Abstract
Eukaryotic gene expression is developmentally regulated, in part by chromatin remodelling, and its dysregulation has been linked to cancer. CHD5 (chromodomain helicase DNA-binding protein 5) is a tumour suppressor gene (TSG) that maps to a region of consistent deletion on 1p36.31 in neuroblastomas (NBs) and other tumour types. CHD5 encodes a protein with chromatin remodelling, helicase and DNA-binding motifs that is preferentially expressed in neural and testicular tissues. CHD5 is highly homologous to CHD3 and CHD4, which are the core subunits of nucleosome remodelling and deacetylation (NuRD) complexes. To determine if CHD5 forms a similar complex, we performed studies on nuclear extracts from NBLS, SY5Y (both with endogenous CHD5 expression), NLF (CHD5 null) and NLF cells stably transfected with CHD5 cDNA (wild-type and V5–histidine-tagged). Immunoprecipitation (IP) was performed with either CHD5 antibody or antibody to V5/histidine-tagged protein. We identified NuRD components both by GST–FOG1 (Friend Of GATA1) pull-down and by IP. We also performed MS/MS analysis to confirm the presence of CHD5 or other protein components of the NuRD complex, as well as to identify other novel proteins. CHD5 was clearly associated with all canonical NuRD components, including metastasis-associated protein (MTA)1/2, GATA zinc finger domain containing 2A (GATAD2A), histone deacetylase (HDAC)1/2, retinoblastoma-binding protein (RBBP)4/7 and methyl DNA-binding domain protein (MBD)2/3, as determined by Western blotting and MS/MS. Our data suggest CHD5 forms a NuRD complex similar to CHD4. However, CHD5–NuRD may also have unique protein associations that confer functional specificity and may contribute to normal development and to tumour suppression in NB and other cancers.
Keywords: chromodomain helicase DNA-binding protein 5 (CHD5), neuroblastoma, nuclear localization, nucleosome remodelling histone deacetylase (NuRD) complex, tumour suppressor
Introduction
Neuroblastoma (NB) is the most common extracranial solid tumour of childhood. This tumour of the sympathetic nervous system accounts for 8%–10% of childhood cancers and 15% of childhood cancer deaths [1,2]. NBs may regress spontaneously, especially in infants, or grow relentlessly despite intensive, multimodality therapy. We and others have identified distinct patterns of genomic changes that underlie these disparate clinical behaviours [1,3]. Deletion of the short arm of chromosome 1 (1p) has been observed in 35% of primary NBs and 70% of NB-derived cell lines [4–6]. We first identified 1p deletion as a characteristic change in advanced stage NBs [7], which presumably reflects loss of one or more tumour suppressor genes (TSGs) from this region. We used DNA-based polymorphisms [6,8] to refine the smallest region of consistent deletion (SRD) of 1p36 to a ∼2 Mb region on 1p36.31 and we identified CHD5 (chromodomain helicase DNA-binding protein 5) as a bona fide TSG from this region in NBs [9–12]. CHD5 expression is low or absent from NB cell lines and most high-risk tumours and low expression is associated with unfavourable features and outcome [9–11,13,14]. Bagchi et al. [15] also identified CHD5 as a TSG on the orthologous region of mouse chromosome 4 using a chromosome engineering approach. Furthermore, CHD5 has been implicated as a TSG in a variety of other cancers, such as gliomas and cancers of the colon, breast, lung, ovary, prostate, stomach, larynx and gall bladder [15–27].
CHD5 is a member of the chromodomain-helicase-DNA-binding (CHD) family [11,28,29]. Currently the CHD family has nine members and they are divided into three subfamilies [27,30]. CHD1 and CHD2 comprise the first subfamily, which contains a classic DNA-binding domain. The second subfamily consists of CHD3 (Mi2α) and CHD4 (Mi2β), which have two PHD-zinc finger motifs and each forms a nucleosome remodelling and deacetylation (NuRD) complex. The third subfamily consists of CHD6–CHD9, which was originally identified based on structural homology to other known CHD members [29,31]. Of all the CHD family members, CHD5 is most homologous to CHD3 and CHD4. Indeed, CHD3–CHD5 are the only members that have two PHD domains near the N-terminus, in addition to paired chromodomains and a split SWItch/Sucrose NonFermentable (SWI/SNF)-like helicase/ATPase domain [32]. Thus, CHD5 belongs most appropriately in the second subfamily. However, CHD5 is expressed almost exclusively in the nervous system and in testis, whereas CHD3 and CHD4 are expressed more ubiquitously [27,33].
Gene regulation is a highly co-ordinated process that involves ordered recruitment of transcriptional machinery to maintain proper chromatin structure and chromatin remodelling proteins play crucial roles in this process [34]. NuRD complexes were first identified more than 15 years ago [34,35] and they alter chromatin structure and gene expression in part by causing ATP-dependent remodelling of nucleosomes [36]. Most studies have focused on the Mi2β/CHD4–NuRD complex, which has both ATPase and histone deacetylase activity [34]. Other canonical NuRD components are: metastasis-associated proteins 1 and 2 (MTA1/2), retinoblastoma-binding proteins (RBBP)4 and RBBP7 (formerly RbAp48 and RbAp46 respectively), GATAD2A and -B (formerly p66α/β) and methyl DNA-binding domain proteins (MBD2/3) [28,37,38].
NuRD complexes have been implicated in regulating gene transcription, genome stability, DNA damage and DNA repair [28,37,39–41]. Altered NuRD function is associated with a variety of cancers, as described above [15–27]. CHD5 has also been shown to regulate the expression of neural-specific genes that are implicated in aging, as well as in Alzheimer's disease [42,43]. However, it has been unclear if CHD5 forms a NuRD complex similar to CHD3 and CHD4 or whether CHD5 forms unique protein associations. In the present study, we provide evidence that CHD5 forms a bona fide NuRD complex with all the same canonical proteins as CHD4. Furthermore, CHD5 forms novel protein associations that may account for the functional differences between CHD4– and CHD5–NuRD complexes.
Experimental
Reagents
Cell culture media [Roswell Park Memorial Institute medium (RPMI-1640)], antibiotics and FBS were obtained from Invitrogen Inc. Parental NB cell lines were maintained in our laboratory but are also available from the A.T.C.C. Restriction enzymes and other molecular biology reagents were purchased from Roche Applied Sciences, Promega Inc. and New England Biolabs Inc. ‘Complete’ protease inhibitor cocktail tablets were obtained from Roche Applied Sciences. Halt protease and phosphatase inhibitor cocktail was obtained from Sigma–Aldrich Company. Nuclear extraction kits were obtained from Thermo Scientific. NuPAGE gels (4%–12%), buffers and prestained Rainbow molecular mass markers were obtained from Invitrogen.
