Abstract
In developed countries, typhoid fever is often associated with persons who travel to endemic areas or immigrate from them. Typhoid fever is a systemic infection caused by Salmonella enterica serovar Typhi. Because of the emergence of antimicrobial resistance to standard first-line drugs, fluoroquinolones are the drugs of choice. Resistance to ciprofloxacin by this Salmonella serovar represents an emerging public health issue. Two S. enterica ser. Typhi strains resistant to ciprofloxacin (CIP) were reported to the Italian surveillance system for foodborne and waterborne diseases (EnterNet-Italia) in 2013. The strains were isolated from two Italian tourists upon their arrival from India. A retrospective analysis of 17 other S. enterica ser. Typhi strains isolated in Italy during 2011–2013 was performed to determine their resistance to CIP. For this purpose, we assayed for susceptibility to antimicrobial agents and conducted PCR and nucleotide sequence analyses. Moreover, all strains were typed using pulsed-field gel electrophoresis to evaluate possible clonal relationships. Sixty-eight percent of the S. enterica ser. Typhi strains were resistant to CIP (MICs, 0.125–16 mg/L), and all isolates were negative for determinants of plasmid-mediated quinolone resistance. Analysis of sequences encoding DNA gyrase and topoisomerase IV subunits revealed mutations in gyrA, gyrB, and parC. Thirteen different clonal groups were detected, and the two CIP-resistant strains isolated from the individuals who visited India exhibited the same PFGE pattern. Because of these findings, the emergence of CIP-resistant S. enterica ser. Typhi isolates in Italy deserves attention, and monitoring antibiotic susceptibility is important for efficiently managing cases of typhoid fever.
Introduction
Typhoid fever is a systemic infection caused mainly by Salmonella enterica serovar Typhi. This disease is a major burden in developing countries and accounts for more than 20 million cases and 200,000–600,000 deaths annually worldwide. Approximately 90% of deaths occur in Asia (http://www.who.int/immunization/sage/SAGE_Background_paper_typhoid_newVaccines.pdf). With the widespread emergence and spread of S. enterica ser. Typhi strains that are resistant to chloramphenicol, ampicillin, and trimethoprim, ciprofloxacin (CIP) is now the first-line drug of choice for treating typhoid fever. However, the frequency of strains resistant to nalidixic acid (NAL) and CIP has increased during the last 10 years [1]. S. enterica ser. Typhi strains isolated from Asia, Latin America, and Africa share this pattern of drug resistance [1]. Clinical evidence indicates a poor response to systemic infections caused by S. enterica ser. Typhi strains that are resistant to CIP, e.g. the minimum inhibitory concentration (MIC) > 0.06 mg/L (EUCAST v.5.0, 2015) [2, 3].
Quinolone resistance among Salmonella spp. is frequently associated with mutations in the genes encoding DNA gyrase (gyrA and gyrB) or topoisomerase (parC and parE) that reside in the quinolone resistance-determining region (QRDR) [4, 5]. Plasmid genes that mediate quinolone resistance (PMQR), including qnr (qnrA, qnrB, qnrS, qnrC, and qnrD), qepA, oqxAB, and aac(6')-Ib-cr, are present in Salmonella species that are resistant to quinolones but only qnrB, qnrS, and aac(6')-Ib-cr genes have been described in S. enterica ser. Typhi [6–8]. The quinolone resistance of Salmonella spp. has also been associated with decreased permeability to quinolones and overexpression of efflux pumps [9].
Typhoid fever is a rare disease in Italy, and approximately 20 confirmed cases are reported yearly to the National Surveillance System for Foodborne and Waterborne Diseases, which is coordinated by the Istituto Superiore di Sanità (EnterNet Italia, www.iss.it/ente). Typhoid fever occurs mainly in individuals who travel to endemic areas as well as in immigrants from these regions. Initial treatment of enteric fever typically employs empirical antimicrobial therapy before the results of drug-sensitivity tests are available. To prevent possible treatment failures, it is important for healthcare providers to possess knowledge of the antibiotic resistance of S. enterica ser. Typhi [10]. In particular, monitoring the antibiotic susceptibility spectrum of this serovar in Italy is important for efficiently managing patients with typhoid fever who contracted the disease in endemic areas and for preventing the spread of the disease.
