Abstract
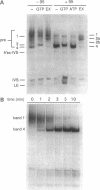
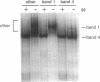
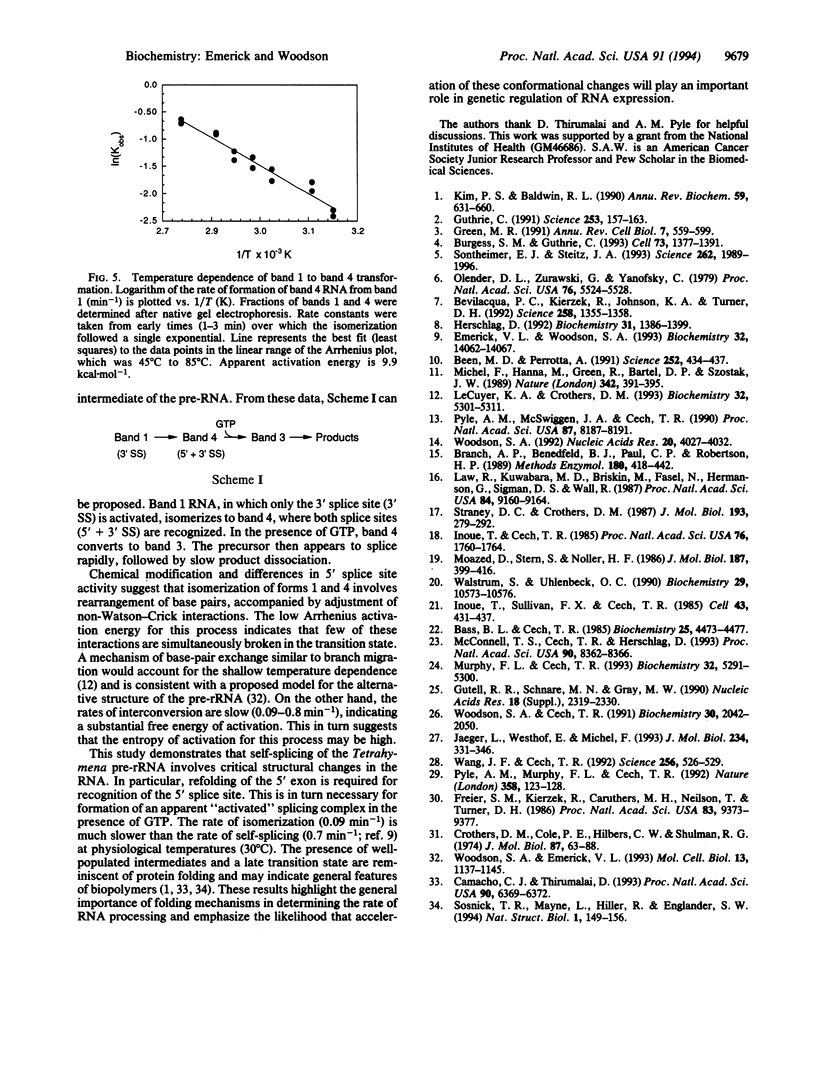
Evidence that folding of the Tetrahymena pre-rRNA follows a defined path and is rate-determining for splicing at physiological temperatures is presented. Structural isomers were separated by native polyacrylamide gel electrophoresis and their splicing activities were compared. GTP binding selectively shifts the active form of the pre-RNA to an electrophoretic band containing both spliced and unspliced RNA. In situ chemical modification provides evidence for base-pair rearrangements in the 5' exon and structural alterations in the intron core of partially and fully active forms. Transition to the fully active precursor requires high temperature, but the activation energy is lower than expected for opening of RNA helices. Implications for control of RNA conformation during splicing are discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bass B. L., Cech T. R. Ribozyme inhibitors: deoxyguanosine and dideoxyguanosine are competitive inhibitors of self-splicing of the Tetrahymena ribosomal ribonucleic acid precursor. Biochemistry. 1986 Aug 12;25(16):4473–4477. doi: 10.1021/bi00364a001. [DOI] [PubMed] [Google Scholar]
- Been M. D., Perrotta A. T. Group I intron self-splicing with adenosine: evidence for a single nucleoside-binding site. Science. 1991 Apr 19;252(5004):434–437. doi: 10.1126/science.2017681. [DOI] [PubMed] [Google Scholar]
- Bevilacqua P. C., Kierzek R., Johnson K. A., Turner D. H. Dynamics of ribozyme binding of substrate revealed by fluorescence-detected stopped-flow methods. Science. 1992 Nov 20;258(5086):1355–1358. doi: 10.1126/science.1455230. [DOI] [PubMed] [Google Scholar]
- Branch A. D., Benenfeld B. J., Paul C. P., Robertson H. D. Analysis of ultraviolet-induced RNA-RNA cross-links: a means for probing RNA structure-function relationships. Methods Enzymol. 1989;180:418–442. doi: 10.1016/0076-6879(89)80115-6. [DOI] [PubMed] [Google Scholar]
- Burgess S. M., Guthrie C. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993 Jul 2;73(7):1377–1391. doi: 10.1016/0092-8674(93)90363-u. [DOI] [PubMed] [Google Scholar]
- Camacho C. J., Thirumalai D. Kinetics and thermodynamics of folding in model proteins. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6369–6372. doi: 10.1073/pnas.90.13.6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crothers D. M., Cole P. E., Hilbers C. W., Shulman R. G. The molecular mechanism of thermal unfolding of Escherichia coli formylmethionine transfer RNA. J Mol Biol. 1974 Jul 25;87(1):63–88. doi: 10.1016/0022-2836(74)90560-9. [DOI] [PubMed] [Google Scholar]
