Abstract
Serum lipids are associated with myocardial infarction and cardiovascular disease in humans. Here we dissected the genetic architecture of blood lipid traits by applying genome-wide association studies (GWAS) in 1,256 pigs from Laiwu, Erhualian and Duroc × (Landrace × Yorkshire) populations, and a meta-analysis of GWAS in more than 2,400 pigs from five diverse populations. A total of 22 genomic loci surpassing the suggestive significance level were detected on 11 pig chromosomes (SSC) for six blood lipid traits. Meta-analysis of GWAS identified 5 novel loci associated with blood lipid traits. Comparison of GWAS loci across the tested populations revealed a substantial level of genetic heterogeneity for porcine blood lipid levels. We further evaluated the causality of nine polymorphisms nearby or within the APOB gene on SSC3 for serum LDL-C and TC levels. Of the 9 polymorphisms, an indel showed the most significant association with LDL-C and TC in Laiwu pigs. But the significant association was not identified in the White Duroc × Erhualian F2 resource population, in which the QTL for LDL-C and TC was also detected on SSC3. This indicates that population-specific signals may exist for the SSC3 QTL. Further investigations are warranted to validate this assumption.
Introduction
Blood lipids reflect lipid metabolism of the whole body and the health status of humans. Abnormal concentrations of blood lipids are associated with familial hypercholesterolemia, cardiovascular disease and diabetes. The clinical tests of total cholesterol (TC), low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein (HDL-C), triglycerides (TG) and atherosclerosis index (AI) are widely used in the cardiovascular disease risk assessment [1, 2]. Identifying causal genes regulating blood lipid levels will contribute to the prevention and treatment of atherosclerosis. So far, more than 100 quantitative trait loci (QTL) have been detected to associate with one or more serum lipid contents in humans. For example, 43 loci were found to associate with plasma lipoprotein concentration and cholesterol content by genome-wide association study (GWAS) in 17,296 women [3]. Several genes, such as GALNT2, TRIB1 and SORT1, are reported to be the causative genes for TG, very low-density lipoprotein (VLDL) and LDL-C, respectively [4–6]. However, these genetic variants together account for ~25-30% of the genetic component of phenotypic variation, suggesting that many serum lipid-associated genetic variants remain to be found [7].
Pigs have been used as a biomedical model of human disease for decades. A pig model for diabetes shows that 12-lipoxygenase and oxidant stress play key roles in accelerating atherosclerosis due to diabetes and hyperlipemia [8]. So far, more than 80 QTL for blood lipid levels have been reported in the porcine QTL database (http://www.animalgenome.org/cgi-bin/QTLdb/SS/index). We previously identified 15 QTL for serum lipids in a White Duroc × Erhualian F2 resource population by a genome-wide scan with 194 microsatellite markers [9]. By applying genome-wide association study (GWAS) with Illumina porcine 60K SNP beadchips, we further detected a total of 18 genomic loci for blood lipid levels in the same F2 resource population and a Sutai population. We found the most significant SNP in the F2 intercross at SSC3: 124.77 Mb for LDL-C and TC, proximal to the APOB gene [10, 11].
In the current study, we performed GWAS and a meta-analysis of GWAS in a total of 2,402 animals from five diverse populations, including a White Duroc × Erhualian F2 resource population, a Chinese synthetic commercial line of Sutai, a commercial Duroc × (Landrace × Large White) line (DLY), two Chinese indigenous pig breeds of Erhualian and Laiwu. We identified novel genomic loci associated with blood lipid contents and evaluated the associations of the APOB polymorphisms with blood lipids in pigs.
Materials and Methods
Ethic statement
All the tested animals are raised in compliance with the care and use guidelines of experimental animals established by the Ministry of Agriculture of China. This study was approved by the ethics committee of Jiangxi Agricultural University.
Experimental populations
The experimental animals used in this study included Laiwu (N = 316), Erhualian (N = 333) and DLY (N = 607) pigs. Besides, previously reported data from the F2 (N = 729) and Sutai (N = 417) pigs were used in the meta-analysis of GWAS. The F2 and Sutai populations were described in detail in our previous studies [9, 10]. In brief, two White Duroc sires and 17 Erhualian dams were mated to produce F1 animals, and then nine F1 boars were randomly crossed with 59 F1 sows to produce F2 individuals [9]. The Sutai pig is a Chinese synthetic commercial line that was originally derived from a cross between Duroc and Erhualian breeds and has experienced artificial selection for over 18 generations. Laiwu is a Chinese indigenous pig breed that is famous for its exceptionally high intramuscular fat content (9 ~ 12%). Erhualian is another Chinese indigenous pig breed that has been known for its high prolificacy. All animals were raised in standard indoor conditions and were fed three times a day with diet containing 16% crude protein, 3100 kJ of digestible energy and 0.78% lysine. Water was available ad libitum from nipple drinkers. F2 and Sutai pigs were slaughtered at age of 240 ± 3 days. Laiwu and Erhualian pigs were slaughtered at 300 ± 3 days. DLY pigs were raised in a commercial company under the same feeding condition and were slaughtered at around 180 days. All experimental pigs were fasted overnight (about 12 hours) but were free access to water before slaughter.
Phenotype recording
All blood samples were collected from the major arteries when the pigs were exsanguinated. After coagulation at room temperature, the clots were centrifuged at 3,000 rpm at 4°C for 20 min to separate serums. All serum samples were deposited at -80°C until utilized. We used the diagnostic kits of Determiner-L LDL-C, Determiner-L HDL-C, Determiner-L TG and Determiner-L TC II (Kyowa Medex, Japan) for measuring LDL-C, HDL-C, TG and TC levels, respectively, according to the manufacturer’s instructions. Atherosclerosis index (AI) was calculated according to the formula: AI = (TC-HDL-C)/HDL-C [1, 2]. All measurements were performed in an AU5421 Automatic Biochemistry Analyzer (Backman-Kelt, USA) at the First Affiliated Hospital of Nanchang University.
Chip SNP genotyping and quality control
All experimental pigs were genotyped using Porcine SNP60 BeadChips following the Infinium HD Assay Ultra protocol (Illumina, USA). The positions of 61,565 SNPs on the chip on the current pig genome assembly (Sscrofa 10.2) were retrieved from the NRSP-8 Community Data Repository (http://www.animalgenome.org/repository/pig/Genome_build_10.2_mappings/). Quality control (QC) procedures were performed by using Plink v1.07 [12]. SNPs with call rate < 95%, minor allele frequency (MAF) < 1%, or showing significant departure from the Hardy Weinberg equilibrium (P < 1×10-5) were removed from further analysis. The same QC criteria were applied to SNP data of all tested pigs. A final set of 47,158, 33,968 and 40,152 SNPs in Laiwu, Erhualian and DLY pigs, respectively, together with 40,790 SNPs in F2 pigs and 49,225 SNPs in Sutai pigs, were used for subsequent statistical analyses.
