Abstract
A three-dimensional (3-D) cell culture system that allows control of both substrate stiffness and integrin binding density was created and characterized. This system consisted of two self-assembling peptide (SAP) sequences that were mixed in different ratios to achieve the desired gel stiffness and adhesiveness. The specific peptides used were KFE ((acetyl)-FKFEFKFE-CONH2), which has previously been reported not to support cell adhesion or MVN formation, and KFE-RGD ((acetyl)-GRGDSP-GG-FKFEFKFE-CONH2), which is a similar sequence that incorporates the RGD integrin binding site. Storage modulus for these gels ranged from ~60 to 6000 Pa, depending on their composition and concentration. Atomic force microscopy revealed ECM-like fiber microarchitecture of gels consisting of both pure KFE and pure KFE-RGD as well as mixtures of the two peptides. This system was used to study the contributions of both matrix stiffness and adhesiveness on microvascular network (MVN) formation of endothelial cells and the morphology of human mesenchymal stem cells (hMSC). When endothelial cells were encapsulated within 3-D gel matrices without binding sites, little cell elongation and no network formation occurred, regardless of the stiffness. In contrast, matrices containing the RGD binding site facilitated robust MVN formation, and the extent of this MVN formation was inversely proportional to matrix stiffness. Compared with a matrix of the same stiffness with no binding sites, a matrix containing RGD-functionalized peptides resulted in a ~2.5-fold increase in the average length of network structure, which was used as a quantitative measure of MVN formation. Matrices with hMSC facilitated an increased number and length of cellular projections at higher stiffness when RGD was present, but induced a round morphology at every stiffness when RGD was absent. Taken together, these results demonstrate the ability to control both substrate stiffness and binding site density within 3-D cell-populated gels and reveal an important role for both stiffness and adhesion on cellular behavior that is cell-type specific.
Keywords: Three-dimensional cell culture, Self-assembling peptide, Microvascular networks, Stiffness, Binding site density
1. Introduction
There is a growing body of literature demonstrating that cell function is affected by substrate stiffness [1]. For cultured fibroblasts, substrate stiffness affects the rate [2] and direction [3] of cell migration, focal adhesion [2] and stress fiber formation [4,5], responsiveness to exogenous growth factors [6] and proliferation [7]. The spreading of smooth muscle cells is dependent on both the density of binding ligands on a surface and the stiffness of the material [8]. Neurons have increased branching densities when cultured on soft substrates, while glial cells—which are normally co-cultured with these neurons—only survive on more rigid substrates [9]. Substrate stiffness influences the differentiation of mesenchymal stem cells, with soft, intermediate and stiff materials being neurogenic, myogenic and osteogenic, respectively [10].
The aforementioned studies cultured cells on top of protein-laminated polyacrylamide gels. Cells do not directly bind to the polyacrylamide, but instead interact with the laminating layer of protein. The concentration of this protein layer can be held constant while changing the stiffness of the polyacrylamide gel, and thus the cells can be exposed to a constant binding site density while the stiffness is varied [8,9]. The most widely recognized limitation of the system, however, is that living cells cannot be embedded within polyacrylamide gels, but instead can only be cultured on top of them (i.e. two-dimensional (2-D) culture) [11,12]. While it is tempting to assume that cells cultured on top of a biomaterial represent a simpler but faithful 2-D analog of a three-dimensional (3-D) environment, this assumption is not well justified by the available literature. For example, relative to cells cultured on top of planar surfaces, cells cultured within 3-D gels exhibit altered morphology [13–15], morphogenesis [16–18], cytoskeletal organization [19,20], matrix adhesion complexes [21–23], proliferation [24], survival [25] and differentiation [20,26]. Thus, there is a growing consensus that a 3-D biomaterial environment is crucial to properly study many key cellular functions [27–29].
The behavior of cells cultured within 3-D extracellular matrix (ECM) gels changes as the hydrogel concentration is altered [30–32]. Since gel stiffness rises with increasing gel concentration, these changes in behavior are often attributed to matrix stiffness [33]. Other factors, however, such as cell–matrix binding site density, pore size, fiber diameter and diffusion rates, can also vary with gel concentration. Thus, decoupling stiffness from these other variables is essential to evaluate cell behavior properly as a function of gel concentration [34]. A number of approaches have been used to isolate the independent contribution of stiffness, such as altering mechanical boundary conditions [30,35,36], crosslinking [37–39] or composition [32,40–42]. While such approaches to control stiffness in natural polymers have many merits, they are often limited by the interdependence of fiber size, pore size and stiffness, as well as an inability to control both matrix stiffness and binding site density.
The self-assembling peptides (SAP) used in this study consist of alternating hydrophilic and hydrophobic amino acids that form a fibrous hydrogel structure when exposed to millimolar salt solutions [43–47]. Jung et al. [48] recently described the co-assembly of a SAP that lacked known binding sites with another SAP of the same base sequence that contained RGD or IKVAV integrin binding sites. Using this system, they demonstrated that endothelial cells adhere to, spread out on and proliferate more when seeded on top of a gel that included integrin binding sites, compared with a SAP lacking an integrin binding sequence. While this system has many merits, it is not useful for studying the effects of matrix stiffness within a 3-D environment because the endothelial cells were cultured as a 2-D monolayer on top of the SAP gels, in a similar way to what has been done with polyacrylamide systems. Herein, a co-assembly system that supports the culture of cells within 3-D gels is described. The present system consists of one SAP that lacks integrin binding sites and another SAP possessing the same base sequence that contains RGD, similar to that of Jung et al. The RGD sequence was chosen because it is the cell recognition site of a large number of ECM and platelet adhesion proteins, including vitronectin, type I collagen, fibrinogen, von Willebrand factor, osteopontin, thrombospondin, other collagens and laminin [49]. Endothelial cells express at least 11 different integrins [50], of which six recognize RGD (α3β1, α5β1, αvβ1, αvβ3, αvβ5 and αvβ8). Furthermore, the significance of RGD to integrin attachment is demonstrated through the inhibition of cell adhesion to surfaces coated with various ECM proteins when soluble RGD containing peptides are present in the medium [51].
Rheology was used to determine the stiffness profile of the SAP system, and its fiber morphology was characterized using atomic force microscopy (AFM). Using this system to control both matrix stiffness and integrin binding site density, the present authors studied the response of both endothelial cells and human mesenchymal stem cells (hMSC) embedded within the 3-D gels to these parameters in terms of microvascular network (MVN) formation and cellular projections, respectively. MVN formation is a useful model system because of its importance in physiology, pathology and tissue engineering. In addition, previous studies that have explored the impact of only matrix stiffness on MVN formation served as comparisons in an effort to understand the contributions of both stiffness and binding site density [30,32,37,39–42,47]. The choice of HUVEC allowed for a more direct comparison with these studies since they are a popular cell type for such experiments [30,32,41,42,47].
2. Materials and methods
2.1. SAP gelation and cell culture
Peptide sequences (acetyl)-GRGDSP-GG-FKFEFKFE-CONH2 (KFE-RGD) and (acetyl)-FKFEFKFE-CONH2 (KFE) from BiomerTechnology (Pleasanton, CA) were dissolved in deionized water to a final concentration of 1% (10 mg ml−1) and sonicated for 20–60 min. Mixtures of the two peptides were also sonicated before use in cell studies.
Human umbilical vein endothelial cells (HUVEC) (Lonza, Walkersville, MD) were cultured in EGM-2 (Lonza) on collagen-coated flasks and used between passages 4 and 7. Cells were suspended in peptide gels as previously reported [42]. Briefly, HUVEC were washed with PBS, trypsin and then Versene (all from Invitrogen, Carlsbad, CA). After they detached, the cells were pelleted, washed in 20% sucrose, pelleted and resuspended in 20% sucrose. The cell–sucrose mixture was mixed 1:1 with peptide (mixtures of KFE and KFE-RGD) to achieve a final concentration of 2.5 × 106 cells ml−1. Cell-peptide mixtures (150 μl) were gently pipetted into 12 mm membrane inserts (PICM-012–50, Millipore, Billerica, MA) pre-wet from the bottom with 300 μl gelation media (complete EGM-2 with 10% serum added). After being allowed to solidify for ~5 min, 700 μl of gelation medium was added to the top of the gel and the outside of the insert. After 1 h, this medium was exchanged for regular medium supplemented with 50 ng ml−1 phorbol-myristate acetate (Sigma–Aldrich, St. Louis, MO) and 50 ng ml−1 vascular endothelial growth factor (R&D Systems, Minneapolis, MN). Supplemented media was replaced after 24 h, and the gels were fixed in 4% paraformaldehyde after 48 h.
