 and 4 provide the building blocks for the additive model automatic structure recovery method. In combination, they enable estimation of the predictors that are not important, as well as those that contribute at the pth degree polynomially.
We let denote the set of predictors that contribute polynomially at degree p and only at degree p, and we let denote the set of predictors that contribute beyond polynomially degree p, which includes higher-degree polynomials as well as nonpolynomial functions. Thus means at degree p, and indicates beyond degree p. With these naming conventions, a predictor that contributes at degree p = 0 is no more informative than a constant and is therefore unimportant. Specifically, , for p = 1,2, … and for p = 0, 1, 2, … Here md(·) = 0 means that md(t) = 0 for any t in its domain of interest, and md(·) ≠ 0 means that md(t) ≠ 0 for some t. Our automatic structure recovery method proceeds in the following steps.
Step 1
Identify important and unimportant predictors.
With appropriately chosen τ0, solve (3) to obtain , and set and , the complement of in the set {1, … D}. Then and are estimates of the sets of unimportant and important predictors, respectively.
Step 2
Identify from the predictors that contribute linearly.
After identifying the set of important predictors, the next step is to identify a subset of consisting of functions that contribute linearly. We remove unimportant predictors Xd with from consideration and apply the method of § 4, namely (4) with p* = 1, to the data , where denotes the subvector of Xi with indices in . We define and to be the backfitting estimates obtained using the data with bandwidths for . Let denote the cardinality of the set . For an appropriately tuned τ1, we solve the optimization problem
and denote its optimizer by , for . The set estimates the set of predictors contributing linearly, and estimates the set of predictors that contribute beyond linearly.
Step 3
Identify from the predictors that contribute quadratically.
In Step 2 the set of linear predictors was identified. The set is global in principle, and this property can be ensured via profiling (Severini & Wong, 1992). For we denote the global fit coefficient for the linear predictor Xd by β1d or, in vector form, . For a given , we Apply the backfitting algorithm of degree pd = 2 with bandwidth to the data and denote the corresponding estimates by and ; we have given a fourth argument to emphasize its dependence on . Using profiling, we solve
to obtain the best estimate for , which we denote by to emphasize its dependence on . With the above notation and definitions, we solve the optimization problem
for appropriately tuned τ2, and denote its optimizer by . Then estimates the set of predictors that contribute quadratically, and estimates the set of predictors that contribute beyond quadratically.
Steps 4 to M + 1
Identify predictors that have kth-degree polynomial effects by using appropriate τk for k 3, 4, ….
After Step 3, we have estimated the set of linear predictors and the set of quadratic predictors. In a straightforward manner Step 3 can be modified to identify the sets of predictors that contribute polynomially at degrees k = 3, k = 4, and so on, up to k = M.
Step M + 2. Fit the final model.
After Step M + 1, we have obtained estimates for k = 0, 1 … , M and . The final model is then estimated by combining the profiling technique and the backfitting technique as in Step 3. In this final step, we couple the backfitting algorithm with local linear smoothing, as we want to estimate only the function md(·) itself for .
6. Theoretical properties
In this section we study the consistency of the proposed structure recovery scheme. We first show that the estimated set of unimportant predictors is consistent.
Proposition 1
Suppose τ0 → ∞ such that as n → ∞, and assume that Conditions A1–A5 in the Appendix hold. Then the solution to (3) satisfies in probability for , and in probability for . Consequently, as n → ∞.
Remark 1
There is a small gap between Proposition 1 and the proposed procedure, which is confounded with the numerical analysis. In our procedure is defined as , but Proposition 1 shows that the solution to (3) satisfies in probability for . Thus there would be closer agreement between the proposition and the algorithm if we had defined as for some small δ > 0. However, based on our numerical experience so far, the optimizer is indeed sparse, returning exact zeros. Therefore, we shall keep the definition of as . This remark also applies to Theorem 1.
The consistency in Proposition 1 is readily extended to the estimated set of predictors that contribute polynomially at different degrees, resulting in the following consistency property for the proposed structure recovery scheme.