APOB SNP identification and genotyping
According to the APOB gene sequence in the pig reference genome assembly at NCBI (http://www.ncbi.nlm.nih.gov/gene), we designed 23 primer pairs (S1 Table) to screen exonic polymorphisms in the APOB gene using Primer 3 (http://primer3.ut.ee). DNA samples of two F0 boars and 13 F0 sows from the white Duroc × Erhualian F2 population were used to identify polymorphisms by Sanger sequencing. Amplification was performed in a 25-μl reaction mixture containing 50 ng of genomic DNA and 2 U of Taq DNA polymerase (Takara, Japan) under the thermocycle condition of 94°C for 4 minutes, 40 × (94°C for 30 sec, annealing temperature for 30 sec and 72°C for 45 sec) and 72°C for 10 minutes on a PE 9700 thermal cycler (Applied Biosystem, USA). SeqMan in the DNAStar software package (DNAStar, USA) was used to align the obtained sequences and the reference sequence. An indel and 8 single base mutations were identified on exons in the pig APOB gene. According to the pig APOB Refseq (GenBank accession: NW_003613573.1), these polymorphisms were all named after HGVS nomenclature.
We then genotyped these APOB polymorphisms in the five experimental populations using primers and probes listed in S2 Table. Eight SNPs were separately genotyped in a 10-μl reaction mixture including 30 ng of genomic DNA, 0.2 μM of each primer, 0.15 μM of each probe and 5 mM of TaqMan Genotyping Master Mix (Applied Biosystems, USA) at 95°C for 10 minutes, 40 × (95°C for 15 sec and 60°C for 1 minute) on an ABI 7900 HT (Applied Biosystems, USA). The indel was genotyped by routine PCR and agarose gel electrophoresis (S1 Fig). PCR products were sequenced after being purified with the QIAquick DNA Purification Kit (Qiagen, Germany) to check the sequence identity.
Statistical analysis
GWAS and Meta-analysis
Heritability of each trait was estimated by using –lmm procedure of GEMMA based on genomic relationship matrix [13]. A mixed model was used to analyze the associations of the eligible SNPs with blood lipid traits: Y = Xb + Sα+ Zμ+ e, where b is the fixed effects that included sex and batch (the batch was 22, 11 and 4 for DLY, Erhualian and Laiwu, respectively); X is the incidence matrix of the fixed effects; α is the SNP substitution effect; S is the incidence matrix for α; μ is the vector of random additive genetic effects that follow the distribution N (0, Gσ2), where G is the kinship matrix derived from SNP markers [14, 15] and σ2 is the additive variance; Z is the identity matrix for μ; e is the residual error. The mmscore function of GenABEL was used to estimate the significance of associations between SNP markers and target traits [16]. A bonferroni correction was applied to determine the genome-wide (P < 0.05/SNP number) and suggestive (P < 1/SNP number) significance thresholds. The meta-analysis of GWAS was performed on the five populations of F2, Sutai, DLY, Laiwu and Erhualian pigs by employing METAL [17]. In brief, for each marker, the same reference allele was selected for all tested populations and a Z-score for evidence of association was calculated. The Z-statistics summarized the magnitude and the direction of allelic effect relative to the reference allele. An overall Z-score and p-value were then calculated from a weighted sum of the individual statistics. Weights were proportional to the square-root of the number of individuals examined in each population. The porcine genome assembly 10.2 was retrieved to characterize functionally plausible candidate genes (http://www.ensembl.org/Sus_scrofa/Location/Genome).
Population stratification affects the validity of genome-wide association study [18]. Population stratification was corrected by fitting the covariance among individuals that was inferred from high density SNP data. Moreover, genomic control (GC) was used to correct the effect of stratification (λ) that was estimated from the null test statistics (under the null hypothesis of no SNP associated with the trait) [19]. Here, we evaluated population stratification by examining the distribution of test statistics in a quantile-quantile (Q-Q) plot [18]. The Q-Q plots were constructed with R software.
Linkage disequilibrium and linkage analysis (LDLA) for SNPs on SSC3
Haplotypes along SSC3 were reconstructed for F2, Laiwu and DLY pigs using the 60K SNP data, pedigree information and a Hidden Markov model [20]. The model simultaneously phased SNP genotypes and assigned the ensuing haplotypes to a predetermined number of ancestral haplotypes. Then, the effects of these ancestral haplotypes were estimated using a mixed model framework: Y = Xb + Zu + e [21], where Y is the vector of phenotypes; b is the estimator of fixed effects including sex and batch; X and Z are the incidence matrices for b and u; u is the random additive genetic effect following the multinormal distribution u ~ N (0, Gσα 2), in which G is the individual-individual similarity matrix which was calculated from SNP information on SSC3 and σα 2 is the polygenetic additive variance; and e is a vector of residual error with a distribution of N(0, Iσe 2). The haplotype-based LDLA analysis can use both within-family linkage information and across-family linkage disequilibrium information resulting from historical recombination events in ancestors of founder animals. The LDLA analysis was conducted using R scripts.
Single marker association analysis and F-drop test
Single marker association test was performed for the nine polymorphisms. The genotypes of these polymorphisms were integrated into the 60K SNP genotype data of F2 and Laiwu pigs, respectively. The associations of these polymorphisms with blood lipids were assessed by the GWAS analysis as described above. Sex and batch were treated as fixed effects, and population substructure was corrected by fitting the kinship matrix derived from the 60K SNP data. The bonferroni correction was used to evaluate the significance threshold of the associations. In the Laiwu population, F-drop test was performed by separately including the genotypes of the indel, NW_003613573.1:g.20713A>G and NW_003613573.1:g.48834A>T as a fixed effect in the single marker association analysis.
Results
Phenotypic values
The phenotypic values of the Sutai and F2 populations have been reported in our previous studies [9, 10]. The phenotypic values measured in the Laiwu, DLY and Erhualian populations are listed in S3 Table. AI, which is set as an index to measure the degree of atherosclerosis by the international medical community, was explored for GWAS in pigs for the first time. The correlations between the six analyzed blood lipid traits are shown in S2 Fig. We estimated the heritability of these blood lipid traits, ranging from 0.12 to 0.57 in the tested populations.