The hMSC (Lonza) were expanded with MSC growth medium (Lonza) in T-75 flasks and used by passage 5. The hMSC were encapsulated within SAP gels in the same manner as HUVEC, with the exception that MSC growth medium was used for gelation and culture.
2.2. Morphology visualization and quantification
After fixation, HUVEC-containing gels were stained with Alexa 633 phalloidin (160 nM, Invitrogen) and DAPI (Sigma), mounted on glass slides and were subsequently imaged on a confocal microscope (405 and 633 lasers; Leica TCS SP5 AOBS confocal system, Bensheim, Germany). Quantification was performed as previously described [30] with slight modification. Briefly, 10 μm confocal stacks were taken (1 μm spacing) and then compressed into a single maximum projection image using ImageJ (NIH, Bethesda, MD). Using a custom-written ImageJ macro, images were thresholded, skeletonized and the average length of skeletonized objects (ALS) was recorded for five images for each gel. Data represent three independent experiments and five to seven gels for each condition.
After 24 h of culture within SAP gels, hMSC were imaged by differential interference contrast (DIC) microscopy (Olympus IX81, Center Valley, PA). The gels were taken directly from the incubator and imaged immediately, and thus fixatives were not used. All images were taken near the middle of the gels to ensure that the imaged cells were in a true 3-D environment rather than growing on the top or bottom of the gel. Multiple images were taken of three separate gel replicates, and a total of 20 cells were randomly selected from these images at each gel condition to be used for quantification. The number of projections for each of these cells was counted manually, and the total length of these projections for each cell was measured manually using ImageJ.
2.3. Rheology
Cell-free SAP gels were made and analyzed using a previously described method [42]. Previous experiments with similar SAP gels indicate that the stiffness values are uniform and stable 16 h after formation of the gels [42]. The rheological measurements used the parallel plate rheometry method and were performed at 24 h after gel formation to ensure the stability of the gels. KFE gels were cast into steel washers (8 mm ID and 900 μm high) and analyzed on an AR1000 rheometer (TA Instruments, New Castle, DE) using an 8 mm plate (n = 2–4). KFE-RGD gels were cast into nylon washers (9.5 mm ID and 1.6 mm high) and analyzed using a 25 mm plate on an Anton Paar MCR300 rheometer (Anton Paar, Graz, Austria) (n = 3–8 gels). The raw storage and loss moduli were adjusted according to the size of the gel, not the size of the plate. Mixtures of the peptides were cast in nylon washers and were made and analyzed using a 25 mm plate on an Anton Paar MCR300 rheometer, with the exception of the final three KFE-RGD gels at 10 mg ml−1, which were analyzed using a 25 mm plate on a TA Instruments ARES-LSII rheometer (TA Instruments, New Castle, Delaware) (n = 3–4). The storage (G′) modulus was obtained at frequencies of 1 and 10 Hz with a sweep from 0.1 to 10% strain. This test was used to select the optimal strain percentage for bulk stiffness measurements (storage and loss (G″) moduli), determined as 1% strain. All gels were analyzed at constant strain (1%) from 1 Hz to 10 Hz, a range in which G′ and G″ were frequency-independent.
2.4. AFM
Two methods were used to prepare peptide solutions for AFM imaging: (1) SAP were dissolved in distilled water to a final concentration of 1.0 mg ml−1, as previously described [46]; or (2) SAP solutions were diluted in PBS to a final concentration of 0.75 mg ml−1 to induce gel formation. AFM samples were prepared by aliquoting 50 μl of the peptide solution onto freshly cleaved mica (Ruby Muscovite, S&J Trading, NJ). All samples were prepared in duplicate. Both types of SAP samples (with and without PBS) were allowed to adsorb onto the mica surface for 30 s at room temperature, after which the samples were rinsed with distilled water, blotted and allowed to air dry in ambient conditions for a minimum of 3 h.
Samples were imaged using a Multimode AFM equipped with a Nanoscope IIIa controller (Digital Instruments, Santa Barbara, CA). Because the samples are soft biopolymers, soft silicon AFM tips (NSG03, NT-MDT, Moscow, Russia) were selected to prevent damage to the samples. Probes with a nominal spring constant of 0.35– 6.06 N m−1 and resonant frequency of 47–150 kHz were used in tapping mode at a scan rate of ~2 Hz. Height and amplitude images were collected from three different regions per sample at scan sizes ranging from 2 × 2 to 20 × 20 μm2. Fiber heights of peptides for samples prepared in distilled water were ascertained from the AFM topographic (height) images for n = 300–400 measurements per sample type, using the section analysis tool of the NanoScope Analysis software, version 1.40 (Bruker Corporation, Santa Barbara, CA).
2.5. Statistics
Fiber height measurements are reported using t-tests to ascertain the differences between KFE and KFE-RGD fibers, helical and non-helical fibers in the mixture, KFE and helical fibers in the mixture, and KFE-RGD and non-helical fibers in the mixture. Differences in MVN formation as well as hMSC projection number and length were calculated using ANOVA followed by Tukey’s Honest Significant Difference test. Significant differences were defined as p < 0.05.
3. Results and discussion
The present study investigated the morphological transformation of endothelial cells and hMSC in response to changes in stiffness and binding site density within a 3-D fibrous matrix that allows for control of both these parameters. The KFE SAP sequence does not support cellular adhesion, while the KFE-RGD sequence does support cell adhesion as a result of the incorporated RGD binding site. Thus, combining KFE and KFE-RGD in the proper ratio allows control over both stiffness and RGD density within a single gel, allowing the study of the effects of these parameters on HUVEC MVN formation and hMSC morphology.
3.1. Rheology of SAP gels
In order to investigate the rheological behavior of KFE-RGD, which has not been characterized previously, strain sweeps from 0.1 to 10% strain were performed on 6 mg ml−1 KFE-RGD at 1 and 10 Hz (Fig. 1A). At 1 Hz, storage modulus (G′) varied by ~10% as strain varied from 0.5 to 5%. Over the same range of strains, G′ varied more dramatically (~40%) at the higher frequency of 10 Hz (Fig. 1A). For 6 mg ml−1 KFE-RGD subjected to 1% strain, G′ varied by ~10% as frequency varied from 1 to 10 Hz (Fig. 1B). Thus, by conducting rheological measurements at 1% strain, G′ is a measure of the stiffness of the gels that is relatively frequency independent over the explored range. The storage (Fig. 1C) and loss (Fig. 1D) moduli of KFE and KFE-RGD obtained by averaging the values over the frequency range tested increased with increasing concentration. At a given concentration, the stiffness of KFE gels was ~7–10-fold greater than KFE-RGD gels, with the stiffness vs. concentration relationship for both materials well described by a power law relationship with a similar slope or exponent (Fig. 1C, Table 1). Mixtures of the peptides were created using a constant KFE-RGD concentration of 1 mg ml−1 with an increasing amount of KFE (Fig. 1C and D). The dashed line represents the predicted value of these mixtures, calculated using a linear combination of KFE and KFE-RGD stiffness values and weighted for the ratio of peptides used at each concentration. The calculation of G′ for mixtures is seen in Eq. (1), whereas the calculation for G″ can be used by substituting the power law relationship values found in Table 1. CTotal represents the total mass concentration of peptides in the mixture, while CKFE and CKFE-RGD are the concentrations of the individual peptides.
| (1) |
Fig. 1.