Theorem 1
Suppose that for k = 0, 1, … , M, τk → ∞ and as n → ∞, and assume that Conditions A1–A5 in the Appendix hold. Then the estimators and satisfy as n → ∞.
The proofs of Proposition 1 and Theorem 1 are given in the Appendix.
7. Simulation studies
We use simulation models adapted from Zhang et al. (2011) to study the finite-sample performance of the proposed method. In each model, predictors are generated as Xj = (Uj + ηU)/ (1 + η) for j = 1, … , D, where U1, … , UD, U are independent and identically distributed Un[0, 1] variables, with η at levels 0 and 0·5, resulting in pairwise correlations of 0 and 0·2. We compare our method with the smoothing spline analysis-of-variance method (Kimeldorf & Wahba, 1971; Gu, 2002), as well as the linear and nonlinear discovery method of Zhang et al. (2011) and its refitted version, in terms of both integrated squared error and predictor-type identification.
For an estimate of the additive model (1), define the integrated squared error , estimated via Monte Carlo using an independent test set of size 1000. The linear and nonlinear discovery methods, original and refitted, are designed to identify predictors having null, linear, nonlinear, or a mixture of linear and nonlinear effects, whereas the goal of the proposed method is to identify predictors that contribute polynomially at different degrees and predictors that contribute beyond polynomially of a specified degree M. In the simulation studies, we fixed M to be 2. We use the Bayesian information criterion for tuning parameter selection in all methods.
Model 1
In the first data-generation model, Y = f1(X1)+ f2(X2)+ f3(X3)+ε with f1(t)=3t, f2(t)=2 sin(2πt) and f3(t)=2(3t – 1)2. Here ε ~ N(0, σ2) is independent of the predictors. Two values of the standard deviation, σ = 1 and σ = 2, and two dimensions, D = 10 and D = 20, are considered. For all settings, X4, … , XD are unimportant. In terms of the linear and nonlinear discovery classification scheme, X1 has a purely linear effect, X2 has a purely nonlinear effect, and X3 has a mixture of linear and nonlinear effects (Zhang et al., 2011). For our method, X1 is a linear predictor, X3 is a quadratic predictor, and X2 contributes beyond quadratically. Training samples of size n =100 are used. The results from 100 simulated datasets are reported in Tables 1 and 2.
Table 1.
Simulation results for Model 1: average integrated squared errors (×102) with standard deviations (×102) in parentheses
| D | σ | η | Oracle 1 | SSANOVA | LAND | LANDr | PSC | Oracle 2 |
|---|---|---|---|---|---|---|---|---|
| 10 | 1 | 0·0 | 12·5 (5·5) | 23·9 (9·0) | 11·9 (7·4) | 14·9 (6·8) | 11·6 (6·2) | 9·8 (4·9) |
| 0·5 | 13·2 (5·4) | 25·5 (8·8) | 58·9 (36·8) | 17·2 (7·3) | 12·3 (5·9) | 10·4 (4·4) | ||
| 2 | 0·0 | 44·0 (22·9) | 92·7 (41·6) | 82·1 (48·0) | 71·2 (42·4) | 52·0 (34·7) | 37·3 (21·9) | |
| 0·5 | 50·3 (26·3) | 100·4 (35·9) | 142·6 (47·9) | 88·1 (38·0) | 67·4 (34·9) | 42·8 (21·7) | ||
| 20 | 1 | 0·0 | 12·8 (5·4) | 45·5 (14·3) | 16·4 (12·6) | 18·7 (9·7) | 14·8 (7·8) | 9·9 (4·4) |
| 0·5 | 13·4 (5·7) | 50·6 (19·6) | 64·4 (36·0) | 24·4 (11·6) | 14·3 (8·9) | 11·2 (5·4) | ||
| 2 | 0·0 | 45·1 (21·0) | 187·0 (65·8) | 135·8 (70·9) | 130·7 (60·1) | 69·3 (40·7) | 37·8 (18·5) | |
| 0·5 | 45·7 (18·9) | 192·8 (62·8) | 169·5 (63·5) | 155·3 (63·8) | 76·2 (41·7) | 40·4 (19·6) |
D, dimension; σ, model error standard deviation; η, predictor correlation parameter; SSANOVA, smoothing spline analysis-of-variance method; LAND, linear and nonlinear discovery method; LANDr, linear and nonlinear discovery method, refitted version; PSC, the proposed polynomial structure classification method; Oracle 1, oracle for LAND and LANDr; Oracle 2, oracle for PSC.