GWAS results for blood lipid levels in the three pig populations
The GWAS results of blood lipid traits in the F2 and Sutai populations have been shown in our previous paper [10]. In this study, we firstly performed the GWAS analysis in the Laiwu, DLY and Erhualian populations using an additive model. The “Q-Q” plots showed that the distribution of observed P values deviated from expected P values in the extreme tail (S3 Fig). However, the inflation factor (λ) values were around 1.0 in the three experimental populations, indicating that population structures were properly corrected.
We identified a total of 105, 9 and 8 SNPs that achieved the suggestive significance level for the measured traits in the Laiwu (P < 1/47,158), DLY (P < 1/40,152) and Erhualian (P < 1/33,968) populations, respectively. We found 67 SNPs that surpassed the genome-wide significance level in Laiwu pigs (P < 0.05/47,158). The most significant SNP across all traits was MARC0083986 (P = 3.25 × 10-11) at 125.21Mb on SSC3 in Laiwu pigs (Table 1). In the DLY population, only one SNP on SSC3 surpassed the genome-wide significance level for AI (P < 0.05/40,152). However, in Erhualian pigs, no SNPs achieved the genome-wide significance level for all six blood lipid traits (P > 0.05/33,968).
Table 1. Genome-wide significance loci identified by GWAS for serum lipids in the three tested populations.
| Trait | Population | NSNP a | Top SNP | Position (bp) | Allele effect b | P value | Candidate gene c |
|---|---|---|---|---|---|---|---|
| LDL-C | Laiwu | 39 | MARC0083986 | 3: 125211999 | 0.20 ± 0.03 | 3.25 × 10-11 | NCOA1, KLHL29, APOB, C2ORF43, TTC32 |
| TC | Laiwu | 27 | H3GA0010711 | 3: 126108723 | 0.21 ± 0.03 | 2.72 × 10-9 | NCOA1, KLHL29, APOB, C2ORF43, TTC32 |
| HDL/LDL | Laiwu | 1 | MARC0010341 | 5: 25185108 | 0.24 ± 0.04 | 4.75 × 10-7 | TAC3, GPR182 |
| AI | DLY | 1 | ALGA0021166 | 3: 123041389 | 0.20 ± 0.04 | 2.78 × 10-7 | NCOA1, KLHL29, APOB |
a. The number of SNPs that achieved genome-wide significance level.
b. Allele substitution effect, the least square mean ± SE was showed for each phenotype; the unit for LDL-C and TC is mmol/L
c. Candidate genes were selected from annotated genes with functional relevance to blood lipids or lipid metabolism in an interval of 5 Mb centered at the top SNP at each significant locus.
Low density lipoproteins
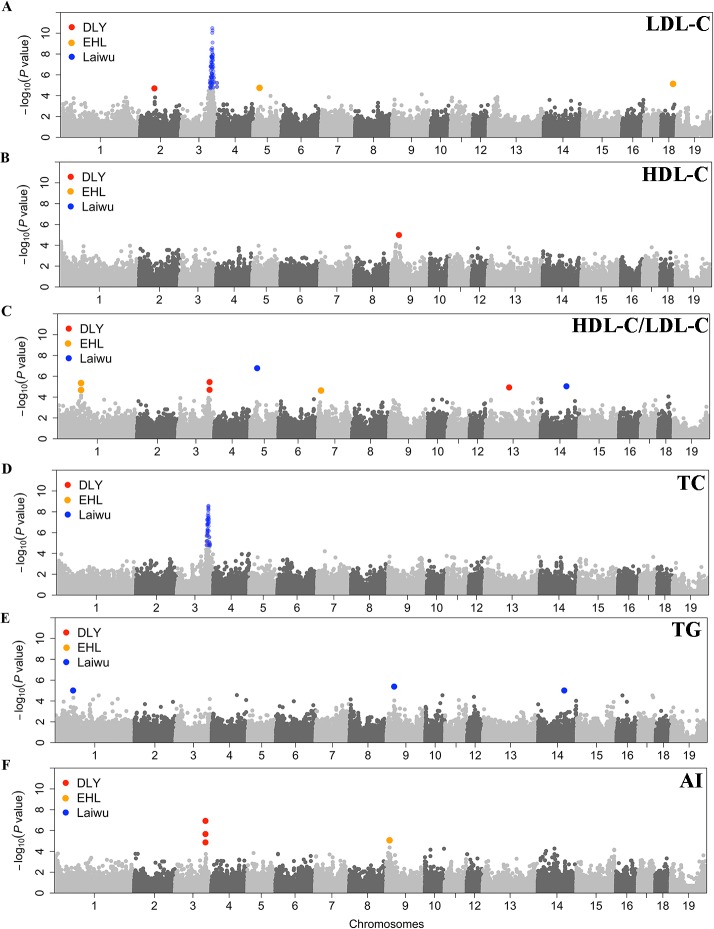
As presented in S4 Table, a total of 61 SNPs located on SSC3 showed association signals for LDL-C in the Laiwu population. All these SNPs are located in a region of 116.38 to 143.79 Mb, which has been reported to harbor QTL for LDL-C in pigs [10, 22]. The most significant SNP was found at SSC3: 125,211,999 bp (P = 3.25 × 10-11, Fig 1A). In the DLY population, only one SNP at SSC2: 60.34 Mb achieved the suggestive significance level for LDL-C (P = 2.15 × 10-5). In the Erhualian population, we identified 4 SNPs surpassing the suggestive significance level, including 3 SNPs on SSC18 and one SNP on SSC5.
Fig 1. Manhattan plots of GWAS for blood lipids in the Laiwu, DLY and Erhualian populations.
SNPs surpassing the threshold of suggestive significance in the three populations are denoted with different colors (red for DLY, orange for EHL and blue for Laiwu). (A-F) for LDL-C, HDL-C, HDL-C/LDL-C, TC, TG and AI, respectively.
High density lipoproteins
We only identified one SNP at SSC9: 18.74 Mb that surpassed the suggestive significance level in the DLY population (P = 1.34 × 10-5, Fig 1B). In Laiwu and Erhualian pigs, no HDL-associated SNP was identified (S4 Table).
HDL-C/LDL-C
As shown in S4 Table, we detected 6 genomic loci that were significantly associated with HDL-C/LDL-C, including 2 loci at SSC5: 25.19 Mb and SSC14: 109.02 Mb in Laiwu pigs, 2 loci at SSC3: 123.04-123.39 Mb and SSC13: 84.66Mb in the DLY population, and 2 loci at SSC1: 94.11-95.10 Mb and SSC7: 7.47 Mb in the Erhualian population (Fig 1C).