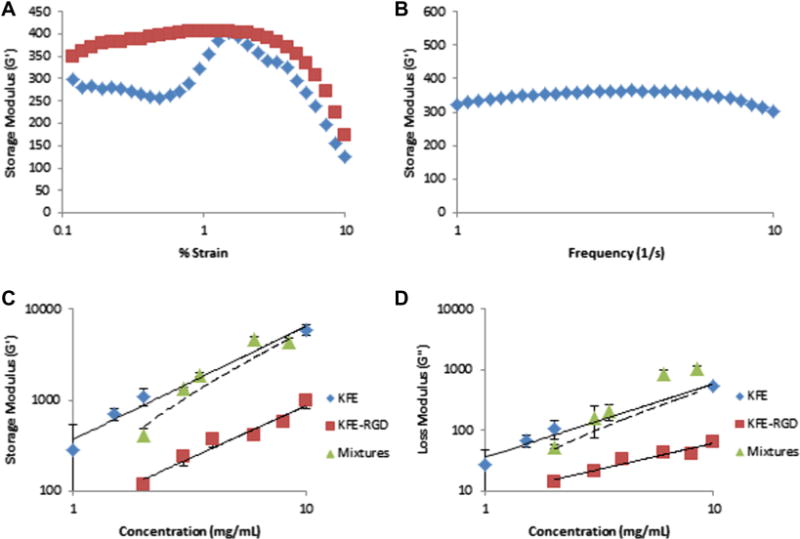
Rheology results of SAP gels used in the study. (A) Strain sweeps of a 6 mg ml−1 KFE-RGD gel at frequencies of 1 Hz (squares) and 10 Hz (diamonds) were used to choose a strain value (1%) for moduli testing. (B) Frequency sweep of a 6 mg ml−1 KFE-RGD gel at a constant 1% strain produced a relatively flat storage modulus reading. (C) Results for storage modulus (G′) of KFE (n = 3–4), KFE-RGD (n = 3–8) and mixtures (n = 3–4). All mixtures had a constant KFE-RGD concentration at 1 mg ml−1 with a varying KFE concentration such that the concentration reported in the x-axis for the mixtures is the sum of KFE and KFE-RGD at each condition. (D) Results for loss modulus (G″) of KFE, KFE-RGD and mixtures. The dashed line (C, D) is the predicted value of the mixtures using a linear combination of KFE and KFE-RGD stiffness weighted for the ratio of peptides used at each concentration.
Table 1.
Storage and loss moduli for SAP used in this study in terms of the power law relationship dependent on concentration (C).
| KFE | KFE + RGD | |
|---|---|---|
| Storage modulus (Pa) | G′ = 370C1.24 | G′ = 60.2C1.16 |
| R2 = 0.97 | R2 = 0.95 | |
| Loss modulus (Pa) | G″ = 36.5C1.20 | G″ = 8.56C0.86 |
| R2 = 0.96 | R2 = 0.93 |
The observation that introducing RGD to the base sequence of the SAP reduced stiffness is consistent with the qualitative observations of Genové et al. [52], who found that introducing integrin binding sites into the RAD-16 SAP sequence also reduced its stiffness, potentially by inhibiting fiber assembly. In contrast, Jung et al. [48] observed that introducing RGD binding sites into the Q11 SAP sequence did not alter stiffness. Jung et al. [48] suggested that the different effects on stiffness were due to the fact that their peptide had a secondary β-turn structure in contrast to the β-sheet secondary structure used by Genové et al. The data support this notion, since a peptide base sequence of KFE was used, which has been shown previously to have a β-sheet structure [46]. While the structure of KFE-RGD was not determined in this study, previous studies have shown that incorporation of a binding site into the base sequence of other SAP does not affect the structural makeup of the resulting peptide [48,52]. Importantly, the data indicate that stiffness can be varied independently of the integrin binding site density by varying the ratio of non-adhesive and RGD-containing peptides.
Measured values of G′ for the KFE-RGD gels covered a range of ~100–1000 Pa, while the G′ for KFE gels ranged from ~285 to 6000 Pa. The threshold of gelation for KFE-RGD is 1 mg ml−1, which, according to extrapolation of the rheological measurements, limits the system to stiffness values above ~60 Pa. Stiffness is controlled in the system by increasing the concentration of KFE while keeping KFE-RGD and thus binding site density constant. This results in a rapidly stiffening gel that approaches the stiffness profile of pure KFE, as indicated by the measurements with mixtures in Fig. 1C and D. To control binding site density at a constant stiffness, in contrast, the concentrations of both peptides must be changed. This concept is further illustrated in Section 3.4 and Fig. 7, where pairs of SAP gels, one pure KFE and another a mixture of KFE and KFE-RGD, are made such that the stiffnesses of the gels are similar.
Fig. 7.
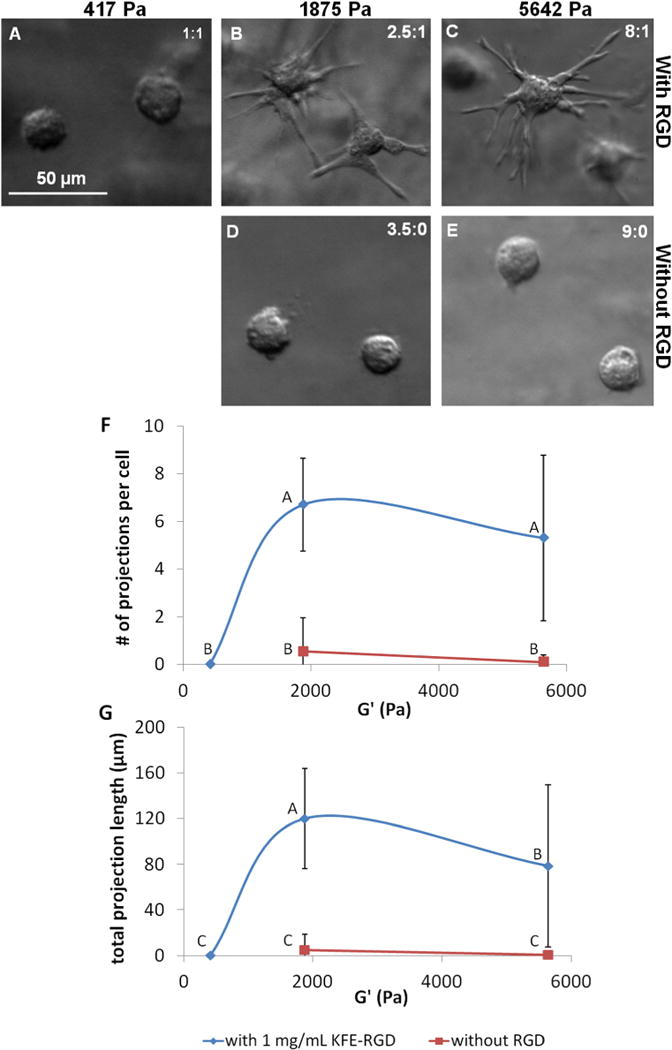
hMSC morphology is influenced by both stiffness and RGD concentration. (A–E) DIC images taken after 24 h of culture within SAP show that hMSC extend projections into their surroundings in gels of high stiffness in the presence of RGD, but remain spherical in the absence of RGD, regardless of stiffness. The stiffness values for images (A, B) were measured with rheology, while the stiffness values for (C–E) were estimated from Fig. 1B. Peptide concentrations are given in the top right corner of each image as mg ml−1 KFE: mg ml−1 KFE-RGD. (F, G) Quantification of the number of projections per cell and the total length of these projections per cell suggests that hMSC morphology is dependent on both stiffness and RGD concentration after 24 h. A total of 20 cells were randomly selected and measured from three replicate gels at each condition. Points not labeled with the same letter are statistically different. Error bars indicate standard deviations.
Gels formed from commercially available collagen extracts typically range in stiffness from 100 to 1400 Pa [34,53–55], though sensitive measurements of an elastic contribution as small as 2 Pa in dilute solutions has been reported [33,40]. Fibrin gels typically have a stiffness ranging from 20 to 2150 Pa [9,56]. Polyethylene glycol (PEG), another system for 3-D culture, does not allow for soft matrices, since PEG-based gels have not been made with a stiffness <1 kPa [11]. While the present authors have not explored the upper stiffness limit that the SAP system can achieve, the demonstrated 60–6000 Pa range covers much of the typical stiffness range used in cell culture experiments (reviewed by Nemir and West [11]) and thus could be useful for investigating stiffness effects on many cell types.
3.2. AFM of SAP fibers
KFE has previously been shown to be fibrous when dissolved in distilled water [46]. The present authors have demonstrated the fibrous nature of gels formed from KFE, KFE-RGD and mixtures of the two peptides using transmission electron microscopy and AFM. Only the results from AFM are shown, since this technique yielded clear images.
AFM images of SAP reconstituted in distilled water revealed that KFE, KFE-RGD and a 50/50 mixture of the two consisted of a network of fibrous structures (Fig. 2). The ultrastructure of individual fibers in these samples differed, however, depending on the type of peptide used. The KFE peptide formed helical fibers (Fig. 2A and Supplemental Figure), while helical patterns were not evident on KFE-RGD fibers (Fig. 2B and Supplemental Figure). A 50/50 mixture of the peptides revealed the presence of both helical and non-helical fibers, suggesting a mixture of discrete KFE and KFE-RGD fibers (Fig. 2C and Supplemental Figure). When comparing fiber heights, KFE fibers were 108% greater than KFE-RGD fibers and, within the mixture, helical fibers were 84% greater than non-helical fibers (Fig. 2H). Also, within the mixture, the mean and distribution of heights for the helical fibers were similar to those in pure KFE (Fig. 2D vs. Fig. 2F), while the mean and distribution of heights for the non-helical fibers were similar to those in pure KFE-RGD (Fig. 2E vs. Fig. 2G). These similarities suggest that mixtures of KFE and KFE-RGD peptides consist of discrete fibers composed of either KFE or KFE-RGD. In support of these observations, Gasiorowski and Collier [57] used immunoelectron microscopy to also observe the existence of independent fibers within a sample that was created from a mixture of two solutions of reconstituted SAP.