Antibodies
Rabbit polyclonal CHD5 antibody and goat primary polyclonal antibodies for MTA-1, MTA-2, RBBP4, RBBP7, histone deacetylase (HDAC)1, HDAC2, MBD2 and MBD3 were from Santa Cruz Biotechnology. CHD4 antibody was from Bethyl Laboratories. Polyclonal antibodies for HDAC1, HDAC2, RBBP7, RBBP4/7 and MDB3 were from Cell Signaling Technology. GATAD2A antibody was from Upstate Biotechnology. Tagged V5–His was from Invitrogen. Secondary antibodies were from GE Healthcare Life Sciences and Santa Cruz.
Plasmid cloning
CHD5–ORF2 was amplified directly from brain total RNA (by cDNA cloning) and subcloned after PCR amplification. Expression plasmids for the CHD5 transcript were generated in a eukaryotic V5–His-tagged pcDNA3.1 expression plasmid at NotI and XhoI sites. DNA sequencing from both ends and restriction enzyme analyses confirmed transcript fidelity. GST and GST– FOG1 [N-terminal 45 amino acids (aa)] plasmids were described previously [44].
Cell culture and transfections
We transfected the NLF NB cell line [9–12] either with a CHD5 expression vector (NLF–CHD5), with or without a V5–His-tag or with empty pcDNA 3.1 vector. NB cell lines (NBLS, SY5Y and NLF) were cultured in RPMI-1640 + 10% FBS and gentamicin, along with the appropriate selection antibiotics. Transfections of NLF cells were performed using Lipofectamine 2000 (Invitrogen). Cells were cultured for 6–8 weeks in the presence of G418 (400 μg/ml) to select single cell clones. Single cell suspensions were cultured in the presence of neomycin to obtain clones derived from single cells.
Immunofluorescence
Parental NLF and NLF–CHD5 were cultured on cover slips in six-well plates. Forty-eight hours after plating, cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) in PBS for 15 min at room temperature. Cells were washed gently twice and permeabilized with 0.2% Triton X-100 in 1% BSA, followed by immunostaining using either rabbit anti-CHD5 (1:1000) or tagged monoclonal anti-mouse V5–His (1:1000; Invitrogen) followed by Alexa Fluor 488, goat anti-rabbit IgG (A11008) and Fluor 546, goat anti-mouse IgG (A11030) as secondary antibodies (1:400–1:2000). Coverslips were mounted and exposed to ProLong®Gold Antifade Reagent with DAPI (0.1 μg/ml, Molecular Probes) for 10 min to stain nuclei. Images were captured using a phase contrast microscope and analysed using Olympus 1×70 with Slidebook4 software (Universal Imaging).
Cell extract preparation
Subcellular fractionated proteins (cytosol, membrane, nuclear and cytoskeleton) were extracted using commercially available kits from ThermoFisher Scientific. Nuclear and cytosolic fractions were prepared with two buffer (low- and high-salt) extraction, as described previously [44]. Protein concentrations were determined using Bradford Protein Assay Reagent with SmartSpec Plus spectrophotometer (BIO-RAD Laboratories).
Immunoprecipitation
Immunoprecipitations (IPs) of CHD5– and CHD4–NuRD complexes were performed using nuclear protein (100–2000 μg) from NBLS, SY5Y, parental NLF, as well as NLF–CHD5 cells. Typical IP buffer contained protease (Sigma P-8340; 1:500) and phosphatase inhibitors (50 mM Na fluoride, 10 mM Na pyrophosphate, 5 mM Na vanadate). Extracts were pre-cleared with agarose beads and incubated overnight using either anti-CHD5 antibody (1–25 μl) or V5–His tag antibody (1–10 μl) in the presence of protease inhibitors. Bound complexes were washed and lysed in 20 μl lysis buffer and aliquots were applied to either 4%–12% gradient or SDS/PAGE (10% gel).
Western analysis and GST–FOG1 pull-downs
Western blot analyses for CHD5 and CHD5–NuRD components were performed following PAGE, as described previously [44,45], using NuPAGE Bis-Tris gels with MOPS-SDS running buffer (Invitrogen). Proteins were transferred to nitrocellulose membranes (GE Healthcare Life Sciences) and probed with antibodies as described above using ECL detection (Invitrogen). We performed in vitro binding studies of GST, GST–FOG1 (45 aa N-terminal fragment) a GATA1 cofactor, to pull down a CHD4 (Mi-2β), as described earlier [44]. We used a GST– FOG1 construct to pull down complete NuRD complexes from NB cells. Twenty micrograms of either GST or GST–FOG1 was incubated with 2 mg of NLF or NLF–CHD5 nuclear extract overnight in buffer containing 150 mM NaCl, 50 mM Tris/HCl (pH 7.5), 0.5% Igepal (Sigma), protease inhibitor cocktail (P-8340, Sigma, 1:500) and 1 mM DTT. Bound proteins were washed twice in the above buffer with 350 mM NaCl, then three times with 650 mM NaCl buffer, followed by an additional wash in 350 mM NaCl, then separated by SDS/PAGE and stained with SimplyBlue (Invitrogen), a modified form of Coomassie Blue. Aliquots were also analysed by Western blot with appropriate NuRD component antibodies.
Protein sequence analysis by LC-MS/MS
LC-MS/MS analysis to identify proteins associated with CHD4 and CHD5 was performed at the Taplin Biological Mass Spectrometry Facility (Harvard Medical School). Nuclear extracts were immunoprecipitated with specific antibody and subjected to SDS/PAGE (10% gel) followed by Coomassie/Simply Blue staining. Excised gel bands were cut into approximately 1 mm3 pieces. Gel pieces were then subjected to a modified in-gel trypsin digestion procedure [46]. Gel pieces were washed and dehydrated with acetonitrile for 10 min followed by removal of acetonitrile. Pieces were then completely dried in a speed-vac. Rehydration of the gel pieces was with 50 mM ammonium bicarbonate solution containing 12.5 ng/μl modified sequencing-grade trypsin (Promega) at 4 °C. After 45 min, the excess trypsin solution was removed and replaced with 50 mM ammonium bicarbonate solution to just cover the gel pieces. Samples were then placed in a 37°C room overnight. Peptides were later extracted by removing the ammonium bicarbonate solution, followed by one wash with a solution containing 50% acetonitrile and 1% formic acid. The extracts were dried in a speed-vac (∼1 h). The samples were then stored at 4°C until analysis.