Total cholesterol
We identified 39 TC-associated SNPs at SSC3: 118.61-131.79 Mb (P = 1.76 × 10-5 to 2.72 × 10-9) in Laiwu pigs (S4 Table). In the DLY population, one SNP MARC0085934 at SSC3: 126,039,599 bp was detected to significantly associate with TC (P = 1.22 × 10-5). However, no TC-associated SNPs were identified in the Erhualian population (Fig 1D).
Triglycerides
We found a total of 3 SNPs that were significantly associated with TG in the Laiwu population, including one SNP at SSC1: 74.02 Mb, one SNP at SSC9: 32.88 Mb and one SNP at SSC14: 107.03 Mb. No TG-associated SNPs were detected in the DLY and Erhualian populations (Fig 1E).
Atherosclerosis index
As presented in S4 Table, we did not detect any significant SNPs for AI in Laiwu pigs. But we identified 3 SNPs at SSC3: 123.04-123.39 Mb that were significantly associated with AI in the DLY population (P = 8.08 × 10-6 to 2.78 × 10-7). Further, we detected another AI-associated SNP at SSC9: 14,306,925 bp in the Erhualian population (P = 1.19 × 10-5, Fig 1F).
Novel loci identified by the meta-analysis
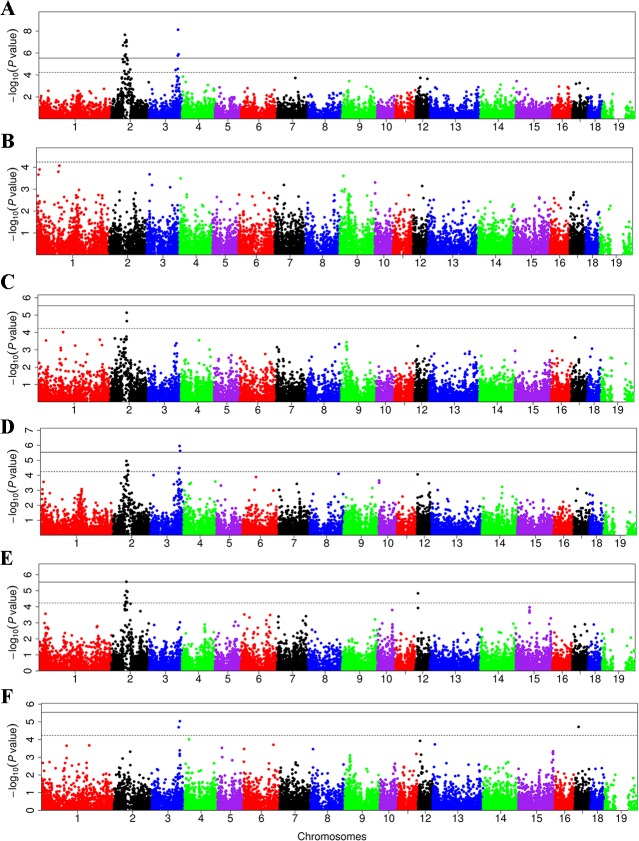
To identify novel significant loci, we performed a meta-analysis in the five pig populations. The results are shown in S5 Table. We detected a total of 51 SNPs within 9 genomic regions that were significantly associated with blood lipid traits (Fig 2). Eighteen out of the 51 SNPs surpassed the genome-wide significance level. Of the 9 genomic loci, to our knowledge, two loci (SSC2: 58.58-69.29 Mb and SSC12: 10.85 Mb) are reported to associate with TG for the first time (Fig 2E). SSC2: 58.58-66.37 Mb, SSC17: 16.77 Mb and SSC2: 70.54-71.04 Mb showed association with TC, AI and HDL-C/LDL-C, respectively. Moreover, the locus on SSC3 was evidenced to associate with LDL-C, TC and AI in the meta-analysis (Fig 2).
Fig 2. Manhattan plots of meta-analysis of GWAS for blood lipid traits in the five experimental populations.
The x-axis shows chromosomal positions, and y-axis shows negative log10 P values. The solid and dashed lines indicate the thresholds of genome-wide and suggestive significance, respectively. Panels (A) to (F) represent the meta-analysis results for LDL-C, HDL-C, HDL-C/LDL-C, TC, TG and AI, respectively.
LDLA mapping result for blood lipid traits with SNPs on SSC3
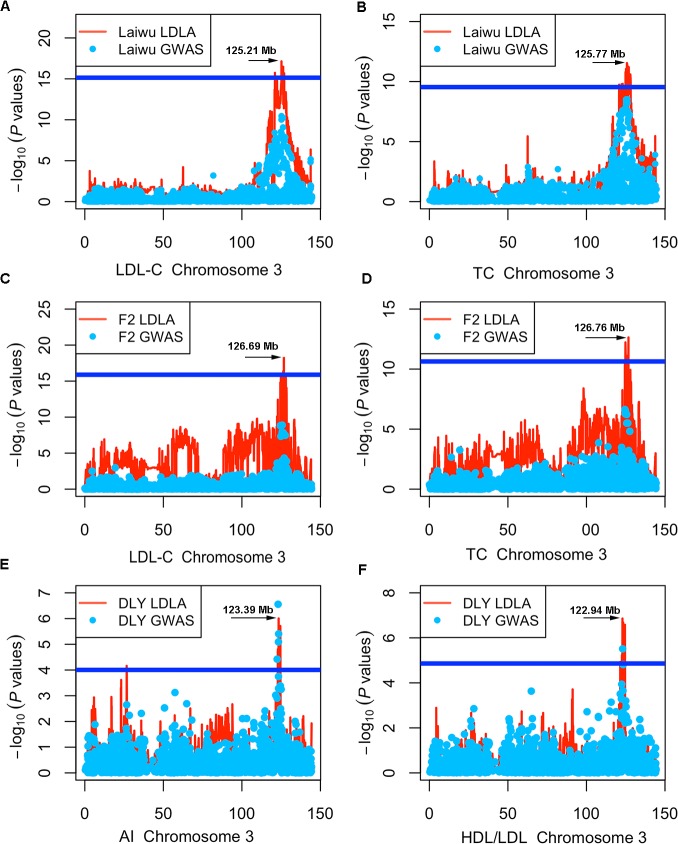
We have previously tested the causality of LDLR, a promising candidate gene for the significant locus on SSC2, for serum lipids [11]. Here, we made a close examination on the SSC3 locus that harbored a cluster of significant SNPs for blood lipids in Laiwu, F2 and DLY pigs (Fig 1). Our haplotype-based association study (LDLA mapping) showed the most significant haplotypes around the 125.0-127.0 Mb region on this chromosome, which encompassed a highly plausible causative gene: APOB (Fig 3). The most prominent SNP (MARC0083986) and haplotype that we identified are all proximal to the APOB gene in Laiwu pigs.