Therefore, the aim of the present study was to characterize S. enterica ser. Typhi strains resistant to CIP that were isolated in Italy from 2011 to 2013. For this purpose, we determined the antimicrobial resistance patterns of these strains, characterized the relationships among strains by using pulse-field gel electrophoresis (PFGE), and performed nucleotide sequence analysis to identify the mutations responsible for drug resistance.
Material and Methods
Bacterial isolates
Over a 3-year period (from 2011 to 2013), only 19 of the 106 S. enterica ser. Typhi strains reported to the Italian surveillance system for enteric pathogens (EnterNet Italia) were received at our Institution. All isolates were confirmed as S. enterica ser. Typhi by using the serotyping Kauffmann–White scheme [11]. The data reported in this study were extracted from an anonymous national surveillance network for human gastrointestinal infections (EnterNet Italia). Because no confidential patient information was used, an ethics statement is not applicable, and Institutional Review Board approval or informed consent of patients was not required.
Assays of antimicrobial susceptibility
Antibiotic susceptibility tests were performed using the disk diffusion method with antimicrobial discs (Becton Dickinson, MD 21152–0999, USA), and the antibiotic concentrations (μg) were as follows: nalidixic acid (NAL, 30), ampicillin (A, 10), cefotaxime (CTX, 5), ceftazidime (CAZ, 10), amoxicillin/clavulanic acid 2:1 (AMC, 30), meropenem (MEM, 10), chloramphenicol (C, 30), gentamicin (G, 10), kanamycin (K, 30), streptomycin (S, 10), sulfonamides (Su, 0.25), tetracycline (T, 30), trimethoprim (TMP, 5), and trimethoprim–sulfamethoxazole (SXT, 1.25/23.75). The reference strain Escherichia coli ATCC 25922 was used as a control for each experiment. The MIC of CIP was determined using Etest strips (AB Biodisk, S-169 56 Solna, Sweden). The data were interpreted using the EUCAST guidelines (http://www.eucast.org/clinical_breakpoints/v.5.0) that define CIP resistance and sensitivity as MICs > 0.06 mg/L and ≤ 0.06 mg/L, respectively.
Identification of genetic determinants of quinolone resistance
The chromosomal quinolone resistance-determining regions (QRDRs) gyrA, gyrB, parC, and parE [5] and plasmid-mediated quinolone resistance regions (PMQRs) qnrA, qnrB, qnrC, qnrD, qnrS, aac(6’)-Ib-cr, qepA, and oqxAB were amplified using PCR [12–14] and the amplicons were subjected to nucleotide sequence analysis.
PFGE
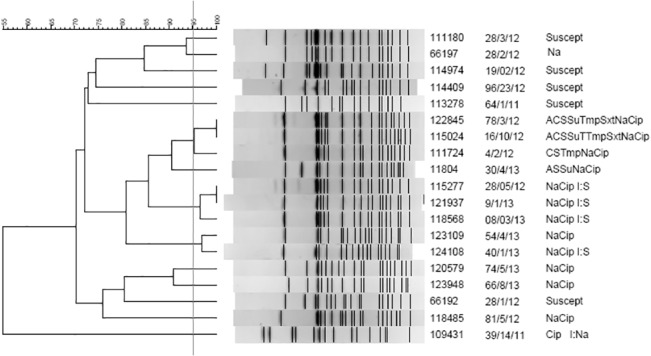
Clonal relationships among the strains were assessed using PFGE according to the PulseNet protocol [15]. Genomic DNA was digested with XbaI (New England Biolabs, Ipswich, MA, USA), and Salmonella enterica serovar Braenderup H9812 DNA was used as the molecular size marker. Dendrogram and cluster analyses were performed using algorithms included in the BioNumerics software package v.6.6 (Applied Maths, Sint-Martens-Latem, Belgium). The percentage similarity between different chromosomal fingerprints was scored using the Dice coefficient. The unweighted pair group method with arithmetic means (UPGMA) with a 1.00% tolerance limit and 1.00% optimization was used to generate the dendrogram. A clonal relationship among strains was defined as a coefficient of similarity ≥ 95%. The clonal groups were named alphabetically from A to N (Fig 1).