On the day of analysis, the samples were reconstituted in 5– 10 μl of HPLC solvent A (2.5% acetonitrile, 0.1% formic acid). A nanoscale reverse-phase HPLC capillary column was created by packing 5 μm of C18 spherical silica beads into a fused silica capillary (100 μm inner diameter × ∼25 cm length) with a flame-drawn tip [47]. After equilibrating the column, each sample was loaded via a Famos auto sampler (LC Packings) on to the column. A gradient was formed and peptides were eluted with increasing concentrations of solvent B (97.5% acetonitrile, 0.1% formic acid).
As peptides eluted, they were subjected to ESI and then entered into an LTQ Orbitrap Velos Pro ion-trap mass spectrometer (ThermoFisher Scientific). Peptides were detected, isolated and fragmented to produce an MS/MS of specific fragment ions for each peptide. Peptide sequences (and hence protein identity) were determined by matching the acquired fragmentation pattern with protein databases using the software program, Sequest (ThermoFisher) [48]. All databases include a reversed version of all the sequences and the data were filtered to between a 1% and 2% false discovery rates (FDRs). Results were tabulated in columns, indicating gene symbol/name, total peptides identified and unique peptides for specific protein.
Results
CHD5 expression is localized to the nucleus
In order to determine the subcellular localization of CHD5, we performed a series of Western analyses using various cell fractions (cytosol, membrane, nuclear and chromatin bound nuclear). Our results indicate that the CHD5 was detected only in the nuclear fractions from NLF–CHD5, but absent from parental NLF cells, whereas CHD4 was detected in both NLF–CHD5 and parental NLF cells (Figures 1A and 1B). CHD5 protein was detected in both nuclear and chromatin-bound nuclear fractions, but it was not detected in appreciable amounts in cytoplasmic or membrane compartments. To validate this subcellular fractionation, blots were stripped and probed with cytosol-specific antibodies such as glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and the larger subunit of NF-kB (nuclear factor kappa-light-chain-enhancer of activated B-cells) p65. As anticipated, we detected GAPDH and NF-kB proteins only in the cytosolic fractions (Figure 1A), indicating that our cell fractions were specific to each subcellular compartment. In order to confirm that these observations were not due to CHD5 overexpression in NLF transfected cells, we performed Western blot analysis using nuclear extracts from NBLS and SY5Y cells that express endogenous CHD5. We observed that both NBLS and SY5Y cells expressed CHD4 and CHD5 only in the nucleus, although the expression level of CHD5 was much lower in NBLS and SY5Y cells when compared with NLF–CHD5 (Figure 1B). Expression patterns of cytosol-specific markers GAPDH and NF-kB p65 were consistent whether the cells expressed CHD5 endogenously or by overexpression (Figure 1B).
Figure 1. Nuclear localization of CHD5: Western analysis.
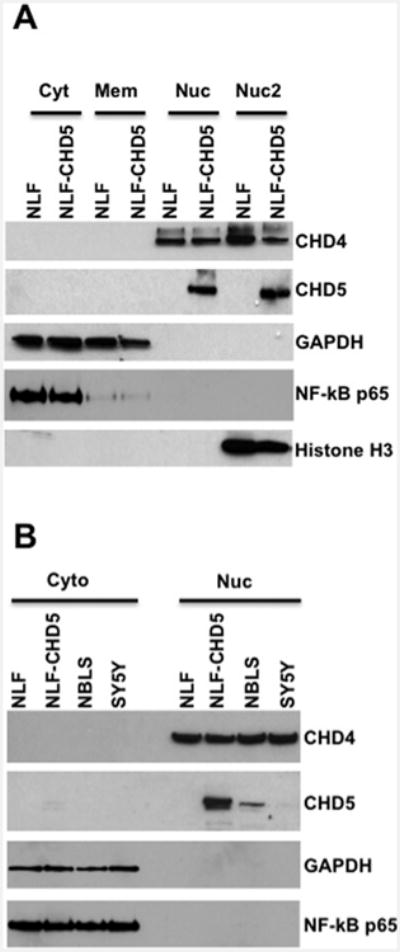
Various NB cell lines were subjected to SDS/PAGE (4 %–12% gels) and probed with fraction-specific antibodies as shown on the right. (A) Subcellular fractions, cytosol, membrane, nuclear (Nuc) and chromatin-bound nuclear (Nuc2), for NLF and NLF–CHD5. (B) Cellular fractions, cytosol and nuclear, for NLF, NLF–CHD5, NBLS and NY5Y. CHD5 (∼250 kDa) was detected only in the nuclear fraction from NLF–CHD5-S, NBLS and SY5Y (very low expression), but not in cytosolic or membrane fractions. CHD4 was present in all nuclear fractions as expected in all tested cell lines. Blots were stripped and probed with GAPDH and p65, the larger subunit of NF-kB, as markers for cytosolic fractions.
To further confirm these observations of nuclear localization of CHD5, we performed immunofluorescence (IF) studies with a CHD5-specific polyclonal antibody. Parental NLF cells did not express CHD5 protein and served as negative control. IF results from transfected NLF–CHD5 cells demonstrated that CHD5 was expressed only in the nucleus (Figure 2A). This was confirmed by analysis of independent NLF clones expressing CHD5 tagged with the V5–His epitope using a V5–His antibody, which also demonstrated exclusive expression in the nucleus (Figure 2B). We also performed parallel experiments with the NBLS cell line that expresses CHD5 endogenously and we obtained similar results (Figure 2C). These results demonstrate that CHD5 is expressed exclusively in the nucleus, consistent with a role in chromatin remodelling, similar to CHD4.
Figure 2. Nuclear localization of CHD5-IF.