Fig 3. LDLA analysis for blood lipid traits with the SNPs on SSC3 in the Laiwu, F2 and DLY populations.
The x-axis indicates the SNP positions on SSC3 (Mb), and y-axis shows negative log10 (P values) from haplotype-based association study. Blue dots show the qualified SNPs in GWAS, and red lines represent the haplotype. The horizontal line indicates the 95% of confidence interval by LOD drop off 2 from the most significant haplotype. (A-B) for LDL-C and TC in the Laiwu population; (C-D) for LDL-C and TC in the F2 population; (E-F) for AI and HDL-C/LDL-C in the DLY population.
Associations of APOB polymorphisms with blood lipid traits
We screened exonic variants of the APOB gene by Sanger sequencing of 15 F0 founder animals of the F2 population. Blasting of the flanking sequence of the nine polymorphisms against the human reference genome at http://asia.ensembl.org/index.html revealed that the indel and NW_003613573.1:g.20713A>G variants have no hits with human APOB gene and the other seven are within the human APOB gene. Furthermore, we blasted the indel and NW_003613573.1:g.20713A>G variants against the pig reference genome sequence in the Ensembl database and the Wuzhishan genome, a Chinese pig genome [23]. The blasting result showed that the two polymorphisms should be in the upstream of the pig APOB. As a result, we identified 8 SNPs and an indel, of which two are tested in the upstream of the APOB gene and the others reside in the coding region of this gene. To assess the associations of these APOB polymorphisms with blood lipids, we firstly performed the standard association test in the Laiwu and F2 populations (Table 2). The indel, NW_003613573.1:g.20713A>G and NW_003613573.1:g.48834A>T variants showed significant associations with LDL-C in Laiwu pigs (P < 2.12 × 10-5). The association was also observed between the indel and NW_003613573.1:g.20713A>G with TC in the Laiwu population. To further evaluate the causality of the indel, NW_003613573.1:g.20713A>G and NW_003613573.1:g.48834A>T variants for LDL-C and TC in Laiwu pigs, we performed the F-drop test. Only in the case of the indel mutation, the association signals for LDL-C and TC vanished completely in this population (Fig 4). None of these variants was associated with LDL-C and TC in the F2 population (P > 2.45 × 10-5).
Table 2. Single marker association test of the 9 polymorphisms with LDL-C and TC in the Laiwu and F2 populations.
| Population | Variant | MAF | HWE a | Single marker association test | |
|---|---|---|---|---|---|
| LDL-C (P value) | TC (P value) | ||||
| Laiwu (n = 300) | NW_003613573.1:g.19863_20286delinsCATCGTACCGA | 0.35 | 6.00 × 10-3 | 8.15 × 10-8 | 1.34 × 10-7 |
| NW_003613573.1:g.20713A>G | 0.44 | 3.00 × 10-3 | 8.05 × 10-7 | 2.05 × 10-5 | |
| NW_003613573.1:g.48236G>C | 0.26 | 0.63 | 0.43 | 0.06 | |
| NW_003613573.1:g.48834A>T | 0.49 | 0.1 | 1.87 × 10-7 | 9.00 × 10-4 | |
| NW_003613573.1:g.48943G>C | 0.17 | 1 | 0.19 | 4.00 × 10-3 | |
| NW_003613573.1:g.50536T>C | 0.05 | 0.98 | 0.39 | 0.58 | |
| NW_003613573.1:g.53468G>A | 0.26 | 0.57 | 0.42 | 0.06 | |
| NW_003613573.1:g.53546G>A | 0.06 | 0.7 | 0.81 | 0.31 | |
| NW_003613573.1:g.58318C>G | 0.15 | 0.64 | 0.75 | 0.14 | |
| F2 (n = 729) | NW_003613573.1:g.19863_20286delinsCATCGTACCGA | 0.27 | 0.03 | 0.18 | 0.22 |
| NW_003613573.1:g.20713A>G | 0.29 | 0.77 | 1.00 × 10-3 | 7.00 × 10-4 | |
| NW_003613573.1:g.48236G>C | 0.13 | 7.00 × 10-3 | 0.01 | 7.00 × 10-3 | |
| NW_003613573.1:g.48834A>T | 0.5 | 6.00 × 10-4 | 0.07 | 0.02 | |
| NW_003613573.1:g.48943G>C | 0.34 | 2.00 × 10-3 | 0.25 | 0.31 | |
| NW_003613573.1:g.50536T>C | 0.26 | 3.00 × 10-3 | 0.32 | 0.38 | |
| NW_003613573.1:g.53468G>A | 0.19 | 0.18 | 0.21 | 0.03 | |
| NW_003613573.1:g.53546G>A | 0.09 | 0.17 | 0.57 | 0.91 | |
| NW_003613573.1:g.58318C>G | 0.44 | 2.00 × 10-4 | 0.01 | 9.00 × 10-3 | |
aHWE, Hardy Weinberg Equilibrium.
Fig 4. The results of F-drop test in Laiwu pigs.