Fig 1. S. enterica ser. Typhi PFGE and Dendrogram.
Results
S. enterica ser. Typhi strains
Nineteen S. enterica ser. Typhi strains isolated in 2011 (n = 6), 2012 (n = 7), and 2013 (n = 6) were deposited in the collection of the Istituto Superiore di Sanità. The strains were isolated from blood (n = 7), urine (n = 1), feces (n = 7), or an unknown source (n = 4). The ages of patients ranged from 0 to 47 years (seven males, six females, and six unknown). The country of birth was known for 15 of the 19 patients, and four of the 19 patients traveled before presenting with typhoid fever (see Table 1). All the patients were residing in Italy at the time of the study. No other epidemiologically relevant information such as recent travels to endemic areas or recent immigration to Italy was available for 15 of these patients.
Table 1. S. enterica ser. Typhi Strains Isolated in Italy from 2011 to 2013.
| Strain | Region | Date | Sample | Gender | Age | Origin | Travel | Resistance pattern | CIP MIC(mg/L) | GyrA | GyrB | ParC | ParE | PMQR | PFGE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 28/1/12 | CAMPANIA | 2011 | unk | unk | unk | unk | unk | Suscept | 0.008 | wt | wt | wt | wt | neg | L |
| 28/4/12 | CAMPANIA | 2012 | feces | unk | unk | Italian | unk | Suscept | 0.008 | wt | wt | wt | wt | neg | C |
| 28/3/12 | CAMPANIA | 2011 | feces | unk | Italian | unk | Suscept | 0.008 | wt | wt | wt | wt | neg | A | |
| 96/23/12 | CAMPANIA | 2012 | unk | unk | unk | unk | unk | Suscept | 0.015 | wt | wt | wt | wt | neg | D |
| 64/1/11 | FRIULI V.G. | 2011 | urine | M | 20 | African | unk | Suscept | 0.015 | wt | wt | wt | wt | neg | E |
| 28/2/12 | CAMPANIA | 2011 | unk | unk | unk | unk | unk | Na | 0.032 | Asp82Asn | wt | wt | wt | neg | B |
| 39/14/11 | MARCHE | 2011 | blood | M | 0 | Indian | unk | Cip I:Na | 0.125 | wt | wt | Thr57Ser | wt | neg | N |
| 81/5/12 | LAZIO | 2012 | feces | M | 14 | Bangladesh | unk | NaCip | 0.15 | Ser83Phe | Gly435Ala | wt | wt | neg | M |
| 66/8/13 | LAZIO | 2013 | blood | F | 13 | Bangladesh | unk | NaCip | 0.19 | Asp87Asn | Gly435Glu | wt | wt | neg | K |
| 30/4/13 | PIEMONTE | 2013 | unk | unk | unk | unk | unk | ASSuNaCip | 0.25 | Ser83Phe | wt | wt | wt | neg | G |
| 4/2/12 | TRENTO | 2011 | blood | F | 16 | Bangladesh | unk | CSTmpNaCip | 0.25 | Ser83Phe | wt | wt | wt | neg | F |
| 78/3/12 | PIEMONTE | 2012 | feces | F | 34 | Bangladesh | Bangladesh | ACSSuTmpSxtNaCip | 0.25 | Ser83Phe | Gly435Glu | wt | wt | neg | F |
| 16/10/12 | LAZIO | 2012 | feces | M | unk | Bangladesh | unk | ACSSuTTmpSxtNaCip | 0.25 | Ser83Phe | wt | wt | wt | neg | F |
| 28/5/12 | CAMPANIA | 2012 | blood | F | 33 | Indian | unk | NaCip I:S | 0.25 | Ser83Phe | wt | wt | wt | neg | H |
| 8/3/13 | TRENTO | 2012 | blood | M | 3 | Bangladesh | unk | NaCip I:S | 0.25 | Ser83Phe | wt | wt | wt | neg | H |
| 9/1/13 | CAMPANIA | 2013 | blood | M | 29 | Italian | India | NaCip I:S | 0.38 | Ser83Tyr | wt | wt | Ser493Phe | neg | H |
| 74/5/13 | MARCHE | 2013 | feces | M | 32 | Indian | unk | NaCip | 0.5 | Ser83Phe | Gly435Val | wt | wt | neg | J |
| 40/1/13 | PIEMONTE | 2013 | feces | F | 40 | Italian | India | NaCip I:S | 12 | Ser83Phe,Asp87Asn | Gly435Glu | Ser80Ile | wt | neg | I |
| 54/4/13 | PIEMONTE | 2013 | blood | F | 47 | Italian | India | NaCip | 16 | Ser83Phe,Asp87Asn | wt | Ser80Ile | wt | neg | I |
I: Intermediate susceptibility; wt: wild-type; unk: unknown; suscept: susceptible to all the antimicrobials tested.