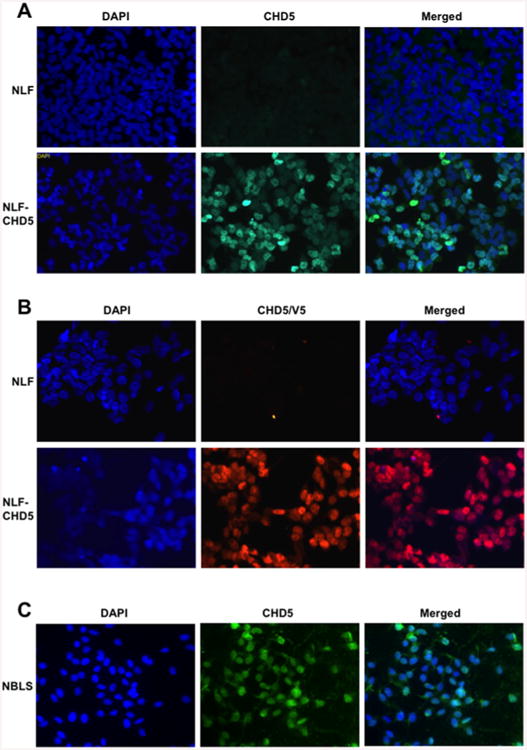
(A) Parental NLF cells (top row) and NLF–CHD5 (second row) were cultured for 48 h. Cells were immunostained with CHD5 as indicated. Nuclei were stained with DAPI (left panels). Nuclear localization of CHD5 was observed when cells overexpressed CHD5 (middle panels), whereas no nuclear staining was observed in control NLF cells (upper middle panel). Merged images are shown on right. (B) IF images indicating nuclear expression with tagged V5–His antibody. (C) Nuclear expression of CHD5 in NBLS cells expressing endogenous CHD5.
CHD5 associates with all NuRD complex proteins
CHD4–NuRD binds to the transcription cofactor FOG1 via a highly conserved short motif at the FOG1 N-terminus [44] and this region is required for the transcription functions of FOG1 [49]. Fusion of this peptide to GST allows purification of the CHD4– NuRD complex to high purity by a single step. To determine if CHD5 forms a NuRD complex similar to CHD4, we performed GST–FOG1 pull-downs with nuclear extracts from parental NLF and NLF–CHD5, followed by stringent washing with 150– 650 mM NaCl. Bound proteins were subjected to SDS/PAGE and stained with SimplyBlue (Figure 3A). Well-separated, stained proteins (Figure 3A) were also excised and identified by MS/MS. All bands were identified as known components of the CHD4–NuRD complex, as described above. Protein complexes were also subjected to Western blot with antibodies to individual NuRD components (Figure 3B). As expected, all NuRD complex proteins were detected, including MTA1 and -2, HDAC1 and -2, GATAD2A and B, RBBP4, RBBP7 and MBD3 with GST– FOG1 pull-down, but none were detected with GST pull-down alone (Figure 3B). Notably, CHD5 also co-purified as part of the complex. Similar results were obtained with the nuclear extracts of NB cell line NBLS that express endogenous CHD5 protein. IP and Western results confirmed the presence of all canonical NuRD components (Figure 3C). Thus, CHD5 associates with all known members of a CHD4–NuRD complex in these NB cell lines, suggesting that it also forms a NuRD-type chromatin-remodelling complex.
Figure 3. Detection of CHD4– and CHD5–NuRD components: GST–FOG1 pull-down.
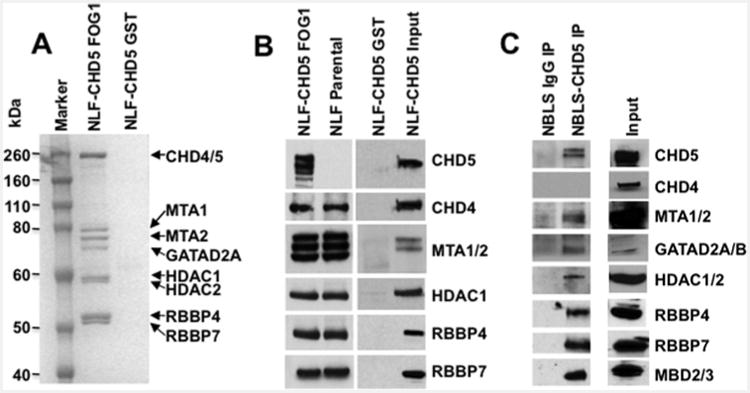
(A) Nuclear extracts of NLF–CHD5 were subjected to either GST alone or GST–FOG1 pull-down followed by PAGE and stained with Simply Blue. All canonical NuRD components from CHD4/5–NuRD complexes were readily visible (marked with arrows). (B) GST–FOG1 bound protein complexes from NLF–CHD5 and NLF parental cells were subjected to 4%–12% PAGE. The identity of individual proteins from the NuRD complex was confirmed by Western blot analysis. CHD5 was detected only in NLF–CHD5-S extracts but absent from NLF extracts. Additional bands in the CHD5 lane are probably due to proteolytic degradation products and additional bands with MTA1/2 probably represent alternate isoform recognition. (C) Identification of individual canonical NuRD components in NBLS cells. Nuclear extracts were immunoprecipitated with either IgG or CHD5 antibody as indicated and complexes were subjected to SDS/PAGE (4%–12% gels) followed by Western blot analysis. MTA1/2, GATAD2A/B, HDAC1/2, RBBP4/7 and MBD2/3 were detected as individual canonical NuRD components along with CHD5.
Since FOG1 recruits NuRD complexes via binding to MTA1/2 and/or RBBP4/7 subunits [44], it was unclear whether CHD5 associates with the entire complex or only select subunits. To test this, we performed IP of NLF cells transfected with V5–His-tagged CHD5 using a V5–His antibody and IP of endogenous CHD4 with a CHD4-specific antibody. Western blotting again demonstrated that CHD5 associated with all known CHD4– NuRD components (Figure 4). In order to further confirm that these interactions are not due to overexpression, we performed IP experiments using nuclear extracts from NBLS cells that express CHD5 endogenously. Our results confirmed the association of CHD5 protein with all NuRD components in this line as well (Figure 3C). Together, these results indicate that CHD5 associates with all members of a canonical NuRD complex and probably forms a similar complex.
Figure 4. Detection of CHD5–NuRD components: IP and Western analysis.