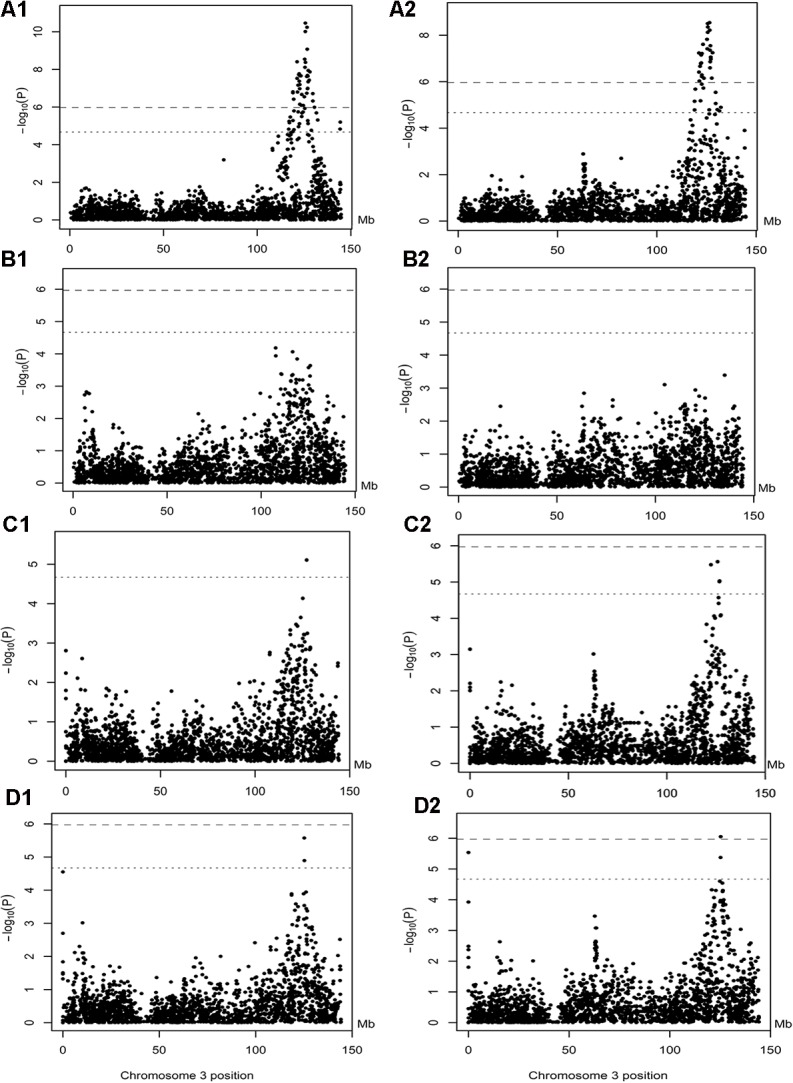
The x-axis shows SNP positions on SSC3 (Mb); the negative log10 (P values) is presented on the y-axis. The horizontal dotted and dashed lines indicate the thresholds of suggestive and genome-wide significance, respectively. A1 and A2 show the GWAS results for LDL-C and TC in the Laiwu population with all qualified SNPs on SSC3, in which the genotypes of 9 polymorphisms were also included; B1, C1 and D1 show the F-drop test results for LDL-C, in which the genotypes of indel, NW_003613573.1:g.20713A>G and NW_003613573.1:g.48834A>T were included as a fixed effect in the model, respectively; B2, C2 and D2 present the F-drop test results for TC.
Discussion
In this study, we investigated the genetic basis of blood lipid traits in more than 2,400 pigs from five populations through GWAS and meta-analysis. To our knowledge, our sample size is the largest one in the GWAS for porcine blood lipid traits. We identified a total of 22 genomic regions that were significantly associated with blood lipid levels in the five populations. The tested populations shared few genomic loci. This provides strong evidence of a substantial level of genetic heterogeneity for pig serum lipids. Of the 22 genomic regions, only 6 were identified in more than one population, and 16 loci were detected in a single population.
The genomic regions associated with blood lipid traits shared by multiple pig populations
Several significantly associated genomic regions identified in this study replicated the findings in the previous reports. SSC3: 124.0-126.0 Mb was associated with LDL-C and TC in Laiwu pigs in this study. This region was also associated with LDL-C and TC in the F2 population [10]. Moreover, Manunza et al. [24] and Gallardo et al. [22] detected significant associations for LDL-C and TC in Duroc pigs in this region. SSC2: 60.34 Mb was associated with LDL-C in the DLY population (P = 2.15 × 10-5). This region has also been shown to be significantly associated with LDL-C in Sutai and F2 pigs [10]. SNP MARC0010324 (SSC9: 18.74 Mb) had significant association with HDL-C in the DLY population. Interestingly, Gallardo et al. [22] also detected a QTL for HDL-C around this region in Duroc pigs. The genomic regions associated with TG on SSC12 and SSC14 were consistent with the findings by Gallardo et al. [22] and Manunza et al. [24], respectively. Further fine-mapping would be needed to examine whether allelic heterogeneity, a frequent feature of polygenic traits in humans [25–27], exists or not in the above-mentioned genomic loci shared by multiple populations.
Population-specific genomic regions associated with porcine blood lipid traits
Of 22 significant loci that we identified, 16 were detected in a single population, suggesting the existence of population heterogeneity (Fig 1). For certain, we cannot rule out the possibility that different sample sizes and phenotypes measured at different ages may cause different GWAS results in each population. At the SSC2 locus, we have observed the association signals for LDL-C and TC in both F2 and Sutai populations. We highlight LDLR as a strong candidate gene at this locus, and further show that different causative variants in this gene likely underlie phenotypic variation in the two populations [11]. Manunza et al. have reported age-specific genetic determinants for porcine serum lipid traits [24]. In humans, most of significant loci for blood lipids were different among different aged individuals or ethnic groups [28, 29]. Therefore, further investigations are warranted to test if population-specific causative variants underlie the significant loci identified in this study.
Possible pleiotropic QTL
We found two genomic regions that were significantly associated with more than one trait in GWAS and meta-analysis. SSC2: 60.0-71.0 Mb was associated with LDL-C, TC, TG and HDL-C/LDL-C in the meta-analysis (S5 Table), and SSC3: 121.0-127.0 Mb was related to LDL-C, TC and AI. The finding can be explained by: 1) QTL with a common variant with pleiotropic effects; 2) Strong correlation between the associated phenotypes (S2 Fig), as previously observed between TC and LDL-C, and between TC and HDL-C [9]; 3) The two regions contain more than one QTL influencing blood lipid traits.
Plausible candidate genes at the identified loci
To identify candidate genes at the genomic loci surpassing the genome-wide significance level, we searched annotated genes with functional relevance to blood lipids or lipid metabolism in an interval of 5 Mb centered at the top SNP. At the SSC3 locus, NCOA1, KLHL29, APOB, C2ORF43 and TTC32 are promising candidate genes. NCOA1 encodes a transcriptional coactivator for steroid. It controls energy balance between white and brown adipose tissues [30]. Both KLHL29 and APOB have been reported as candidate genes for blood lipids [31]. The function of KLHL29 is not yet known, but this gene has been implicated in coronary heart disease [31]. APOB encodes the main apolipoprotein of chylomicrons and low density lipoproteins. It appears to be a strong candidate gene affecting LDL-C and TC in both humans and pigs [9, 22, 32–34]. Several polymorphisms at the APOB locus, including the 3611 MspI polymorphism in the promoter region and a truncating mutation, have been shown to associate with dyslipidemia in humans [32, 35]. C2ORF43 is functionally related to defective apolipoprotein b-100 and coronary heart disease [36]. Lu et al. reported that TTC32 is a strong candidate gene for coronary artery disease by GWAS [37]. SMARCA4, LDLR and INSR play a role in lipid metabolism [26, 38]. These genes reside in the genomic region of SSC2: 64.97-70.25 Mb that was significantly associated with LDL-C and TC in this study. We have investigated that LDLR is a causative gene for LDL-C and TC in the Sutai population, but allelic heterogeneity exists between different populations [11]. TAC3 and GPR182 have been investigated as candidate genes for the locus on SSC5 [39, 40], where MARC0010341 at 25.19 Mb (P = 4.75 × 10-7) was significantly associated with HDL-C/LDL-C in this study.