Table 2.
Summary of predictor classification performance of the linear and nonlinear discovery method and the proposed method for Model 1
| LAND | PSC | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D | σ | η | L | NL | L-N | Nil | CC | L | Q | BQ | Nil | CC |
| 10 | 1 | 0·0 | 0·9 | 0·8 | 1·0 | 6·5 | 54 | 0·9 | 1·0 | 1·0 | 7·0 | 91 |
| 0·5 | 1·0 | 0·2 | 0·9 | 5·9 | 10 | 1·0 | 1·0 | 1·0 | 7·0 | 92 | ||
| 2 | 0·0 | 1·0 | 0·4 | 1·0 | 4·8 | 5 | 0·9 | 1·0 | 0·9 | 7·0 | 74 | |
| 0·5 | 1·0 | 0·0 | 0·7 | 3·8 | 0 | 0·6 | 0·9 | 0·6 | 7·0 | 35 | ||
| Average SD | 0·1 | 0·4 | 0·2 | 1·5 | 0·3 | 0·2 | 0·2 | 0·0 | ||||
| 20 | 1 | 0·0 | 1·0 | 0·7 | 1·0 | 15·6 | 34 | 1·0 | 1·0 | 1·0 | 17·0 | 90 |
| 0·5 | 1·0 | 0·2 | 0·9 | 14·8 | 4 | 1·0 | 1·0 | 1·0 | 17·0 | 93 | ||
| 2 | 0·0 | 1·0 | 0·4 | 1·0 | 11·0 | 1 | 0·9 | 0·9 | 0·9 | 17·0 | 72 | |
| 0·5 | 0·9 | 0·1 | 0·7 | 10·1 | 0 | 0·6 | 0·9 | 0·6 | 17·0 | 33 | ||
| Average SD | 0·1 | 0·4 | 0·2 | 2·5 | 0·3 | 0·3 | 0·2 | 0·0 | ||||
LAND, linear and nonlinear discovery method; PSC, the proposed polynomial structure classification method; D, dimension; σ, model error standard deviation; η, predictor correlation parameter; L, linear; NL, nonlinear; L-N, linear-nonlinear mixture; Nil, noise; CC, proportion (%) of models with fully correct variable classifications; Q, quadratic; BQ, beyond quadratic; Average SD, block-averaged standard deviation.
Table 1 summarizes the performance of the smoothing spline method, the two linear and nonlinear discovery methods, the proposed polynomial structure classification method, and two oracle methods, which are included to assess the utility of the underlying classification schemes irrespective of estimation error. Oracle 1 is the linear and nonlinear discovery oracle that makes use of information on which predictors are noise, purely linear, purely nonlinear, or a mixture of linear and nonlinear. Oracle 2 is the oracle for our method and uses information on which predictors are noise, linear, quadratic, or beyond quadratic. The polynomial structure classification method has the smallest integrated squared error values among all methods for all generating distributions, and oracle 2 dominates oracle 1.
Table 2 summarizes the predictor classification performance of the methods under comparison. For the linear and nonlinear discovery method, the table entries are the average and standard deviation of the numbers of predictors identified as purely linear (X1), purely nonlinear (X2), a linear-nonlinear mixture (X3), and noise (X4, … , XD). Similarly, for the proposed method, the table entries are the average and standard deviation of the numbers of predictors identified as linear (X1), quadratic (X3), beyond quadratic (X2), and noise (X4, … , XD). Also reported are the percentages of times that all variables were correctly classified according to each method’s underlying classification scheme.