Fig. 2.
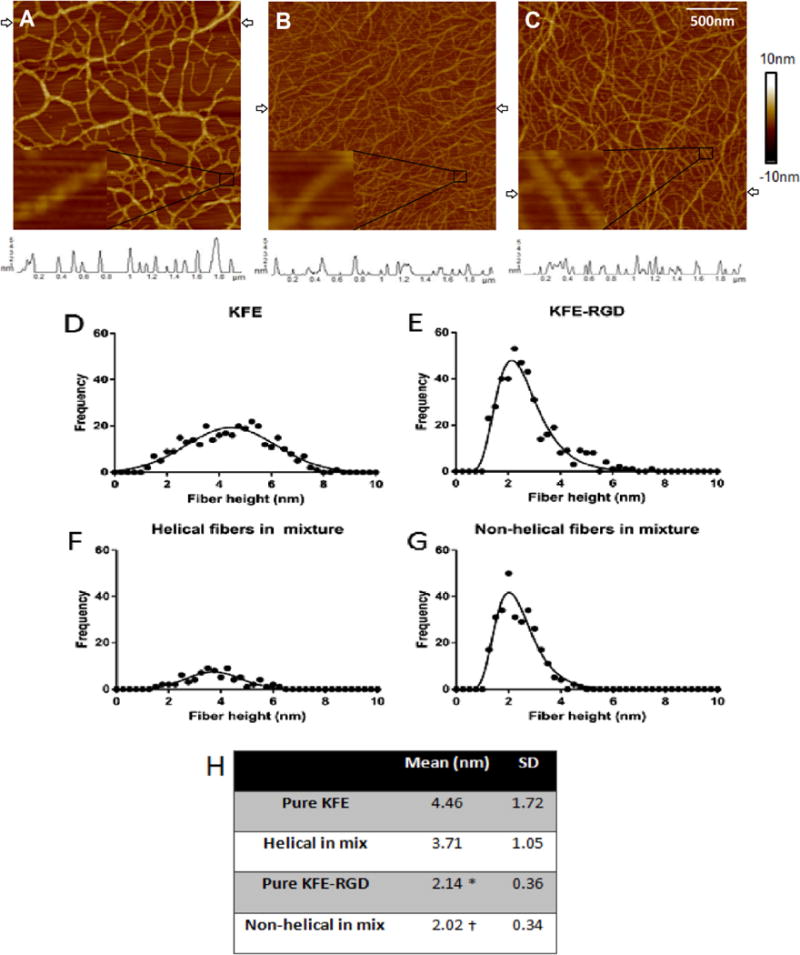
AFM images of SAP with height profiles (trace plots below AFM images) corresponding to the scan line between the arrows in each image. The peptides were prepared in distilled water. Images are of a 2 × 2 μm scan area, each with an inset showing finer detail (A–C): (A) 2 mg ml−1 KFE showing helical fibers; (B) 2 mg ml−1 KFE-RGD showing seemingly non-helical fibers; (C) 50/50 mixture 2 mg ml−1 total, which shows a mixture of the two types of fibers. Fibers were measured across three scan lines per image. Histograms of fiber heights for (D) KFE, (E) KFE-RGD, (F) helical fibers in the mix and (G) non-helical fibers in the mix show similar profiles between KFE and helical fibers in the mix, and (H) KFE-RGD and non-helical fibers in the mix. (D–G) Statistical analysis of the histogram profiles.*indicates significantly different from “pure KFE” and † indicates significantly different from “helical in mix” (p < 0.05).
Millimolar concentrations of ions alter the structure of SAP during the gelation process, and therefore the fiber structure was analyzed when SAP were exposed to a solution of physiological ionic strength [58]. SAP solutions were diluted in PBS to induce gelation before adsorption to the mica surface, and the peptides were imaged over a 20 × 20 μm region. While both SAP sequences formed fibers in the presence of PBS, KFE-RGD appeared to form thicker fibers than KFE (Fig. 3A and B). A 50/50 mixture of the peptides in the presence of PBS revealed a mixture of the two fiber types, suggesting that individual fibers were assembled from either KFE or KFE-RGD peptides, but not mixtures of the two (Fig. 3C). At a 10-fold higher magnification (2 × 2 μm region), KFE appeared as individual fibers (Fig. 3D), while KFE-RGD appeared as braided bundles of fibers (Fig. 3E). A 50/50 mixture of the peptides appeared as a mixture of individual fibers and braided bundles (Fig. 3F). The presence of several overlapping fibers and clumps of fibers prevented the present authors from analyzing the height of individual fibers in these samples, as was done for SAP prepared in distilled water. Even without these quantitative data, the images qualitatively support the notion that the SAP gels consist of an assembly of two distinct fiber types, most likely composed of either KFE or KFE-RGD.
Fig. 3.
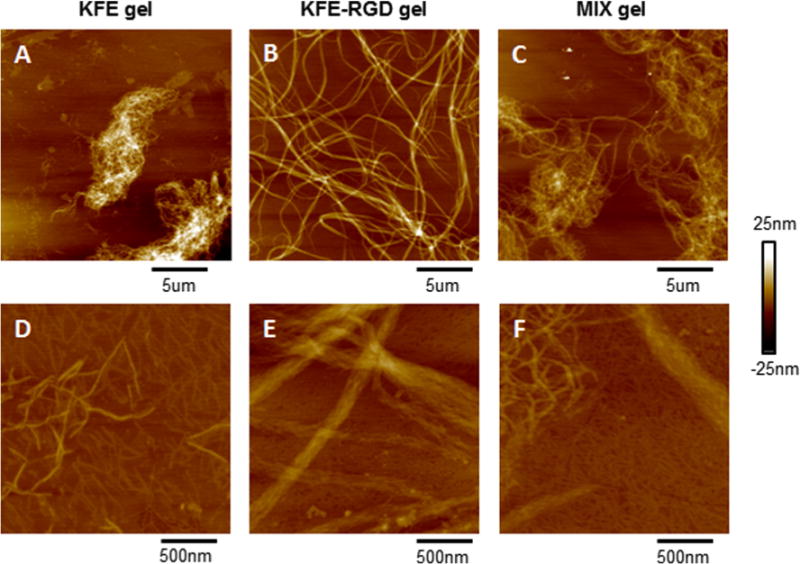
Images obtained after diluting SAP with PBS to induce SAP gelation before AFM imaging, with scan area sizes of (A–C) 20 × 20 μm m and (D, E) 2 × 2 μm m. Peptide concentrations after dilution with PBS are (A, D) 0.75 mg ml−1 KFE, (B, E) 0.75 mg ml−1 KFE-RGD, and (C, F) 50/50 mixture of the peptides at 0.75 mg ml−1. (A–C) Appearance of small fibers for KFE, larger strands for KFE-RGD, and a mixture of the two fiber types in mix gels indicate that each fiber may be exclusively made of one type of SAP. (D, E) KFE appears as individual fibers, while KFE-RGD fibers appear as a braided bundle. Mix gels show discrete types of fibers, indicating that the individual KFE and KFE-RGD fibers may not form hybrid fibers in a simulated gel structure.
3.3. MVN formation in SAP
It is well established that endothelial cells cultured within natural 3-D matrices, such as collagen [30,37,40,59], fibrin [36,41] or collagen–fibrin mixtures [32], form interconnected networks resembling MVN. In addition to natural ECM matrices, some 3-D SAP gels (RAD16-I and RAD16-II) allow MVN formation in vitro, while others do not (KFE-8 and KLD-12); the ability to form MVN is dependent on cell–substrate binding [47]. Work from several of these natural and synthetic systems supports the notion that MVN formation is modified over a certain stiffness range, though this range is dependent on the type of both endothelial cell [60] and matrix [32]. Taken together, these results suggest a role for both cell–substrate adhesion and matrix stiffness in regulating MVN formation within 3-D gels.