Associations of APOB polymorphisms with porcine blood lipid levels
We identified 9 polymorphisms nearby or within the APOB gene in the founders of F2 intercross. Standard association test and F-drop test strongly suggested that the indel is a strong candidate causative mutation for LDL-C and TC at the SSC3 locus in Laiwu pigs. This conclusion was strengthened by the observation that the indel was not found or was segregated at very low frequency in the Sutai, Erhualian and DLY populations (S6 Table), in which the effect of the SSC3 locus was not identified. Unexpectedly, no significant association was observed between the indel and LDL-C or TC in the F2 population in which this variant was segregating at a MAF of 0.27 (Table 2). This observation could be explained by 1) population heterogeneity exists between the two populations. A line of supporting evidence comes from a finding that the most significant haplotype for LDL-C and TC was not identified in the same region on SSC3 in the Laiwu (125.21 Mb and 125.77 Mb) and F2 (126.69 Mb and 126.76 Mb) populations. So variants in different genes may cause the association signal on this chromosome; 2) the indel is not the causative mutation for LDL-C and TC. It is just in high linkage-disequilibrium with the real causative mutation.
Conclusions
In conclusion, we performed GWAS to identify genomic loci associated with blood lipids in five diverse pig populations. Our results highlight a substantial level of population heterogeneity for genetic components of porcine blood lipid traits. Our previous and current studies collectively suggest that population-specific variants may cause the QTL effect on SSC2 and SSC3 identified in multiple pig breeds. APOB is a promising candidate gene for LDL-C and TC at the SSC3 locus. Further fine mapping and functional assays are required to confirm the causality of the indel variant in the APOB gene for LDL-C and TC.
Supporting Information
QQ, Qq and qq stand for homozygote of normal, heterozygote and homozygote of indel, respectively.
(TIF)
The dots indicate the significant correlation coefficients between each pair of traits. Their sizes and colors represent the degree and direction (positive and negative) of the correlations, respectively.
(TIF)
(A1-F1) For LDL-C, HDL-C, HDL-C/LDL-C, TC, TG and AI in the Laiwu population; (A2-F2) For LDL-C, HDL-C, HDL-C/LDL-C, TC, TG and AI in DLY pigs; (A3-F3) For LDL-C, HDL-C, HDL-C/LDL-C, TC, TG and AI in the Erhualian population.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLS)
(XLS)
(XLSX)
Acknowledgments
We are grateful to colleagues in Key Laboratory for Animal Biotechnology of Jiangxi Province and the Ministry of Agriculture of China, Jiangxi Agricultural University for sample collection. We also thank Caicheng Zhang (First Affiliated Hospital of Nanchang University) for his help in measuring blood lipids.
Data Availability
All genotype and phenotype files will be available from the Dryad database (URL: http://datadryad.org/review?doi=doi:10.5061/dryad.4gh70).
Funding Statement
This study was supported by the Natural Science Foundation of China (31160225) and Jiangxi Provincial project for cultivating young scientists (2010BQ01500). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kayamori F, Igarashi K. Effects of dietary nasunin on the serum cholesterol level in rats. Bioscience, biotechnology, and biochemistry. 1994;58(3):570–1. 10.1271/bbb.58.570 [DOI] [Google Scholar]
- 2. Benson MK, Devi K. Influence of omega-6/omega-3 rich dietary oils on lipid profile and antioxidant enzymes in normal and stressed rats. Indian journal of experimental biology. 2009;47(2):98–103. . [PubMed] [Google Scholar]
- 3. Chasman DI, Pare G, Mora S, Hopewell JC, Peloso G, Clarke R, et al. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS genetics. 2009;5(11):e1000730 10.1371/journal.pgen.1000730 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Holleboom AG, Karlsson H, Lin RS, Beres TM, Sierts JA, Herman DS, et al. Heterozygosity for a loss-of-function mutation in GALNT2 improves plasma triglyceride clearance in man. Cell metabolism. 2011;14(6):811–8. 10.1016/j.cmet.2011.11.005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burkhardt R, Toh SA, Lagor WR, Birkeland A, Levin M, Li X, et al. Trib1 is a lipid- and myocardial infarction-associated gene that regulates hepatic lipogenesis and VLDL production in mice. The Journal of clinical investigation. 2010;120(12):4410–4. 10.1172/JCI44213 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466(7307):714–9. 10.1038/nature09266 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Willer CJ, Mohlke KL. Finding genes and variants for lipid levels after genome-wide association analysis. Current opinion in lipidology. 2012;23(2):98–103. 10.1097/MOL.0b013e328350fad2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Natarajan R, Gerrity RG, Gu JL, Lanting L, Thomas L, Nadler JL. Role of 12-lipoxygenase and oxidant stress in hyperglycaemia-induced acceleration of atherosclerosis in a diabetic pig model. Diabetologia. 2002;45(1):125–33. 10.1007/s001250200014 . [DOI] [PubMed] [Google Scholar]
- 9. Chen R, Ren J, Li W, Huang X, Yan X, Yang B, et al. A genome-wide scan for quantitative trait loci affecting serum glucose and lipids in a White Duroc x Erhualian intercross F(2) population. Mammalian genome: official journal of the International Mammalian Genome Society. 2009;20(6):386–92. 10.1007/s00335-009-9190-9 . [DOI] [PubMed] [Google Scholar]
- 10. Chen C, Yang B, Zeng Z, Yang H, Liu C, Ren J, et al. Genetic dissection of blood lipid traits by integrating genome-wide association study and gene expression profiling in a porcine model. BMC genomics. 2013;14:848 10.1186/1471-2164-14-848 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zeng Z, Chen R, Liu C, Yang H, Chen C, Huang L. Evaluation of the causality of the low-density lipoprotein receptor gene (LDLR) for serum lipids in pigs. Animal genetics. 2014;45(5):665–73. 10.1111/age.12183 . [DOI] [PubMed] [Google Scholar]
- 12. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81(3):559–75. 10.1086/519795 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nature genetics. 2012;44(7):821–4. 10.1038/ng.2310 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hayes BJ, Goddard ME. Technical note: prediction of breeding values using marker-derived relationship matrices. Journal of animal science. 2008;86(9):2089–92. 10.2527/jas.2007-0733 . [DOI] [PubMed] [Google Scholar]
- 15. Mateus JC, Eding H, Penedo MC, Rangel-Figueiredo MT. Contributions of Portuguese cattle breeds to genetic diversity using marker-estimated kinships. Animal genetics. 2004;35(4):305–13. 10.1111/j.1365-2052.2004.01168.x . [DOI] [PubMed] [Google Scholar]