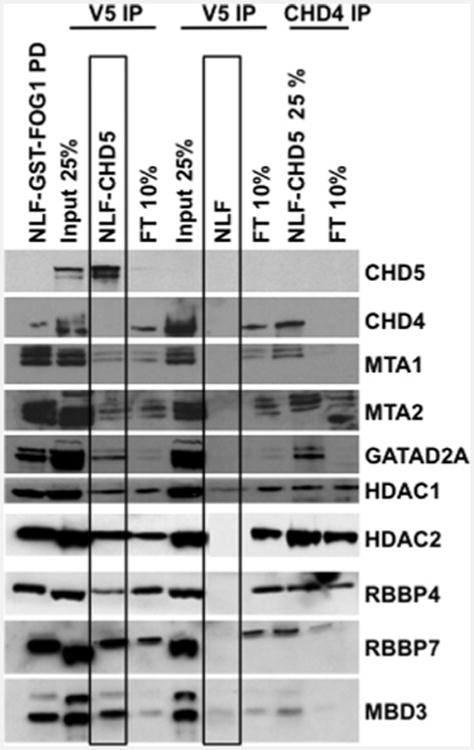
Nuclear extracts from NLF–CHD5 and NLF were immunoprecipitated as indicated. Subsequent proteins from IP and GST–FOG1 pull-down (left lane) were subjected to 4%–12% PAGE. Extracts from NLF following GST–FOG1 pull-down served as negative control. Individual NuRD components were detected by Western blotting with antibodies specific to each NuRD component. The boxes around the NLF–CHD5 and NLF lanes highlight the identification of all canonical NuRD components in NLF–CHD5 and not in NLF as a negative control, as these cells do not express CHD5 endogenously. FT, flow through.
In order to further confirm the association of CHD5 with other NuRD complex proteins, we analysed gel fractions from CHD4 IP and CHD5 IP, (as described in the ‘Experimental’ section) using LC-MS/MS. Nuclear extracts from NLF and NLF– CHD5 cells were isolated and subjected to IP with CHD4-and CHD5-specific antibodies independently. Immune complexes were separated on SDS/PAGE gels and stained with Simply Blue. Stained proteins were excised as single bands or regions of the gel and subjected to MS/MS analysis. We identified all known canonical NuRD components of CHD4–NuRD: MTA1/2, GATAD2A/B, HDAC1/2, RBBP4/7 and MBD2/3. All of the same components were also identified in the CHD5 IP (Table 1), strongly suggesting that CHD4 forms a NuRD complex similar or identical with CHD4. Additional proteins unique to either CHD4 or CHD5 complexes were also identified, which we are investigating.
Table 1. IP/MS identified known canonical NuRD components.
Nuclear extracts expressing CHD5 were immunoprecipitated with CHD5 (A) or CHD4 (B) antibodies independently followed by SDS/PAGE analysis. Gels were stained with Simply Blue and excised gel bands were sent for MS analysis. MS analysis revealed the list of known canonical NuRD components as shown below. Total, number of total peptides present in a given sample application; Unique, number of peptides that are unique and specific to the indicated gene/protein description.
| A. CHD5–NuRD | |||
|---|---|---|---|
|
| |||
| Gene symbol | Description | Unique | Total |
| CHD5 | Chromodomain helicase DNA-binding protein 5 | 56 | 97 |
| MTA1 | Metastasis-associated 1 | 18 | 21 |
| MTA2 | Metastasis-associated 2 | 27 | 34 |
| GATAD2A/p66α | GATA zinc finger domain containing 2A | 23 | 41 |
| GATAD2B/p66β | GATA zinc finger domain containing 2B | 15 | 26 |
| HDAC1 | Histone deacetylase 1 | 11 | 16 |
| HDAC2 | Histone deacetylase 2 | 7 | 11 |
| RBBP4/RbAp48 | Retinoblastoma-binding protein 4 | 14 | 20 |
| RBBP7/RbAp46 | Retinoblastoma-binding protein 7 | 4 | 5 |
| MBD2 | Methyl-CpG-binding domain protein 2 | 4 | 4 |
| MBD3 | Methyl-CpG-binding domain protein 3 | 7 | 8 |
| B. CHD4–NuRD | |||
|
| |||
| Gene symbol | Description | Unique | Total |
|
| |||
| CHD4 | Chromodomain helicase DNA-binding protein 4 | 91 | 275 |
| MTA1 | Metastasis-associated 1 | 20 | 30 |
| MTA2 | Metastasis-associated 2 | 28 | 45 |
| GATAD2A/p66α | GATA zinc finger domain containing 2A | 27 | 77 |
| GATAD2B/p66β | GATA zinc finger domain containing 2B | 25 | 37 |
| HDAC1 | Histone deacetylase 1 | 15 | 28 |
| HDAC2 | Histone deacetylase 2 | 10 | 26 |
| RBBP4/RbAp48 | Retinoblastoma-binding protein 4 | 18 | 42 |
| RBBP7/RbAp46 | Retinoblastoma-binding protein 7 | 5 | 6 |
| MBD2 | Methyl-CpG-binding domain protein 2 | 7 | 8 |
| MBD3 | Methyl-CpG-binding domain protein 3 | 9 | 14 |
Discussion
CHD4/Mi2β is highly conserved across species and it is expressed abundantly in most tissues [28]. However, CHD5 expression was recently localized to neurons in the brain of rodents and so it may have a role in neural development as well as in neurological diseases, such as aging and Alzheimer's disease [42,43,50]. CHD5 may play different roles in different cell types, so the suppression of cell growth and facilitating chromatin condensation are only two aspects of this protein's chromatin-remodelling functions. Egan et al. [42] showed that neuronal differentiation requires direct binding of CHD5 to H3K27me3. Moreover, it has been reported in a mouse model system that depletion of CHD5 in the developing neocortex blocks neuronal differentiation, which leads to an accumulation of undifferentiated neural progenitors [42]. We demonstrated that CHD5 was also expressed at high levels in the testis and CHD5 deficiency causes a failure of developmentally-regulated chromatin condensation during spermatogenesis [33]. This finding has been confirmed recently by others [51]. In addition, high expression of chromatin remodelling factors, including CHD5, are associated with normal spermatogenesis, whereas decreased expression of these genes is closely associated with round spermatid arrest [52].
CHD5 inactivation may contribute to the failure of chromatin condensation of selected domains of DNA, which may contribute to tumorigenesis [42]. Furthermore, CHD5 has also been implicated in the pathogenesis of a variety of cancers in adults and children, including NB [15–27] so its dysregulation by deletion and/or epigenetic modification may affect other tissues as well. Relatively little is known about its function, but our results strongly suggest that CHD5 functions as part of a NuRD-type chromatin-remodelling complex. Nevertheless, the exact mechanism by which CHD5 functions as a TSG in NBs or other cancers is still unclear. Paul et al. [53] recently reported that PHD-mediated histone 3 binding, involving chromatin mediated transcriptional regulation, is required for CHD5-mediated tumour suppression. Whether CHD5 functions independently of a NuRD-type complex is unknown at present.