To investigate the contribution of binding sites on MVN formation, endothelial cells were cultured within 3-D gels composed of either pure KFE or pure KFE-RGD. KFE gels, which do not contain any known cell binding sites, did not appear to support cellular elongation, though thin processes were sometimes observed (Fig. 4A). KFE-RGD gels contain known integrin binding sites and supported HUVEC elongation and MVN formation (Fig. 4B). Thus, incorporation of an integrin binding site significantly impacts endothelial morphology, which is consistent with the work of others using co-assembled SAP for 2-D culture [48,52]. A 1 mg ml−1 KFE gel has greater stiffness than a 1 mg ml−1 KFE-RGD gel, however. Since several studies report that increased stiffness reduces MVN formation [30,32,39,41,42], these data alone cannot exclude the possibility that differences in stiffness might account for the observed differences in MVN formation.
Fig. 4.
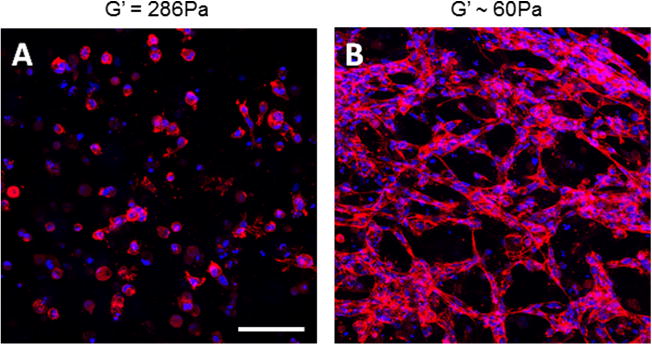
(A) Endothelial cells do not elongate or form MVN in 1 mg ml−1 KFE gels, which do not contain integrin binding sites, but (B) do form MVN in 1 mg ml−1 KFE-RGD gels, which do contain integrin binding sites. Cells are stained with Alexa 633 phalloidin (red), and nuclei are stained with DAPI (blue) to highlight networks and allow for quantification. Stiffness of (A) was measured by rheology, while (B) was calculated using Eq. (1). Scale bar is 100 μm.
To investigate the contribution of stiffness to MVN formation, endothelial cells were cultured within 3-D gels with a fixed concentration of KFE-RGD and various concentrations of KFE. The KFE/KFE-RGD mixtures provided a matrix where cellular binding sites were held constant, while the stiffness of the matrix was modulated by altering the KFE concentration. As the concentration of KFE and thus the stiffness of the gel increased, MVN formation decreased (Fig. 5A–D). Quantification of MVN formation using the average length of the structure (ALS) incorporated contributions of cell elongation and the formation of multicellular structures. Among gels containing 1 mg ml−1 of KFE-RGD, ALS significantly decreased as the amount of KFE increased, suggesting an impact of stiffness on MVN formation (Fig. 6). Notably, the extent of MVN formation in gels consisting of 1 mg ml−1 KFE-RGD and 1 mg ml−1 of KFE (Figs. 5C and 6, ALS = 66.3) was significantly greater than in 1 mg ml−1 KFE gels (Figs. 4A and 6, ALS = 30.9, p = 0.002) even though the stiffness of the mixture was actually higher, again suggesting that the concentration of integrin binding sites in the matrix also impacts MVN formation. Additionally, gels that lack the RGD binding site and containing only KFE were not compacted by cells at any stiffness (data not shown). In contrast, cells were able to compact gels containing KFE-RGD in a stiffness-dependent manner. Soft (1 mg ml−1 KFE-RGD) gels compacted extensively, and some level of compaction was observed up to 0.5 mg ml−1 KFE with 1 mg ml−1 KFE-RGD (data not shown). Although this compaction most likely affected the stiffness of the gels during the experiment, MVN formation was compared with the initial stiffness.
Fig. 5.
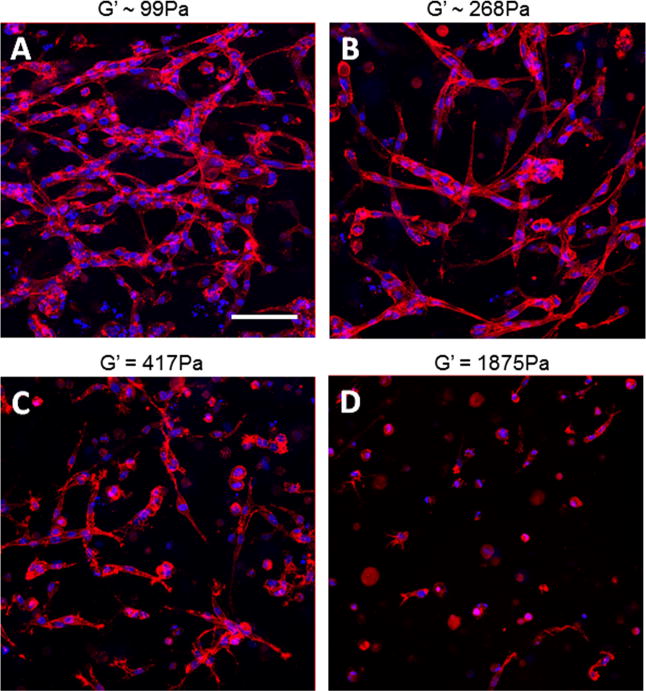
(A–D) Images of phalloidin (red) and DAPI (blue) stained HUVEC showing decreased elongation and MVN formation of endothelial cells as stiffness is increased by increasing KFE, while binding site density and thus KFE-RGD are held constant. Holding KFE-RGD concentration constant at 1 mg ml−1, (A) 0.1, (B) 0.5 (B), (C) 1 or (D) 2.5 mg ml−1 KFE was added to increase stiffness. Stiffnesses of (A) and (B) were estimated using Eq. (1), while (C) and (D) were measured by rheology. Scale bar is 100 μm.
Fig. 6.
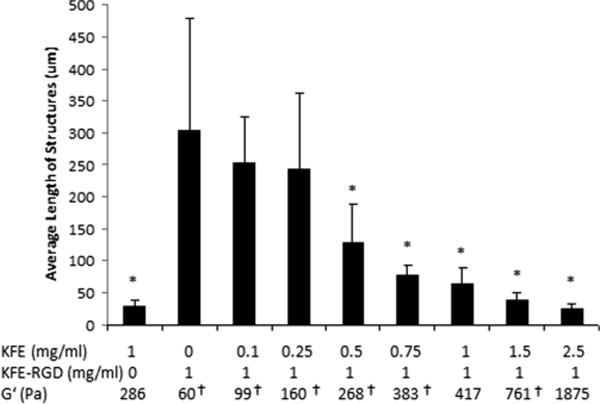
Quantification of MVN showing a decrease in cell elongation as matrix stiffness is increased (n = 5–7). Error bars are standard deviations.*Indicates significantly different from 0 mg ml−1 KFE: 1 mg ml−1 KFE-RGD gels (p < 0.05). G′ values were measured unless indicated with † to signify that they were calculated using Eq. (1).
The above results suggest that both stiffness and the presence of RGD binding sites are important regulators of MVN formation within 3-D SAP gels. Previous reports have often attributed changes in MVN formation within 3-D gels to stiffness effects [30,32,37,39–42], but it is often difficult to definitively exclude the potential contributions of other factors that also change with stiffness. For example, in previous work with endothelial cells within collagen gels, the extent of MVN formation appeared to decrease with increasing stiffness [30]. In initial studies, stiffness was altered by changing the collagen concentration, which potentially changes a number of factors, including fiber diameter, pore size and binding site density. In follow-up studies, the effective stiffness of the matrix was changed, while the collagen concentration was kept constant by altering the mechanical boundary conditions [30]. While this approach eliminates potential artifacts due to changing collagen concentration and demonstrates the same effects on MVN formation, it is difficult to quantify the changes in effective stiffness [30]. Approaches used by others to delineate the contribution of stiffness to observed changes in MVN formation are also limited. For example, several groups have explored the behavior of endothelial cells in response to the non-enzymatic modification of collagen, which changes matrix stiffness independent of collagen concentration [32,37,39,40]. While these results can be attributed to stiffness changes, effects due to variations in fiber structure or function cannot be fully excluded. For example, approaches that use crosslinking to affect stiffness at a constant ECM concentration can result in a change in the ECM fiber diameter and organization [37]. The crosslinking agent adds biological complexity to the system and might affect cellular behavior independent of the bulk ECM (e.g. via advanced glycation end product mediated signaling [39]). Altering the molecular weight of collagen oligomers used for gel formation varies stiffness at a constant collagen concentration, but this approach also changes the fiber diameter and pore size [40]. Combining ECM proteins can also generate gels of different stiffness at a constant overall ECM density [32]. Since ECM proteins affect cells in different ways, however, it is difficult to distinguish which effects are mechanically derived and which are due to composition.