- Emerick V. L., Woodson S. A. Self-splicing of the Tetrahymena pre-rRNA is decreased by misfolding during transcription. Biochemistry. 1993 Dec 21;32(50):14062–14067. doi: 10.1021/bi00213a040. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Gutell R. R., Schnare M. N., Gray M. W. A compilation of large subunit (23S-like) ribosomal RNA sequences presented in a secondary structure format. Nucleic Acids Res. 1990 Apr 25;18 (Suppl):2319–2330. doi: 10.1093/nar/18.suppl.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991 Jul 12;253(5016):157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- Herschlag D. Evidence for processivity and two-step binding of the RNA substrate from studies of J1/2 mutants of the Tetrahymena ribozyme. Biochemistry. 1992 Feb 11;31(5):1386–1399. doi: 10.1021/bi00120a015. [DOI] [PubMed] [Google Scholar]
- Inoue T., Sullivan F. X., Cech T. R. Intermolecular exon ligation of the rRNA precursor of Tetrahymena: oligonucleotides can function as 5' exons. Cell. 1985 Dec;43(2 Pt 1):431–437. doi: 10.1016/0092-8674(85)90173-4. [DOI] [PubMed] [Google Scholar]
- Jaeger L., Westhof E., Michel F. Monitoring of the cooperative unfolding of the sunY group I intron of bacteriophage T4. The active form of the sunY ribozyme is stabilized by multiple interactions with 3' terminal intron components. J Mol Biol. 1993 Nov 20;234(2):331–346. doi: 10.1006/jmbi.1993.1590. [DOI] [PubMed] [Google Scholar]
- Kim P. S., Baldwin R. L. Intermediates in the folding reactions of small proteins. Annu Rev Biochem. 1990;59:631–660. doi: 10.1146/annurev.bi.59.070190.003215. [DOI] [PubMed] [Google Scholar]
- Law R., Kuwabara M. D., Briskin M., Fasel N., Hermanson G., Sigman D. S., Wall R. Protein-binding site at the immunoglobulin mu membrane polyadenylylation signal: possible role in transcription termination. Proc Natl Acad Sci U S A. 1987 Dec;84(24):9160–9164. doi: 10.1073/pnas.84.24.9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCuyer K. A., Crothers D. M. The Leptomonas collosoma spliced leader RNA can switch between two alternate structural forms. Biochemistry. 1993 May 25;32(20):5301–5311. doi: 10.1021/bi00071a004. [DOI] [PubMed] [Google Scholar]
- McConnell T. S., Cech T. R., Herschlag D. Guanosine binding to the Tetrahymena ribozyme: thermodynamic coupling with oligonucleotide binding. Proc Natl Acad Sci U S A. 1993 Sep 15;90(18):8362–8366. doi: 10.1073/pnas.90.18.8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Hanna M., Green R., Bartel D. P., Szostak J. W. The guanosine binding site of the Tetrahymena ribozyme. Nature. 1989 Nov 23;342(6248):391–395. doi: 10.1038/342391a0. [DOI] [PubMed] [Google Scholar]
- Moazed D., Stern S., Noller H. F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986 Feb 5;187(3):399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Murphy F. L., Cech T. R. An independently folding domain of RNA tertiary structure within the Tetrahymena ribozyme. Biochemistry. 1993 May 25;32(20):5291–5300. doi: 10.1021/bi00071a003. [DOI] [PubMed] [Google Scholar]