The proposed polynomial structure classification method performs well for all generative models, although both it and the linear and nonlinear discovery method generally perform less well for larger values of D, σ and η. Because the two methods are based on different classification schemes, predictor classification is not directly comparable, except with respect to the noise variables. The proposed method identified all noise variables for all 100 simulated datasets for all generative models, whereas the linear and nonlinear discovery method missed some noise predictors, more in cases where σ, η and especially D were large.
For one simulated dataset with σ = 1, η = 0·5 and D = 10, we plot in Fig. 1 the nonzero optimizers of (3) in the first variable selection step as functions of τ0. The optimizers corresponding to the important predictors, X1, X2 and X3, change to nonzero values quickly as τ0 increases and before any unimportant predictors show changes. Only four lines are visible in the graph because the optimizers corresponding to the other six predictors are zero for 0 ≤ τ0 ≤ 45.
Fig. 1.
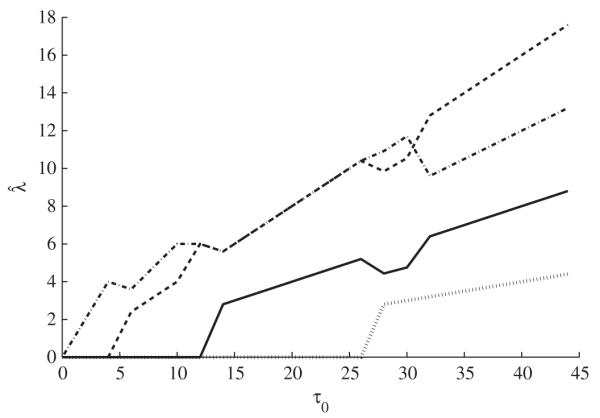
Solution paths to optimization problem (3) for one simulated dataset from Model 1, plotted over 0 < τ0 ≤ 45. At τ0 = 40 the paths are, from bottom to top, for variables X9, X1, X3 and X2.
Model 2
In the second data-generation model, with f1(t)=3t, f2(t)=−4t, f3(t)=2t, f4(t)=2 sin(2πt), f5(t)=3 sin(2πt)/{2 – sin(2πt)}, f6(t)=5[0·1 sin(2πt) + 0·2 cos(2πt) + 0·3{sin(2πt)}2 + 0·4{cos(2πt)}3 + 0·5{sin(2πt)}3] + 2t and f7(t)=2(3t – 1)2. Here ε ~ N(0, σ2) is independent of the predictors, with σ controlling the noise level; we take σ = 1 or 2. The dimension is D = 20 and so there are 13 noise predictors, X8, … , X20. For the linear and nonlinear discovery methods, X1, X2 and X3 are linear, X4 and X5 are nonlinear, and X6 and X7 are mixed linear-nonlinear. For our method, X1, X2 and X3 are linear, X7 is quadratic, and X4, X5 and X6 are beyond quadratic. Training samples of size n = 250 are used. Tables 3 and 4 present the results from 100 simulated datasets in the same format as for Model 1. Conclusions about the performance of the methods are similar to those for Model 1.
Table 3.
Simulation results for Model 2: average integrated squared errors (×102) with standard deviations (×102) in parentheses
| σ | η | Oracle 1 | SSANOVA | LAND | LANDr | PSC | Oracle 2 |
|---|---|---|---|---|---|---|---|
| 1 | 0·0 | 15 (4) | 25 (6) | 16 (5) | 17 (5) | 14 (4) | 14 (4) |
| 0·5 | 15 (4) | 25 (6) | 16 (5) | 17 (6) | 15 (6) | 13 (4) | |
| 2 | 0·0 | 52 (14) | 91 (22) | 58 (20) | 66 (19) | 56 (17) | 48 (13) |
| 0·5 | 47 (16) | 86 (24) | 77 (34) | 73 (31) | 64 (26) | 44 (15) |
σ, model error standard deviation; η, predictor correlation parameter; SSANOVA, smoothing spline analysis-of-variance method; LAND, linear and nonlinear discovery method; LANDr, linear and nonlinear discovery method, refitted version; PSC, the proposed polynomial structure classification method; Oracle 1, oracle for LAND and LANDr; Oracle 2, oracle for PSC.