We demonstrate, in the present study, by both GST–FOG1 pulldown experiments and IP studies, that CHD5 is associated with all canonical members of a CHD4–NuRD complex (MTA1/2, HDAC1/2, GATAD2A/B, RBBP4/7, MBD2/3). Thus, based on its homology with CHD4 (Supplementary Table S1), its nuclear localization and its association with all typical CHD4– NuRD components, it is extremely likely that CHD5 forms a NuRD-type chromatin-remodelling complex. This is consistent with the report by Potts et al. [43] who also found an association of CHD5 with several NuRD components. It is unknown whether CHD5 competes with CHD4 or CHD3 for NuRD components, but all NuRD components appear to be expressed abundantly. Our data provide evidence that CHD5 must have different functions than CHD4 in NBs as well as other tissues.
First, CHD4 is expressed ubiquitously in almost all tissues in the body, whereas CHD5 expression is very restricted, with the highest expression in the nervous system and testis [11,33,42,43]. Second, NBs with 1p deletion and virtually no CHD5 expression have abundant expression of CHD4, yet they grow readily both in vitro and in vivo [9,10,13]. When these same NBs were transfected with CHD5, clonigenicity and tumorigenicity were dramatically suppressed [9]. Indeed, CHD5 transfection has a similar effect on other cancers with 1p36 deletion and CHD5 transcriptional silencing [19–24,54–56]. Thus, CHD4– NuRD cannot simply substitute for CHD5–NuRD in these cells.
One formal possibility was that CHD5 did not associate with all the same proteins as did CHD4 in a NuRD complex. However, our data demonstrate by both Western analysis and MS analysis that all components known to be associated with CHD4–NuRD (MTA1/2, GATAD2A/B, HDAC1/2, RBBP4/7 and MBD2/3) were also associated with CHD5 (Table 1). Nevertheless, CHD4 is expressed abundantly in NLF, yet cell growth, colony formation and tumour formation is inhibited only when CHD5 is introduced [9]. This suggests that CHD5 may have protein interactions with other proteins or functions that are entirely independent of its role as part of a NuRD complex and we are exploring both possibilities.
The functional characterization of CHD5, alone or as part of a CHD5–NuRD complex, would provide insight into the role of CHD5 in epigenetic modification of gene expression in normal development and in cancer, including NB. Targeting CHD5 for up-regulation would be theoretically possible, because even in tumours with 1p deletion and loss of one copy, the remaining allele is rarely if ever mutated. Instead, the promoter of the remaining allele is frequently methylated, resulting in epigenetic silencing [9,14] and transcription could be silenced by other mechanisms, such as histone modification. Therefore, reversing the epigenetic modifications that result in transcriptional silencing of CHD5 [19– 23] should restore expression of an intact allele and presumably restore growth control. Thus, it will be important to identify the genes and proteins that regulate CHD5 expression, the genes that CHD5 regulates and the proteins with which CHD5 interacts, to fully understand the role of CHD5 in normal development as well as malignant transformation.
Supplementary Material
Supplementary Table S1: Shown here is the schematic diagram of amino acid alignment map for CHD3, CHD4 and CHD5 proteins indicating the sequence homology. Consensus sequence is shown in the bottom lane and the color description is as follows based on the nature of the R-group of amino acid.
Acknowledgments
Authors thank Dr Linda Gonzales from the Division of Neonatology for her expert help in fluorescence microscopy. We also thank Ross Tomaino at Taplin Biological Mass Spectrometry Facility, at Harvard Medical School, Boston, MA for peptide analysis.
Funding: This work was supported by the National Institutes of Health [grant number R01-CA39771]; the Alex's Lemonade Stand Foundation; and the Audrey E. Evans Chair in Molecular Oncology (to G.M.B).
Abbreviations
- aa
amino acids
- CHD
chromodomain-helicase-DNA binding
- CHD4/5
chromodomain helicase DNA-binding protein4/5
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HDAC
histone deacetylase complex
- IF
immunofluorescence
- IP
immunoprecipitation
- MBD
methyl DNA-binding protein
- MTA
metastasis-associated protein
- NB
neuroblastoma
- NF-kB
nuclear factor kappa-light-chain-enhancer of activated B-cells
- NuRD
nucleosome remodelling and deacetylation
- RBBP
retinoblastoma-binding protein
- RPMI
Roswell Park Memorial Institute medium
- TSG
tumour suppressor gene
Footnotes
Author Contribution: Venkatadri Kolla designed and performed all experiments. Koumudi Naraparaju conducted and assisted Western blot analysis. Tiangang Zhuang, Mayumi Higashi and Sriharsha Kolla assisted in conducting experiments and preparation of manuscript. Gerd Blobel gave valuable suggestions for the present study and provided the GST–FOG1 reagent for these analyses. Garrett Brodeur conceived of the initial experimental design, supervised the experiments, analysed the data, interpreted results and helped write and edit the manuscript to its final form.