- Oxender D. L., Zurawski G., Yanofsky C. Attenuation in the Escherichia coli tryptophan operon: role of RNA secondary structure involving the tryptophan codon region. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5524–5528. doi: 10.1073/pnas.76.11.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle A. M., McSwiggen J. A., Cech T. R. Direct measurement of oligonucleotide substrate binding to wild-type and mutant ribozymes from Tetrahymena. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8187–8191. doi: 10.1073/pnas.87.21.8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle A. M., Murphy F. L., Cech T. R. RNA substrate binding site in the catalytic core of the Tetrahymena ribozyme. Nature. 1992 Jul 9;358(6382):123–128. doi: 10.1038/358123a0. [DOI] [PubMed] [Google Scholar]
- Sontheimer E. J., Steitz J. A. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993 Dec 24;262(5142):1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- Sosnick T. R., Mayne L., Hiller R., Englander S. W. The barriers in protein folding. Nat Struct Biol. 1994 Mar;1(3):149–156. doi: 10.1038/nsb0394-149. [DOI] [PubMed] [Google Scholar]
- Straney D. C., Crothers D. M. Comparison of the open complexes formed by RNA polymerase at the Escherichia coli lac UV5 promoter. J Mol Biol. 1987 Jan 20;193(2):279–292. doi: 10.1016/0022-2836(87)90219-1. [DOI] [PubMed] [Google Scholar]
- Walstrum S. A., Uhlenbeck O. C. The self-splicing RNA of Tetrahymena is trapped in a less active conformation by gel purification. Biochemistry. 1990 Nov 20;29(46):10573–10576. doi: 10.1021/bi00498a022. [DOI] [PubMed] [Google Scholar]
- Wang J. F., Cech T. R. Tertiary structure around the guanosine-binding site of the Tetrahymena ribozyme. Science. 1992 Apr 24;256(5056):526–529. doi: 10.1126/science.1315076. [DOI] [PubMed] [Google Scholar]
- Woodson S. A., Cech T. R. Alternative secondary structures in the 5' exon affect both forward and reverse self-splicing of the Tetrahymena intervening sequence RNA. Biochemistry. 1991 Feb 26;30(8):2042–2050. doi: 10.1021/bi00222a006. [DOI] [PubMed] [Google Scholar]
- Woodson S. A., Emerick V. L. An alternative helix in the 26S rRNA promotes excision and integration of the Tetrahymena intervening sequence. Mol Cell Biol. 1993 Feb;13(2):1137–1145. doi: 10.1128/mcb.13.2.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson S. A. Exon sequences distant from the splice junction are required for efficient self-splicing of the Tetrahymena IVS. Nucleic Acids Res. 1992 Aug 11;20(15):4027–4032. doi: 10.1093/nar/20.15.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]