Table 4.
Summary of predictor classification performance of the linear and nonlinear discovery method and the proposed method for Model 2
| LAND | PSC | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| σ | η | L | NL | LN | Nil | CC | L | Q | BQ | Nil | CC |
| 1 | 0·0 | 3·0 | 1·6 | 2·0 | 12·5 | 47 | 3·0 | 1·0 | 3·0 | 13·0 | 99 |
| 0·5 | 3·0 | 1·1 | 2·0 | 11·8 | 16 | 2·9 | 1·0 | 3·0 | 13·0 | 89 | |
| 2 | 0·0 | 3·0 | 1·5 | 1·9 | 11·2 | 14 | 2·8 | 1·0 | 3·0 | 13·0 | 74 |
| 0·5 | 3·0 | 0·8 | 1·8 | 9·7 | 1 | 2·2 | 1·0 | 2·8 | 13·0 | 23 | |
| Average SD | 0·2 | 0·6 | 0·2 | 2·0 | 0·3 | 0·1 | 0·1 | 0·1 | |||
LAND, linear and nonlinear discovery method; PSC, the proposed polynomial structure classification method; σ, model error standard deviation; η, predictor correlation parameter; L, linear; NL, nonlinear; L-N, linear-nonlinear mixture; Nil, noise; CC, proportion (%) of models with fully correct variable classifications; Q, quadratic; BQ, beyond quadratic; Average SD, block-averaged standard deviation.
8. Extension to partially linear models
The partially linear model is an extension of the additive model where, in addition to predictors X1, … , XD, there are predictors Z1, … , Zq known to contribute linearly, so that
| (5) |
The Zj commonly include indicators for categorical variables such as gender, race and location.
We illustrate the extension of our method to the partially linear model (5) via profiling by analysing the diabetes data from Willems et al. (1997). The goal of that study was to understand the prevalence of obesity, diabetes and other cardiovascular risk factors. The data include 18 variables on each of 403 African American subjects from central Virginia, with some missing values. The response variable in our analysis is glycosolated haemoglobin, which is of interest because a value greater than 7·0 is usually regarded as giving a positive diagnosis of diabetes.
We exclude two variables, the second systolic blood pressure and second diastolic blood pressure, as they are missingness-prone replicates of two other included variables, the first systolic and first diastolic blood pressures. Doing so leaves 15 candidate predictors. Twelve of these 15 variables are continuous and are rescaled to the unit interval: X1 = cholesterol, X2 = stabilized glucose, X3 = high density lipoprotein (hdl), X4 = cholesterol to hdl ratio, X5 = age, X6 = height, X7 = weight, X8 = first systolic blood pressure, X9 = first diastolic blood pressure, X10 = waist circumference, X11 = hip circumference, and X12 = postprandial time when samples were drawn. The remaining three variables are categorical variables for location, gender and frame. Location is a factor with two levels, Buckingham and Louisa; we set Z1 = 0 for Buckingham and Z1 = 1 otherwise. For gender we set Z2 =0 for female and Z2 =1 for male. The frame variable has three levels; we set Z3 = 0 and Z4 = 0 for small frames, Z3 = 1 and Z4 = 0 for medium frames, and Z3 = 0 and Z4 = 1 for large frames.
We fit the partially linear model (5) to study the dependence of glycosolated haemoglobin on the variables X1, … , X12, Z1, … , Z4, using the natural extension of the automatic structure recovery algorithm to discern the effects of X1, … , X12. We use the Bayesian information criterion to select tuning parameters. Our method identified X1, X3, X6, … , X12 as unimportant predictors; X5 was selected to have a linear effect, X4 a quadratic effect, and X2 a beyond-quadratic effect.