- 16. Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23(10):1294–6. 10.1093/bioinformatics/btm108 . [DOI] [PubMed] [Google Scholar]
- 17. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. 10.1093/bioinformatics/btq340 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA: the journal of the American Medical Association. 2008;299(11):1335–44. 10.1001/jama.299.11.1335 . [DOI] [PubMed] [Google Scholar]
- 19. Devlin B, Roeder K, Wasserman L. Genomic control, a new approach to genetic-based association studies. Theoretical population biology. 2001;60(3):155–66. [DOI] [PubMed] [Google Scholar]
- 20. Druet T, Georges M. A hidden markov model combining linkage and linkage disequilibrium information for haplotype reconstruction and quantitative trait locus fine mapping. Genetics. 2010;184(3):789–98. 10.1534/genetics.109.108431 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Z, Guillaume F, Sartelet A, Charlier C, Georges M, Farnir F, et al. Ancestral haplotype-based association mapping with generalized linear mixed models accounting for stratification. Bioinformatics. 2012;28(19):2467–73. 10.1093/bioinformatics/bts348 . [DOI] [PubMed] [Google Scholar]
- 22. Gallardo D, Pena RN, Amills M, Varona L, Ramirez O, Reixach J, et al. Mapping of quantitative trait loci for cholesterol, LDL, HDL, and triglyceride serum concentrations in pigs. Physiological genomics. 2008;35(3):199–209. 10.1152/physiolgenomics.90249.2008 . [DOI] [PubMed] [Google Scholar]
- 23. Fang X, Mou Y, Huang Z, Li Y, Han L, Zhang Y, et al. The sequence and analysis of a Chinese pig genome. GigaScience. 2012;1(1):16 10.1186/2047-217X-1-16 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Manunza A, Casellas J, Quintanilla R, Gonzalez-Prendes R, Pena RN, Tibau J, et al. A genome-wide association analysis for porcine serum lipid traits reveals the existence of age-specific genetic determinants. BMC genomics. 2014;15:758 10.1186/1471-2164-15-758 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu Y, Waite LL, Jackson AU, Sheu WH, Buyske S, Absher D, et al. Trans-ethnic fine-mapping of lipid loci identifies population-specific signals and allelic heterogeneity that increases the trait variance explained. PLoS genetics. 2013;9(3):e1003379 10.1371/journal.pgen.1003379 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–13. 10.1038/nature09270 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–8. 10.1038/nature09410 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kamatani Y, Matsuda K, Okada Y, Kubo M, Hosono N, Daigo Y, et al. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nature genetics. 2010;42(3):210–5. 10.1038/ng.531 . [DOI] [PubMed] [Google Scholar]
- 29. Dumitrescu L, Brown-Gentry K, Goodloe R, Glenn K, Yang W, Kornegay N, et al. Evidence for age as a modifier of genetic associations for lipid levels. Annals of human genetics. 2011;75(5):589–97. 10.1111/j.1469-1809.2011.00664.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Santa-Maria CA, Dantzer J, Li L, Skaar T, Oesterreich S, Rae JM, et al. Association of variants in candidate genes on lipid profiles in women with early breast cancer on adjuvant aromatase inhibitor therapy. Cancer Res. 2013;73 10.1158/0008-5472. SABCS13-P1-08-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gottesman O, Drill E, Lotay V, Bottinger E, Peter I. Can genetic pleiotropy replicate common clinical constellations of cardiovascular disease and risk? PloS one. 2012;7(9):e46419 10.1371/journal.pone.0046419 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Al-Bustan SA, Alnaqeeb MA, Annice BG, Ebrahim GA, Refai TM. Genetic association of APOB polymorphisms with variation in serum lipid profile among the Kuwait population. Lipids in health and disease. 2014;13(1):157 10.1186/1476-511X-13-157 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakazone MA, De Marchi MA, Pinhel MA, Barros CF, Julio MA, Pinheiro A, et al. Effects of APOE, APOB and LDLR variants on serum lipids and lack of association with xanthelasma in individuals from Southeastern Brazil. Genetics and molecular biology. 2009;32(2):227–33. 10.1590/S1415-47572009005000028 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chasman DI, Pare G, Zee RY, Parker AN, Cook NR, Buring JE, et al. Genetic loci associated with plasma concentration of low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, triglycerides, apolipoprotein A1, and Apolipoprotein B among 6382 white women in genome-wide analysis with replication. Circulation Cardiovascular genetics. 2008;1(1):21–30. 10.1161/CIRCGENETICS.108.773168 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang LS, de Graaf J, Breslow JL. ApoB gene MspI RFLP in exon 26 changes amino acid 3611 from Arg to Gln. Journal of lipid research. 1988;29(1):63–7. . [PubMed] [Google Scholar]
- 36. Shen H, Damcott CM, Rampersaud E, Pollin TI, Horenstein RB, McArdle PF, et al. Familial defective apolipoprotein B-100 and increased low-density lipoprotein cholesterol and coronary artery calcification in the old order amish. Archives of internal medicine. 2010;170(20):1850–5. 10.1001/archinternmed.2010.384 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lu X, Wang L, Chen S, He L, Yang X, Shi Y, et al. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nature genetics. 2012;44(8):890–4. 10.1038/ng.2337 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xu S. Theoretical basis of the Beavis effect. Genetics. 2003;165(4):2259–68. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Steinhoff MS, Mentzer BV, Geppetti P, Pothoulakis C, Bunnett NW. Tachykinins and their receptors: contributions to physiological control and the mechanisms of disease. American physiological society. 2014;Vol.94 no.1, 265–301. 10.1152/physrev.00031.2013 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kwon DN, Chang BS, Kim JH. Gene expression and pathway analysis of effects of the CMAH deactivation on mouse lung, kidney and heart. PloS one. 2014;9(9): e107559 10.1371/journal.pone.0107559 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
QQ, Qq and qq stand for homozygote of normal, heterozygote and homozygote of indel, respectively.
(TIF)
The dots indicate the significant correlation coefficients between each pair of traits. Their sizes and colors represent the degree and direction (positive and negative) of the correlations, respectively.
(TIF)
(A1-F1) For LDL-C, HDL-C, HDL-C/LDL-C, TC, TG and AI in the Laiwu population; (A2-F2) For LDL-C, HDL-C, HDL-C/LDL-C, TC, TG and AI in DLY pigs; (A3-F3) For LDL-C, HDL-C, HDL-C/LDL-C, TC, TG and AI in the Erhualian population.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLS)
(XLS)
(XLSX)
Data Availability Statement
All genotype and phenotype files will be available from the Dryad database (URL: http://datadryad.org/review?doi=doi:10.5061/dryad.4gh70).