The use of SAP to explore the contributions of adhesion and stiffness possesses several attractive features that address these concerns. First, synthetic matrices have been described as a “blank slate” to which select functions can be added [61]. In contrast, ECM proteins exhibit natural heterogeneity and are rich in biological complexity. For example, while focal adhesions provide sites for cell–substrate binding, they also serve in many signaling pathways that influence cell behavior [62,63]. Collagen has many distinct binding sites that uniquely affect cell function, and these motifs can interact with other ECM proteins and be subject to glycosylation, which can affect protein structure [64,65]. Thus, changing ECM concentration, type or crosslinking can potentially alter many complex processes. Secondly, the modular nature of the SAP system allows for incorporation of desired sequences, such as the RGD binding site, in order to study their effects on cellular function in a 3-D environment. This is especially attractive, given recent work showing that, at least for cells grown on 2-D surfaces, stiffness effects are strongly modulated by the composition and adhesiveness of the surface [48]. In addition, work with MSC has highlighted that, even on 2-D substrates of comparable stiffness, differentiation depends on the ligands presented to the cells [66]. Thirdly, in those SAP gels studied in detail thus far, their microarchitecture, including pore size and fiber diameter, are only weak functions of SAP concentration, suggesting that large changes in stiffness would be associated with only modest changes in other properties [34,42]. It has been shown that KFE and KFE-RGD fibers form independently within a given gel, and thus changing the peptide ratio to modify stiffness and binding density also alters the balance of fiber concentration. Although morphological differences between these two fibers have been shown, the effect on average fiber size by changing this peptide ratio is modest. Additionally, the fibrous structure and pore size of KFE gels have been shown to be similar to a 3.0 mg ml−1 collagen gel, illustrating that SAP are an excellent structural analogue of natural ECM gels [47]. While there is no ideal system to decouple stiffness and binding site density in 3-D cell culture, the KFE/KFE-RGD system provides these unique advantages over natural ECM systems.
Though no SAP or natural ECM matrix system completely isolates the potential contribution of stiffness from all other parameters, the results of a number of studies of endothelial cells within natural and synthetic matrices suggest that matrix stiffness is an important modulator of MVN formation. Francis-Sedlak et al. [39] used glucose-6-phosphase to crosslink collagen, which increased matrix stiffness from ~46 Pa to ~111 Pa. In a capillary sprouting model, there was a decrease in sprout length at day 3, as crosslinking and thus stiffness was increased. In a similar capillary sprouting model by Ghajar et al. [41] that used a fibrin gel where stiffness was modulated by increasing matrix density (~20 Pa to ~350 Pa [56]), sprouting decreased as stiffness increased. In a system by Rao et al. [32] using collagen and fibrin mixtures, MVN formed best at the lowest investigated stiffness (~50 Pa) and decreased as stiffness rose, until very little network formation was observed at the highest reported stiffness (~250 Pa). This effect was observed when the relative amounts of collagen and fibrin were varied as well as when crosslinking was used to stiffen the matrix. Sieminski et al. [30] reported that, relative to free floating gels, the extent of MVN formation decreased in collagen gels adhering to a solid surface. While it is difficult to quantify how much the effective stiffness is changed by altering the mechanical boundary conditions of the gel at a given collagen concentration, free floating gels are less stiff than their constrained counterparts. SAP sequences (RAD16-I and RAD16-II) that facilitate cell adhesion have also been investigated for MVN formation by Sieminski et al. [42]. Gels with stiffness of 46 Pa exhibited robust MVN formation that decreased as stiffness rose to 735 Pa, at which point, most MVN formation ceased. In each of these individual studies, it is possible to make a case that some factor other than stiffness mediates the observed effects on MVN formation. Nevertheless, when taken together, these studies where stiffness is changed by different methods (i.e. changing concentration, crosslinking, composition or mechanical boundary conditions) strongly suggest that matrix stiffness is an important regulator of MVN formation where increasing stiffness decreases the extent of formation. A recent report by Mason et al. [37] showed the opposite trend, where capillary sprouts increased as collagen gels were stiffened from 175 Pa to 515 Pa using ribose crosslinking. The reason for this exception to the trend is not clear, but illustrates the importance of having several unique systems for studying the impact of matrix stiffness within 3-D environments.
3.4. hMSC morphology within SAP
In order to illustrate the potential use of the SAP system with other cell types, hMSC were cultured within SAP gels of various stiffness and RGD concentrations, and the morphology was quantified after 24 h by counting the number of projections per cell and measuring the total length of these projections. Contrary to the trend seen with HUVEC, hMSC adopted a spread morphology as matrix stiffness increased in the presence of RGD (Fig. 7A–C). At a relatively low stiffness of 417 Pa, the cells remained spherical. When gel stiffness was raised to 1875 Pa, however, the hMSC extended 6.7 ± 1.9 projections that had an average total length of 120.1 ± 44.1 μm per cell. When the stiffness was further increased to 5642 Pa, the total projection length per cell decreased to 78.6 ± 71.1 μm, even though the number of projections per cell was not statistically different from that at 1875 Pa. In the absence of RGD, however, cells remained spherical at both 1875 and 5642 Pa (Fig. 7D and E). Although data for 417 Pa gels without RGD were unavailable for this particular experiment, hMSC in these gels have always exhibited a spherical morphology in other experiments. Thus, it seems that both the presence of RGD and threshold stiffness are required for cellular extensions to appear, illustrating the important interplay between stiffness and adhesiveness.
Stiffness as well as binding site type and density have been shown to be crucially important to the morphology and differentiation of hMSC in 2-D culture [10,66,67]. In a study by Huebsch et al. [68], the stiffness and presence of RGD within 3-D alginate gels was also shown to affect hMSC lineage specification, but the cells remained spherical at all investigated stiffness values and RGD concentrations. In contrast, the SAP gels allow cells to spread out into their environment, potentially due to the fact that they have a fibrous structure similar to collagen. Therefore, SAP might be better able to mimic the in vivo environment, and thus they can potentially capture a more faithful representation of cellular behavior. Furthermore, Fig. 7A–E shows that a particular stiffness can be achieved with different RGD concentration that is within the range limited by the stiffness profiles given by pure KFE and pure KFE-RGD in Fig. 1B. Thus, the SAP system is well suited to studying the effects of both stiffness and adhesiveness on hMSC morphology and differentiation in 3-D culture in future studies.
The difference between the observations with HUVEC and hMSC can most likely be attributed to the difference in their cell-specific functions and native in vivo environment. For example, hMSC have been shown to be quiescent on soft substrates that have elasticity similar to the bone marrow in which they normally reside, while they become active and differentiate on stiffer matrices [69]. Therefore, it is important to study the effect of both stiffness and adhesiveness for each specific cell type, so that a scaffold can be tailored to induce the desired cellular behavior for a particular application.
4. Conclusions
This paper described a 3-D cell culture system that can control both stiffness and binding site density. This system is composed of two SAP sequences, which contain the same base sequence, but one of these peptides incorporates the RGD sequence that facilitates cellular adhesion. These peptides form fibers with a structure similar to typical ECM gels used in 3-D cell culture and can be modulated to cover a wide stiffness range. This system was used to investigate MVN formation and hMSC morphology in matrices with or without the RGD binding site at various matrix stiffness values. MVN formation in the system correlates well with other ECM systems, where increased stiffness inhibits MVN formation. In contrast to the observations with HUVEC, hMSC morphology exhibited increased spreading on stiffer matrices when RGD was present. Similar to the HUVEC experiments, however, stiffness had no effect on hMSC morphology when RGD was absent. This system can be used in the future to investigate behavioral changes of other cell types in response to matrix stiffness and binding site density, leading to better design of matrix materials for specific cell types and applications.
Supplementary Material
Acknowledgments
This work was supported by NSF (CMMI-0928739, CBET-1067481, and R01 HL-09652).
Appendix A. Figures with essential color discrimination
Certain figures in this article, particularly Figs. 1–5 and 7, are difficult to interpret in black and white. The full color images can be found in the online version at http://dx.doi.org/10.1016/j.actbio.2013.04.002.
Appendix B. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.actbio.2013.04.002.