References
- 1.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 2.Brodeur GM, Hogarty MD, Mosse YP, Maris JM. Neuroblastoma. In: Pizzo PA, Poplack DG, editors. Principles and Practice of Pediatric Oncology. Lippincott, Williams and Wilkins; Philadelphia: 2011. pp. 886–922. [Google Scholar]
- 3.Mosse YP, Greshock J, Margolin A, Naylor T, Cole K, Khazi D, Hii G, Winter C, Shahzad S, Asziz MU, et al. High-resolution detection and mapping of genomic DNA alterations in neuroblastoma. Genes Chromosomes Cancer. 2005;43:390–403. doi: 10.1002/gcc.20198. [DOI] [PubMed] [Google Scholar]
- 4.Maris JM, White PS, Beltinger CP, Sulman EP, Castleberry RP, Shuster JJ, Look AT, Brodeur GM. Significance of chromosome 1p loss of heterozygosity in neuroblastoma. Cancer Res. 1995;55:4664–4669. [PubMed] [Google Scholar]
- 5.Attiyeh EF, London WB, Mosse YP, Wang Q, Winter C, Khazi D, McGrady PW, Seeger RC, Look AT, Shimada H, et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353:2243–2253. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- 6.Fong CT, Dracopoli NC, White PS, Merrill PT, Griffith RC, Housman DE, Brodeur GM. Loss of heterozygosity for the short arm of chromosome 1 in human neuroblastomas: correlation with N-myc amplification. Proc Natl Acad Sci USA. 1989;86:3753–3757. doi: 10.1073/pnas.86.10.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brodeur GM, Sekhon G, Goldstein MN. Chromosomal aberrations in human neuroblastomas. Cancer. 1977;40:2256–2263. doi: 10.1002/1097-0142(197711)40:5<2256::aid-cncr2820400536>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 8.White PS, Maris JM, Beltinger C, Sulman E, Marshall HN, Fujimori M, Kaufman BA, Biegel JA, Allen C, Hilliard C, et al. A region of consistent deletion in neuroblastoma maps within human chromosome 1p36.2–36.3. Proc Natl Acad Sci USA. 1995;92:5520–5524. doi: 10.1073/pnas.92.12.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujita T, Igarashi J, Okawa ER, Gotoh T, Manne J, Kolla V, Kim J, Zhao H, Pawel BR, London WB, et al. CHD5, a tumor suppressor gene deleted from 1p36.31 in neuroblastomas. J Natl Cancer Inst. 2008;100:940–949. doi: 10.1093/jnci/djn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okawa ER, Gotoh T, Manne J, Igarashi J, Fujita T, Silverman KA, Xhao H, Mosse YP, White PS, Brodeur GM. Expression and sequence analysis of candidates for the 1p36.31 tumor suppressor gene deleted in neuroblastomas. Oncogene. 2008;27:803–810. doi: 10.1038/sj.onc.1210675. [DOI] [PubMed] [Google Scholar]
- 11.Thompson PM, Gotoh T, Kok M, White PS, Brodeur GM. CHD5, a new member of the chromodomain gene family, is preferentially expressed in the nervous system. Oncogene. 2003;22:1002–1011. doi: 10.1038/sj.onc.1206211. [DOI] [PubMed] [Google Scholar]
- 12.White PS, Thompson PM, Gotoh T, Okawa ER, Igarashi J, Kok M, Winter C, Gregory SG, Hogarty MD, Maris JM, Brodeur GM. Definition and characterization of a region of 1p36.3 consistently deleted in neuroblastoma. Oncogene. 2005;24:2684–2694. doi: 10.1038/sj.onc.1208306. [DOI] [PubMed] [Google Scholar]
- 13.Garcia I, Mayol G, Rodriguez E, Sunol M, Gershon TR, Rios J, Cheung NK, Kieran MW, George RE, Perez-Atayde AR, et al. Expression of the neuron-specific protein CHD5 is an independent marker of outcome in neuroblastoma. Mol Cancer. 2010;9:277. doi: 10.1186/1476-4598-9-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koyama H, Zhuang T, Light JE, Kolla V, Higashi M, McGrady PW, London WB, Brodeur GM. Mechanisms of CHD5 Inactivation in neuroblastomas. Clin Cancer Res. 2012;18:1588–1597. doi: 10.1158/1078-0432.CCR-11-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bagchi A, Papazoglu C, Wu Y, Capurso D, Brodt M, Francis D, Bredel M, Vogel H, Mills AA. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128:459–475. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 16.Law ME, Templeton KL, Kitange G, Smith J, Misra A, Feuerstein BG, Jenkins RB. Molecular cytogenetic analysis of chromosomes 1 and 19 in glioma cell lines. Cancer Genet Cytogenet. 2005;160:1–14. doi: 10.1016/j.cancergencyto.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 17.Lang J, Tobias ES, Mackie R. Preliminary evidence for involvement of the tumour suppressor gene CHD5 in a family with cutaneous melanoma. Br J Dermatol. 2011;164:1010–1016. doi: 10.1111/j.1365-2133.2011.10223.x. [DOI] [PubMed] [Google Scholar]
- 18.Ng D, Yang XR, Tucker MA, Goldstein AM. Mutation screening of CHD5 in melanoma-prone families linked to 1p36 revealed no deleterious coding or splice site changes. BMC Res Notes. 2008;1:86. doi: 10.1186/1756-0500-1-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu X, Zhu Z, Li W, Fu X, Su D, Fu L, Zhang Z, Luo A, Sun X, Fu L, Dong JT. Chromodomain helicase DNA binding protein 5 plays a tumor suppressor role in human breast cancer. Breast Cancer Res. 2012;14:R73. doi: 10.1186/bcr3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mokarram P, Kumar K, Brim H, Naghibalhossaini F, Saberi-firoozi M, Nouraie M, Green R, Lee E, Smoot DT, Ashktorab H. Distinct high-profile methylated genes in colorectal cancer. PLoS One. 2009;4:e7012. doi: 10.1371/journal.pone.0007012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai C, Ashktorab H, Pang X, Zhao Y, Sha W, Liu Y, Gu X. MicroRNA-211 expression promotes colorectal cancer cell growth in vitro and in vivo by targeting tumor suppressor CHD5. PLoS One. 2012;7:e29750. doi: 10.1371/journal.pone.0029750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Lau KK, So LK, Lam YW. CHD5 is down-regulated through promoter hypermethylation in gastric cancer. J Biomed Sci. 2009;16:95. doi: 10.1186/1423-0127-16-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao R, Yan Q, Lv J, Huang H, Zheng W, Zhang B, Ma W. CHD5, a tumor suppressor that is epigenetically silenced in lung cancer. Lung Cancer. 2012;76:324–331. doi: 10.1016/j.lungcan.2011.11.019. [DOI] [PubMed] [Google Scholar]
- 24.Gorringe KL, Choong DY, Williams LH, Ramakrishna M, Sridhar A, Qiu W, Bearfoot JL, Campbell IG. Mutation and methylation analysis of the chromodomain-helicase-DNA binding 5 gene in ovarian cancer. Neoplasia. 2008;10:1253–1258. doi: 10.1593/neo.08718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong RR, Chan LK, Tsang TP, Lee CW, Cheung TH, Yim SF, Siu NS, Lee SN, Yu MY, Chim SS, et al. chd5 downregulation associated with poor prognosis in epithelial ovarian cancer. Gynecol Obstet Invest. 2011;72:203–207. doi: 10.1159/000323883. [DOI] [PubMed] [Google Scholar]