As Fig. 2(a) shows, on the original scale X4 has one outlier, 19·3, far outside the range of the other observations, which are between 1·5 and 12·2. Upon removing the outlier and reapplying our method, X1, X3, X4, X6, … , X12 are identified as unimportant, X5 as having a linear effect, and X2 as having a beyond-quadratic effect. The nonlinear fit for X2 is shown in Fig. 2(b).
Fig. 2.
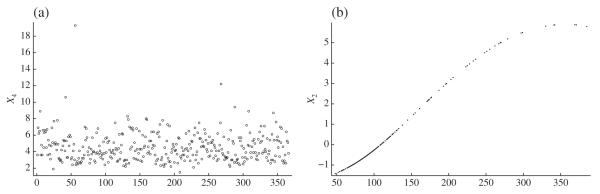
Results of applying our method to the diabetes data: (a) index plot of variable X4, with an outlier visible in the upper left corner; (b) the beyond-quadratic fit for X2 (stabilized glucose) after removing the X4 outlier.
9. Discussion
We have proposed and studied the properties of a new automatic structure recovery and estimation method for the additive model. The method is readily generalizable and can be extended to generalized additive models (Hastie & Tibshirani, 1990) and survival data models (Cheng & Lee, 2009).
Classifying predictors into those that are unimportant and those that contribute through polynomials of specified degree is accomplished by using a nonparametric variable selection method based on the results of Stefanski et al. (2014). Like the nonparametric classification method proposed by Stefanski et al. (2014), each step of our new method uses regularization by bounding a sum of inverse kernel bandwidths. Other choices of penalty, such as for fixed γ ≥ 0, are possible and worthy of study. Based on our limited experience, γ = 1 gives good overall performance. However, in light of the results in Stefanski et al. (2014), it is expected that γ < 1 would increase sparsity whereas γ > 1 would decrease sparsity but yield regression function estimators that have better mean-squared error properties.
As noted by a referee, modifications to our method and additional development of the theory to accommodate the case of a diverging dimension D would be useful directions to pursue. Alternatively, for data with large D, one could precede the use of our method with application of one of the screening procedures recently developed by Fan et al. (2011) and Cheng et al. (2014).
Acknowledgement
We thank two reviewers, an associate editor, and the editor for their most helpful comments. This research was supported by the U.S. National Institutes of Health and National Science Foundation.
Appendix
For d = 1, … , D, denote the density of Xd by fd(·). We assume the following technical conditions from Fan & Jiang (2005), as our method is based on backfitting coupled with local polynomial smoothing.
Condition A1
The kernel function K(·) is bounded and Lipschitz continuous with bounded support.
Condition A2
The densities fd(·) are Lipschitz continuous and bounded away from zero over their bounded supports.
Condition A3
For all pairs d and d’, the joint density functions of Xd and Xd are Lipschitz continuous on their supports.
Condition A4
For all d = 1, … , D, the (pd + 1)th derivatives of md(·) exist and are bounded and continuous.
Condition A5
The error has finite fourth moment, E(∣ε∣4) < ∞.
For the backfitting estimate coupled with local polynomial smoothing as defined in § 3, we have the following asymptotic result.
Lemma A1
Assume that Conditions A1–A5 hold and hd → 0 such that as n n → ∞, for d = 1, … , D. Then
| (A1) |
Proof
Lemma A1 is a straightforward consequence of the third step in the proof of Theorem 1 in Fan & Jiang (2005). The term on the left-hand side of (A1) is rss1/n in their notation, which they show converges in probability to σ2. Fan & Jiang (2005) did not explicitly state the rate of convergence, but it is readily deduced from their proof. We use their notation in the rest of this proof.
Note that rss1 = εAn2ε + BTB + 2BT(WM – In)ε and An2 = In + STS – S – ST + Rn2, according to Fan & Jiang (2005, p. 903). Thus it is enough to track the convergence rates of the different terms. Also, almost surely, according to (B.4) in Fan & Jiang (2005), and by definition. Consequently, . By line 6 from the bottom of the right-hand column of p. 901 in Fan & Jiang (2005), . By (B.11)–(B.24), . As Rn2 = O(1T1/n) uniformly over its elements, . Combining these terms and noting that completes the proof.