References
- 1.Peyton SR, Ghajar CM, Khatiwala CB, Putnam AJ. The emergence of ECM mechanics and cytoskeletal tension as important regulators of cell function. Cell Biochem Biophys. 2007 May 2;47:300–20. doi: 10.1007/s12013-007-0004-y. [DOI] [PubMed] [Google Scholar]
- 2.Pelham Y-L, Jr, Wang RJ. Cell locomotion and focal adhesions are regulated by substrate flexibility. PNAS USA. 1997 Dec;94:13661–5. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000 Jul 1;79:144–52. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halliday JT, Tomasek NL. Mechanical properties of the extracellular matrix influence fibronectin fibril assembly in vitro. Exp Cell Res. 1995;217:109–17. doi: 10.1006/excr.1995.1069. [DOI] [PubMed] [Google Scholar]
- 5.Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, et al. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell motil Cytoskeleton. 2005 Jan 1;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 6.Arora PD, McCulloch CA. The deletion of transforming growth factor-beta-induced myofibroblasts depends on growth conditions and actin organization. Am J Pathol. 1999 Dec 6;155:2087–99. doi: 10.1016/s0002-9440(10)65527-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein EA, Castagnino P, Kothapalli D, Yin L, Byfield FJ, Xu T, et al. Cell cycle control by physiological matrix elasticity and in vivo tissue stiffening. Curr Biol. 2009;19(18):1511–8. doi: 10.1016/j.cub.2009.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher D. Substrate compliance versus ligand density in cell on gel responses. Biophys J. 2004 Jan 1;86(1 Pt):617–28. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flanagan L, Ju Y, Marg B, Osterfield M, Jamney P. Neurite branching on deformable substrates. NeuroReport. 2002;13(18):2411–5. doi: 10.1097/01.wnr.0000048003.96487.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006 Aug 4;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 11.Nemir S, West JL. Synthetic materials in the study of cell response to substrate rigidity. Ann Biomed Eng. 2010 Jan 1;38:2–20. doi: 10.1007/s10439-009-9811-1. [DOI] [PubMed] [Google Scholar]
- 12.Saunders R, Hammer D. Assembly of human umbilical vein endothelial cells on compliant hydrogels. Cell Mol Bioeng. 2010;3(1):60–7. doi: 10.1007/s12195-010-0112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmeichel K, Bissell M. Modeling tissue-specific signaling and organ function in three dimensions. J Cell Sci. 2003 Pt. 12;116:2377–88. doi: 10.1242/jcs.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petersen OW, Rønnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci USA. 1992 Oct 19;89:9064–8. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grinnell F. Fibroblast biology in three-dimensional collagen matrices. Trends Cell Biol. 2003 May 5;13:264–9. doi: 10.1016/s0962-8924(03)00057-6. [DOI] [PubMed] [Google Scholar]
- 16.Yamada KM, Cukierman E. Modeling tissue morphogenesis and cancer in 3-D. Cell. 2007 Aug 4;130:601–10. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Sieminski AL, Padera RF, Blunk T, Gooch KJ. Systemic Delivery of Human Growth Hormone Using Genetically Modified Tissue-Engineered Microvascular Inclusion of Nonendothelial Cells. Tissue Eng. 2002;8(6):1057–69. doi: 10.1089/107632702320934155. [DOI] [PubMed] [Google Scholar]
- 18.Boretti MI, Gooch KJ. Effect of extracellular matrix and 3-D morphogenesis on islet hormone gene expression by Ngn3-infected mouse pancreatic ductal epithelial cells. Tissue Eng Part A. 2008 Dec 12;14:1927–37. doi: 10.1089/ten.tea.2007.0338. [DOI] [PubMed] [Google Scholar]
- 19.Pontes Soares C, Midlej V, de Oliveira MEW, Benchimol M, Costa ML, Mermelstein C. 2-D and 3-D-organized cardiac cells shows differences in cellular morphology, adhesion junctions, presence of myofibrils and protein expression. PLoS One. 2012 Jan 5;7:e38147. doi: 10.1371/journal.pone.0038147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiron S, Tomczak C, Duperray A, Lainé J, Bonne G, Eder A, et al. Complex interactions between human myoblasts and the surrounding 3-D fibrin-based matrix. PLoS One. 2012 Jan 4;7:e36173. doi: 10.1371/journal.pone.0036173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell–matrix adhesions to the third dimension. Science (New York, NY) 2001 Nov;294(5547):1708–12. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 22.Zschenker O, Streichert T, Hehlgans S, Cordes N. Genome-wide gene expression analysis in cancer cells reveals 3-D growth to affect ECM and processes associated with cell adhesion but not DNA repair. PLoS One. 2012 Jan 4;7:e34279. doi: 10.1371/journal.pone.0034279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang F, Weaver V, Peterson O, Larabell C, Dedhar S, Briand P, et al. Reciprocal interactions between B1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Cell Biol. 1998;95(December):14821–6. doi: 10.1073/pnas.95.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chitcholtan K, Asselin E, Parent S, Sykes PH, Evans JJ. Differences in growth properties of endometrial cancer in three dimensional (3-D) culture and 2-D cell monolayer. Exp Cell Res. 2013 Jan 1;319:75–87. doi: 10.1016/j.yexcr.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 25.Weaver VM, Lelièvre S, Lakins JN, Chrenek MA, Jones CR, Giancotti F, et al. β4 Integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell. 2009;2(3):205–16. doi: 10.1016/s1535-6108(02)00125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagamoto Y, Tashiro K, Takayama K, Ohashi K, Kawabata K, Sakurai F, et al. The promotion of hepatic maturation of human pluripotent stem cells in 3-D co-culture using type I collagen and Swiss 3T3 cell sheets. Biomaterials. 2012 Jun 18;33:4526–34. doi: 10.1016/j.biomaterials.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 27.Maltman DJ, Przyborski SA. Developments in three-dimensional cell culture technology aimed at improving the accuracy of in vitro analyses. Biochem Soc Trans. 2010 Aug 4;38:1072–5. doi: 10.1042/BST0381072. [DOI] [PubMed] [Google Scholar]
- 28.Tibbitt MW, Anseth KS. Hydrogels as extracellular matrix mimics for 3-D cell culture. Biotechnol Bioeng. 2009 Jul 4;103:655–63. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baker BM, Chen CS. Deconstructing the third dimension: how 3-D culture microenvironments alter cellular cues. J Cell Sci. 2012 Jul 13;125:3015–24. doi: 10.1242/jcs.079509. Pt. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sieminski AL, Hebbel RP, Gooch KJ. The relative magnitudes of endothelial force generation and matrix stiffness modulate capillary morphogenesis in vitro. Exp Cell Res. 2004 Jul 2;297:574–84. doi: 10.1016/j.yexcr.2004.03.035. [DOI] [PubMed] [Google Scholar]
- 31.Wang L-S, Chung JE, Kurisawa M. Controlling fibroblast proliferation with dimensionality-specific response by stiffness of injectable gelatin hydrogels. J Biomater Sci Polym Ed. 2011;23(September 14):1793–806. doi: 10.1163/156856211X598193. [DOI] [PubMed] [Google Scholar]
- 32.Rao R, Peterson A, Ceccarelli J, Putnam A, Stegemann J. Matrix composition regulates three-dimensional network formation by endothelial cells and mesenchymal stem cells in collagen/fibrin materials. Angiogenesis. 2012;15:253–64. doi: 10.1007/s10456-012-9257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kreger S, Bell B, Bailey J, Stites E, Kuske J, Waisner B, et al. Polymerization and matrix physical properties as important design considerations for soluble collagen formulations. Biopolymers. 2010;93(8):690–707. doi: 10.1002/bip.21431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miroshnikova Y, Jorgens D, Spirio L, Auer M, Sarang-Sieminski A, Weaver V. Engineering strategies to recapitulate epithelial morphogenesis within synthetic three-dimensional extracellular matrix with tunable mechanical properties. Phys Biol. 2011;8(2):026013. doi: 10.1088/1478-3975/8/2/026013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halliday NL, Tomasek JJ. Mechanical properties of the extracellular matrix influence fibronectin fibril assembly in vitro. Exp Cell Res. 1995;217(1):109–17. doi: 10.1006/excr.1995.1069. [DOI] [PubMed] [Google Scholar]