- 26.Robbins CM, Tembe WA, Baker A, Sinari S, Moses TY, Beckstrom-Sternberg S, Beckstrom-Sternberg J, Barrett M, Long J, Chinnaiyan A, et al. Copy number and targeted mutational analysis reveals novel somatic events in metastatic prostate tumors. Genome Res. 2011;21:47–55. doi: 10.1101/gr.107961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolla V, Zhuang T, Higashi M, Naraparaju K, Brodeur GM. Role of CHD5 in human cancers: 10 years later. Cancer Res. 2014;74:652–658. doi: 10.1158/0008-5472.CAN-13-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai AY, Wade PA. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer. 2011;11:588–596. doi: 10.1038/nrc3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marfella CG, Imbalzano AN. The Chd family of chromatin remodelers. Mutat Res. 2007;618:30–40. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall JA, Georgel PT. CHD proteins: a diverse family with strong ties. Biochem Cell Biol. 2007;85:463–476. doi: 10.1139/O07-063. [DOI] [PubMed] [Google Scholar]
- 31.Ramirez J, Dege C, Kutateladze TG, Hagman J. MBD2 and multiple domains of CHD4 are required for transcriptional repression by Mi-2/NuRD complexes. Mol Cell Biol. 2012;32:5078–5088. doi: 10.1128/MCB.00819-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woodage T, Basrai MA, Baxevanis AD, Hieter P, Collins FS. Characterization of the CHD family of proteins. Proc Natl Acad Sci USA. 1997;94:11472–11477. doi: 10.1073/pnas.94.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhuang T, Hess RA, Kolla V, Higashi M, Raabe TD, Brodeur GM. CHD5 is required for spermiogenesis and chromatin condensation. Mech Dev. 2014;131:35–46. doi: 10.1016/j.mod.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 35.Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 36.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- 38.Allen HF, Wade PA, Kutateladze TG. The NuRD architecture. Cell Mol Life Sci. 2013;70:3513–3524. doi: 10.1007/s00018-012-1256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, He S, Tu Y, Ji P, Zong J, Zhang J, Feng F, Zhao J, Gao G, Zhang Y. Downregulation of chromatin remodeling factor CHD5 is associated with a poor prognosis in human glioma. J Clin Neurosci. 2013;20:958–963. doi: 10.1016/j.jocn.2012.07.021. [DOI] [PubMed] [Google Scholar]
- 40.Stanley FK, Moore S, Goodarzi AA. CHD chromatin remodelling enzymes and the DNA damage response. Mutat Res. 2013;750:31–44. doi: 10.1016/j.mrfmmm.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 41.Li DQ, Kumar R. Mi-2/NuRD complex making inroads into DNA-damage response pathway. Cell Cycle. 2010;9:2071–2079. doi: 10.4161/cc.9.11.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Egan CM, Nyman U, Skotte J, Streubel G, Turner S, O'Connell DJ, Rraklli V, Dolan MJ, Chadderton N, Hansen K, et al. CHD5 is required for neurogenesis and has a dual role in facilitating gene expression and polycomb gene repression. Dev Cell. 2013;26:223–236. doi: 10.1016/j.devcel.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 43.Potts RC, Zhang P, Wurster AL, Precht P, Mughal MR, Wood WH, III, Zhang Y, Becker KG, Mattson MP, Pazin MJ. CHD5, a brain-specific paralog of Mi2 chromatin remodeling enzymes, regulates expression of neuronal genes. PLoS One. 2011;6:e24515. doi: 10.1371/journal.pone.0024515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hong W, Nakazawa M, Chen YY, Kori R, Vakoc CR, Rakowski C, Blobel GA. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J. 2005;24:2367–2378. doi: 10.1038/sj.emboj.7600703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 46.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 47.Peng J, Gygi SP. Proteomics: the move to mixtures. J Mass Spectrom. 2001;36:1083–1091. doi: 10.1002/jms.229. [DOI] [PubMed] [Google Scholar]
- 48.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 49.Miccio A, Wang Y, Hong W, Gregory GD, Wang H, Yu X, Choi JK, Shelat S, Tong W, Poncz M, Blobel GA. NuRD mediates activating and repressive functions of GATA-1 and FOG-1 during blood development. EMBO J. 2010;29:442–456. doi: 10.1038/emboj.2009.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vestin A, Mills AA. The tumor suppressor Chd5 is induced during neuronal differentiation in the developing mouse brain. Gene Expr Patterns. 2013;13:482–489. doi: 10.1016/j.gep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li W, Wu J, Kim SY, Zhao M, Hearn SA, Zhang MQ, Meistrich ML, Mills AA. Chd5 orchestrates chromatin remodelling during sperm development. Nat Commun. 2014;5:3812. doi: 10.1038/ncomms4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steilmann C, Cavalcanti MC, Bergmann M, Kliesch S, Weidner W, Steger K. Aberrant mRNA expression of chromatin remodelling factors in round spermatid maturation arrest compared with normal human spermatogenesis. Mol Hum Reprod. 2010;16:726–733. doi: 10.1093/molehr/gaq054. [DOI] [PubMed] [Google Scholar]
- 53.Paul S, Kuo A, Schalch T, Vogel H, Joshua-Tor L, McCombie WR, Gozani O, Hammell M, Mills AA. Chd5 requires PHD-mediated histone 3 binding for tumor suppression. Cell Rep. 2013;3:92–102. doi: 10.1016/j.celrep.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mulero-Navarro S, Esteller M. Chromatin remodeling factor CHD5 is silenced by promoter CpG island hypermethylation in human cancer. Epigenetics. 2008;3:210–215. doi: 10.4161/epi.3.4.6610. [DOI] [PubMed] [Google Scholar]
- 55.Qu Y, Dang S, Hou P. Gene methylation in gastric cancer. Clin Chim Acta. 2013;424:53–65. doi: 10.1016/j.cca.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 56.Wang J, Chen H, Fu S, Xu ZM, Sun KL, Fu WN. The involvement of CHD5 hypermethylation in laryngeal squamous cell carcinoma. Oral Oncol. 2011;47:601–608. doi: 10.1016/j.oraloncology.2011.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1: Shown here is the schematic diagram of amino acid alignment map for CHD3, CHD4 and CHD5 proteins indicating the sequence homology. Consensus sequence is shown in the bottom lane and the color description is as follows based on the nature of the R-group of amino acid.