For k = 0, 1, … , let πk be the set of all polynomial functions of degree k. Define to be the set of predictors with corresponding additive component functions which are polynomial of degree at most that of the corresponding local polynomial used in the backfitting algorithm. Denote its complement by .
Proposition A1
Suppose that Conditions A1–A5 hold, that hd ≥ c0 > 0 for and some constant c > 0, and that hd’ → 0 and as n → ∞ for . Then
| (A2) |
Proof
Opsomer & Ruppert (1999) studied a backfitting estimator for semiparametric additive models and showed that the estimator of the parametric component is n1/2-consistent. The condition hd ≥ c0 > 0 for and some c0 > 0 implies that the smoothing bandwidth hd is bounded away from zero for . As md(·) ∈ πpd for , there is no approximation bias in using local polynomial smoothing to estimate the corresponding component in the backfitting algorithm. Thus the techniques of Opsomer & Ruppert (1999) can be used to show that the estimator of md(·) for is n1/2-consistent as hd > c0 > 0. The n1/2-consistent rate is faster than the consistency rate for smoothing other components.
Proof of Proposition 1
It is easy to show by contradiction that in probability for . If is bounded for some , then the objective function of (3) converges to the sum of σ2 plus an additional positive term due to smoothing bias. The additional bias term is caused by bounded , as the corresponding smoothing bandwidth does not shrink to zero. This is suboptimal, as the smallest limit of the objective is σ2. This proves that in probability for .
In (3), local constant smoothing is used for every component function. Thus pd = 0 for d 1, … , D. The second term on the right-hand side of (A1) or (A2) is due to bias, and the third term is due to variance when using local polynomial smoothing for every component. The condition as n → ∞ ensures that the variance term is dominated by the bias term. At the same time, note that a bounded and small λd for does not introduce any extra term.
If as n → ∞, consider for and for . In this case, diverges to infinity at a faster rate than for . Consequently, the asymptotic bias term on the right-hand side of (A2) using the values is smaller than the bias term on the right-hand side of (A1) using values. Here, smaller is in the sense of asymptotic order if , and in the sense of the constant multiplying the asymptotic order if is bounded. Thus the set of values with as n → ∞ is suboptimal, as we are solving a minimization problem (3). This shows that in probability for and therefore completes the proof.
Proof of Theorem 1
From Proposition 1, we have that as n → ∞. Conditional on , we can prove that as n → ∞ using arguments similar to those in the proof of Proposition 1. This process is iterated to complete the proof of the theorem.
References
- Cheng M-Y, Lee C-Y. Local polynomial estimation of hazard rates under random censoring. J. Chin. Statist. Assoc. 2009;47:19–38. [Google Scholar]
- Cheng M-Y, Honda T, Li J, Peng H. Nonparametric independence screening and structure identification for ultra-high dimensional longitudinal data. Ann. Statist. 2014;42:1819–49. [Google Scholar]
- Daubechies IB, Defrise M, De Mol C. An iterative thresholding algorithm for linear inverse problems with a sparsity constraint. Commun. Pure Appl. Math. 2004;57:1413–57. [Google Scholar]
- Engle RF, Granger CWJ, Rice J, Weiss A. Nonparametric estimates of the relation between weather and electricity sales. J. Am. Statist. Assoc. 1986;81:310–20. [Google Scholar]
- Fan J, Gijbels I. Local Polynomial Modelling and Its Applications. Chapman & Hall; London: 1996. [Google Scholar]
- Fan J, Jiang J. Nonparametric inference for additive models. J. Am. Statist. Assoc. 2005;100:890–907. [Google Scholar]