Antimicrobial susceptibility and analysis of QRDRs and PMQRs
Sixty-eight percent (13/19) of the S. enterica ser. Typhi strains were resistant to CIP. All isolates were susceptible to third-generation cephalosporins. PMQR genes were not detected, and five isolates were susceptible to all the antibiotics tested (MICs for NAL and CIP MIC ranged between 0.008–0.015 mg/L) (Table 1). Further, mutations in the QRDR region were not detected. One strain resistant only to NAL harbored the Asp82Asn GyrA mutation (CIP MIC = 0.032 mg/L). One CIP-resistant strain (MIC = 0.125 mg/L) with intermediate susceptibility to NAL had one ParC mutation (Thr57Ser). Twelve NAL–CIP-resistant strains (MICs 0.15–16 mg/L) harbored mutations in GyrA, GyrB, ParC, and ParE that conferred different levels of resistance to CIP. The two strains isolated from two Italian patients who traveled together to India were highly resistant to CIP (MICs = 12 and 16 mg/L). The GyrA mutations Ser83Phe-Asp87Asn and ParC Ser80Ile were detected in the isolates from each person. The strain with CIP MIC = 12 mg/L harbored the GyrB Gly435Glu point mutation (Table 1). The NAL-CIP resistant strains with CIP MIC values of 0.15 to 0.5 mg/L harbored single mutations in GyrA (Ser83Phe, Ser83Val, or Asp87Asn), some of which were associated with single mutations of GyrB (Gly435Ala, Gly435Glu, or Gly435Val) or a single ParE mutation (Ser493Phe). Four of these NAL–CIP resistant strains were multidrug-resistant (MDR) (Table 1).
Clonal relationships
PFGE analysis revealed 14 different restriction patterns among the 19 isolates. Only three clonal groups were established with a maximum of three isolates (Fig 1). Three of the four MDR strains were included in the same group (F) and were isolated from three natives of Bangladesh (one of whom had traveled to Bangladesh before presenting with illness) at three regions from Italy in different years. Clonal group H comprised three strains that were resistant to NAL and CIP, with intermediate susceptibility to streptomycin. These strains were isolated from one native of Bangladesh, one native of India, and one Italian who had traveled to India. Clonal group I consisted of two CIP-resistant strains isolated from the two Italian patients who had traveled together to India.
Discussion
Certain diseases that are not endemic or completely eradicated can reoccur sporadically. Because of the increase in the immigrant population in Italy and travel by people from countries with endemic typhoid fever, S. enterica ser. Typhi is occasionally isolated in Italy. From 2011 through 2013, the Italian surveillance system for enteric pathogens (EnterNet Italia), reported the isolation of 193 106 S. enterica ser. Typhi strains isolated from blood, urine, feces, or combinations of these sources. Only 19 of these strains were received at our institution. Ten strains were isolated from three natives of India, six were from Bangladesh, one was from Africa, and five were isolated from Italian patients. Three of the five Italian patients had traveled to India and whether the other two had traveled is unknown. No clonal relationships were detected that indicated the possibility of a typhoid fever outbreak in Italy.