- 36.Gassman AA, Kuprys T, Ucuzian AA, Brey E, Matsumura A, Pang Y, et al. Three dimensional 10% cyclic strain reduces bovine aortic endothelial cell angiogenic sprout length and augments tubulogenesis in tubular fibrin hydrogels. J Tissue Eng Regen Med. 2011;5(5):375–83. doi: 10.1002/term.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mason BN, Starchenko A, Williams RM, Bonassar LJ, Reinhart-King CA. Tuning three-dimensional collagen matrix stiffness independently of collagen concentration modulates endothelial cell behavior. Acta Biomater. 2013 Jan 1;9:4635–44. doi: 10.1016/j.actbio.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Francis-Sedlak ME, Uriel S, Larson JC, Greisler HP, Venerus DC, Brey EM. Characterization of type I collagen gels modified by glycation. Biomaterials. 2009 Mar 9;30:1851–6. doi: 10.1016/j.biomaterials.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 39.Francis-Sedlak ME, Moya ML, Huang J-J, Lucas SA, Chandrasekharan N, Larson JC, et al. Collagen glycation alters neovascularization in vitro and in vivo. Microvasc Res. 2010 Jul 1;80:3–9. doi: 10.1016/j.mvr.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 40.Bailey J, Critser P, Whittington C, Kuske J, Yoder M, Voytik-Harbin S. Collagen oligomers modulate physical and biological properties of three-dimensional self-assembled matrices. Biopolymers. 2011;95(2):77–93. doi: 10.1002/bip.21537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghajar CM, Chen X, Harris JW, Suresh V, Hughes CCW, Jeon NL, et al. The effect of matrix density on the regulation of 3-D capillary morphogenesis. Biophys J. 2008 Mar 5;94:1930–41. doi: 10.1529/biophysj.107.120774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sieminski AL, Was AS, Kim G, Gong H, Kamm RD. The stiffness of three-dimensional ionic self-assembling peptide gels affects the extent of capillary-like network formation. Cell Biochem Biophys. 2007 Aug 2;49:73–83. doi: 10.1007/s12013-007-0046-1. [DOI] [PubMed] [Google Scholar]
- 43.Leon E, Verma N, Zhang S. Mechanical properties of a self-assembling oligopeptide matrix. J Biomater Sci Polym Ed. 1998;9(3):297–312. doi: 10.1163/156856298x00668. [DOI] [PubMed] [Google Scholar]
- 44.Zhang S, Holmes T, Lockshin C, Rich A. Spontaneous assembly of a self-complementary oligopeptide to form a stable macroscopic membrane. Proc Natl Acad Sci USA. 1993 Apr 8;90:3334–8. doi: 10.1073/pnas.90.8.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang S, Lockshin C, Cook R, Rich A. Unusually stable β-sheet formation in an ionic self-complementary oligopeptide stability of the structure in dilute solution. Biopolymers. 1994;34(5):663–72. doi: 10.1002/bip.360340508. [DOI] [PubMed] [Google Scholar]
- 46.Marini DM, Hwang W, Lauffenburger DA, Zhang S, Kamm RD. Left-handed helical ribbon intermediates in the self-assembly of a B-sheet peptide. Nano Lett. 2002;2(4):295–9. [Google Scholar]
- 47.Sieminski AL, Semino CE, Gong H, Kamm RD. Primary sequence of ionic self-assembling peptide gels affects endothelial cell adhesion and capillary morphogenesis. J Biomed Mater Res Part A. 2008 Nov 2;87:494–504. doi: 10.1002/jbm.a.31785. [DOI] [PubMed] [Google Scholar]
- 48.Jung J, Nagaraj A, Fox E, Rudra J, Devgun J, Collier J. Co-assembling peptides as defined matrices for endothelial cells. Biomaterials. 2009;30(12):2400–10. doi: 10.1016/j.biomaterials.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruoslahti E, Pierschbacher MD. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238(4826):491–7. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 50.Bouvard D, Brakebusch C, Gustafsson E, Aszodi A, Bengtsson T, Berna A, et al. Functional consequences of integrin gene mutations in mice. Circ Res. 2001 Jul 3;89:211–23. doi: 10.1161/hh1501.094874. [DOI] [PubMed] [Google Scholar]
- 51.Pierschbacher MD, Ruoslahti E. Influence of stereochemistry of the sequence Arg-Gly-Asp-Xaa on binding specificity in cell adhesion. J Biol Chem. 1987 Dec 36;262:17294–8. [PubMed] [Google Scholar]
- 52.Genové E, Shen C, Zhang S, Semino CE. The effect of functionalized self-assembling peptide scaffolds on human aortic endothelial cell function. Biomaterials. 2005 Jun 16;26:3341–51. doi: 10.1016/j.biomaterials.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 53.Byfield F, Reen R, Shentu T, Levitan I, Gooch K. Endothelial actin and cell stiffness is modulated by substrate stiffness in 2-D and 3-D. J Biomech. 2010;42(8):1114–9. doi: 10.1016/j.jbiomech.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Y, Motte S, Kaufman LJ. Pore size variable type I collagen gels and their interaction with glioma cells. Biomaterials. 2010 Jul 21;31:5678–88. doi: 10.1016/j.biomaterials.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 55.Van Goethem E, Poincloux R, Gauffre F, Maridonneau-Parini I, Le Cabec V. Matrix architecture dictates three-dimensional migration modes of human macrophages: differential involvement of proteases and podosome-like structures. J Immunol. 2010 Jan 2;184:1049–61. doi: 10.4049/jimmunol.0902223. [DOI] [PubMed] [Google Scholar]
- 56.Kotlarchyk MA, Shreim SG, Alvarez-Elizondo MB, Estrada LC, Singh R, Valdevit L, et al. Concentration independent modulation of local micromechanics in a fibrin gel. PLoS One. 2011 Jan 5;6:e20201. doi: 10.1371/journal.pone.0020201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gasiorowski JZ, Collier JH. Directed intermixing in multicomponent self-assembling biomaterials. Biomacromolecules. 2011 Oct 10;12:3549–58. doi: 10.1021/bm200763y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caplan MR, Moore PN, Zhang S, Kamm RD, Lauffenburger DA. Self-assembly of a b-sheet protein governed by relief of electrostatic repulsion relative to van der Waals attraction. Biomacromolecules. 2000;1:627–31. doi: 10.1021/bm005586w. [DOI] [PubMed] [Google Scholar]
- 59.Krishnan L, Hoying JB, Nguyen H, Song H, Weiss JA. Interaction of angiogenic microvessels with the extracellular matrix. Am J Physiol Heart Circ Physiol. 2007 Dec 6;293:H3650–8. doi: 10.1152/ajpheart.00772.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sieminski A, Hebbel R. Improved microvascular network in vitro by human blood outgrowth endothelial cells relative to vessel-derived endothelial cells. Tissue Eng. 2005;11(9):1332–45. doi: 10.1089/ten.2005.11.1332. [DOI] [PubMed] [Google Scholar]
- 61.Slaughter BV, Khurshid SS, Fisher OZ, Khademhosseini A, Peppas NA. Hydrogels in regenerative medicine. Adv Mater (Deerfield Beach, Fla) 2009 Sep;21(32–33):3307–29. doi: 10.1002/adma.200802106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu Q, Rounds S. Focal adhesion kinase and endothelial cell apoptosis. Microvasc Res. 2012 Jan 1;83:56–63. doi: 10.1016/j.mvr.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu MH. Endothelial focal adhesions and barrier function. J Physiol. 2005 Dec 2;569:359–66. doi: 10.1113/jphysiol.2005.096537. Pt. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heino J. The collagen family members as cell adhesion proteins. BioEssays. 2007 Oct 10;29:1001–10. doi: 10.1002/bies.20636. [DOI] [PubMed] [Google Scholar]
- 65.Sweeney SM, Orgel JP, Fertala A, McAuliffe JD, Turner KR, Di Lullo GA, et al. Candidate cell and matrix interaction domains on the collagen fibril, the predominant protein of vertebrates. J Biol Chem. 2008 Jul 30;283:21187–97. doi: 10.1074/jbc.M709319200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rowlands AS, George PA, Cooper-White JJ. Directing osteogenic and myogenic differentiation of MSCs: interplay of stiffness and adhesive ligand presentation. Am J Physiol Cell Physiol. 2008 Oct 4;295:C1037–44. doi: 10.1152/ajpcell.67.2008. [DOI] [PubMed] [Google Scholar]
- 67.Leong WS, Tay CY, Yu H, Li A, Wu SC, Duc D-H, et al. Thickness sensing of hMSCs on collagen gel directs stem cell fate. Biochem Biophys Res Commun. 2010 Oct 2;401:287–92. doi: 10.1016/j.bbrc.2010.09.052. [DOI] [PubMed] [Google Scholar]
- 68.Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, et al. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat Mater. 2010 Jun 6;9:518–26. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winer JP, Janmey PA, McCormick ME, Funaki M. Bone marrow-derived human mesenchymal stem cells become quiescent on soft substrates but remain responsive to chemical or mechanical stimuli. Tissue Eng Part A. 2009 Jan 1;15:147–54. doi: 10.1089/ten.tea.2007.0388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