- Fan J, Li R. Variable selection via nonconcave penalized likelihood and its oracle properties. J. Am. Statist. Assoc. 2001;96:1348–60. [Google Scholar]
- Fan J, Lv J. A selective overview of variable selection in high dimensional feature space. Statist. Sinica. 2010;20:101–48. [PMC free article] [PubMed] [Google Scholar]
- Fan J, Feng Y, Song R. Nonparametric independence screening in sparse ultra-high dimensional additive models. J. Am. Statist. Assoc. 2011;106:544–57. doi: 10.1198/jasa.2011.tm09779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Ma Y, Dai W. Nonparametric independence screening in sparse ultra-high dimensional varying coefficient models. J. Am. Statist. Assoc. 2014;109:1270–84. doi: 10.1080/01621459.2013.879828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JH, Stuetzle W. Projection pursuit regression. J. Am. Statist. Assoc. 1981;76:817–23. [Google Scholar]
- Friedman JH, Hastie TJ, Hofling H, Tibshirani RJ. Pathwise coordinate optimization. Ann. Appl. Statist. 2007;1:302–32. [Google Scholar]
- Fu WJ. Penalized regressions: The bridge versus the lasso. J. Comp. Graph. Statist. 1998;7:397–416. [Google Scholar]
- Gu C. Smoothing Spline ANOVA Models. Springer; New York: 2002. [Google Scholar]
- Hastie TJ, Tibshirani RJ. Generalized Additive Models. Chapman & Hall; London: 1990. [Google Scholar]
- Huang J, Horowitz JL, Wei F. Variable selection in nonparametric additive models. Ann. Statist. 2010;38:2282–313. doi: 10.1214/09-AOS781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wei F, Ma S. Semiparametric regression pursuit. Statist. Sinica. 2012;22:1403–26. doi: 10.5705/ss.2010.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimeldorf GS, Wahba G. Some results on Tchebycheffian spline functions. J. Math. Anal. Applic. 1971;33:82–5. [Google Scholar]
- Opsomer JD, Ruppert D. A root-n consistent backfitting estimator for semiparametric additive modeling. J. Comp. Graph. Statist. 1999;8:715–32. [Google Scholar]
- Ravikumar PK, Lafferty JD, Liu H, Wasserman L. Sparse additive models. J. R. Statist. Soc. B. 2009;71:1009–30. [Google Scholar]
- Severini TA, Wong WH. Profile likelihood and conditionally parametric models. Ann. Statist. 1992;20:1768–802. [Google Scholar]
- Shen X, Pan W, Zhu Y, Zhou H. On constrained and regularized high-dimensional regression. Ann. Inst. Statist. Math. 2013;65:807–32. doi: 10.1007/s10463-012-0396-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanski LA, Wu Y, White K. Variable selection in nonparametric classification via measurement error model selection likelihoods. J. Am. Statist. Assoc. 2014;109:574–89. doi: 10.1080/01621459.2013.858630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani RJ. Regression shrinkage and selection via the lasso. J. R. Statist. Soc. B. 1996;58:267–88. [Google Scholar]
- Willems JP, Saunders JT, Hunt DE, Schorling JB. Prevalence of coronary heart disease risk factors among rural blacks: A community-based study. Southern Med. J. 1997;90:814–20. doi: 10.1097/00007611-199708000-00008. [DOI] [PubMed] [Google Scholar]
- Wu TT, Lange K. Coordinate descent procedures for lasso penalized regression. Ann. Appl. Statist. 2008;2:224–44. [Google Scholar]
- Xia Y, Zhang W, Tong H. Efficient estimation for semivarying-coefficient models. Biometrika. 2004;91:661–81. [Google Scholar]
- Zhang HH, Lu W. Adaptive lasso for Cox’s proportional hazards model. Biometrika. 2007;94:691–703. [Google Scholar]
- Zhang HH, Cheng G, Liu Y. Linear or nonlinear? Automatic structure discovery for partially linear models. J. Am. Statist. Assoc. 2011;106:1099–112. doi: 10.1198/jasa.2011.tm10281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H. The adaptive lasso and its oracle properties. J. Am. Statist. Assoc. 2006;101:1418–29. [Google Scholar]