While uncommon in Italy, typhoid fever is highly endemic in developing countries in Africa, Asia (especially Southeast Asia and the Indian subcontinent), and Central and South America. PFGE typing of all the S. enterica ser. Typhi strains isolated in the 2011–2013 period produced only three clonal groups. Identical clones were associated with natives of different endemic countries (India and Bangladesh) (clone F), suggesting the distribution of the same clone. Clone H was composed of three identical clones from three unrelated patients from Bangladesh, which indicates that this particular clone could be widespread in Bangladesh. Clonal group I included two CIP-resistant strains isolated from two Italian patients who had traveled together to India.
With the global emergence of S. enterica ser. Typhi isolates that are resistant to chloramphenicol, ampicillin, and trimethoprim-sulfamethoxazole, as well as others, CIP is the first-line drug for treating typhoid fever [16]. After the introduction of fluoroquinolones as therapy for typhoid fever, treatment failure was reported using CIP [17]. The widespread use of fluoroquinolones is associated with the dissemination of CIP-resistant strains [18]. Here, we show that five (26%) of the S. enterica ser. Typhi isolates were susceptible to all the antimicrobials tested, 12 (63%) were resistant to NAL and CIP, and only four were MDR strains.
With the recent reduction of the MIC breakpoints for resistance to CIP recommended by the EUCAST (http://www.eucast.org/clinical_breakpoints/v.5.0), the NAL–CIP-resistant strains predominated in the present study. A decrease in the number of MDR strains and an increase in the number of fluoroquinolone-resistant strains of S. enterica ser. Typhi isolated from patients residing in the Indian subcontinent and certain regions of Southeast Asia were observed [19–21]. Therefore, it was proposed that drugs other than fluoroquinolones such as oral azithromycin or intravenous ceftriaxone might serve as an effective initial therapy for patients with enteric fever from those countries [19, 22]. Fortunately, resistance to third-generation cephalosporins is rarely encountered [19]. Moreover, the World Health Organization recommends the programmatic use of licensed vaccines (Vi and Ty21a) for controlling endemic disease in specific areas where the burden of disease is great and rates of antibiotic resistance are high (http://www.who.int/immunization/diseases/typhoid/en/).
Fluoroquinolones target DNA gyrase and topoisomerase IV, which comprise subunits encoded by gyrA–gyrB and parC–parE, respectively. Single point mutations in gyrA confer resistance to NAL, and the cumulative effects of mutations in gyr and par may confer resistance to CIP [23]. In the present study, the NAL-CIP-resistant strains harbored different mutations in GyrA, four in GyrB, and one in ParE (Table 1). A single amino acid residue substitution in S. enterica ser. Typhi GyrA conferred resistance to NAL and cinoxacin. In contrast, resistance to CIP and other fluoroquinolones requires two substitutions in GyrA and one in ParC [24–26]. In the strains tested here, according to the new EUCAST CIP breakpoints, we showed that a single GyrA mutation conferred resistance to CIP. Further, these GyrA mutations were frequently detected in other studies and include Ser83Phe, which is one of the most frequent mutations [27–28]; ParC mutations were also reported [21].
The two NAL–CIP-resistant strains (MICs of 12 and 16 mg/L) harbor three point mutations as follows: GyrA (Ser83Phe, Asp87Asn) and ParC (Ser80Ile). The high levels of resistance observed for the triple point mutants indicate that the amino acid residue substitutions protect GyrA and ParC from relatively high concentrations of fluoroquinolones [24]. The identical triple GyrA–ParC mutations identified here in two NAL–CIP resistant strains (CIP MIC = 8 mg/L) were detected in one S. enterica ser. Typhi isolate from Nepal in 2012 [25] and in two S. enterica ser. Typhi strains (CIP MIC > 32 mg/L) from Taiwan in 2011 [26]. Further, two NAL-CIP resistant strains (CIP MICs 64 and 6 mg/L) with the same triple mutation were isolated in India during 2005–2009, although these strains expressed an efflux pump that contributed to their fluoroquinolone resistance [29].
There is a re-emergence of susceptibility to first-line antibiotics and the decline of MDR S. enterica ser. Typhi strains. Our strains were mostly resistant to NAL and CIP due to mutations in the QRDR locus and were susceptible to third-generation cephalosporins. Because this is the first report of the isolation of S. enterica ser. Typhi strains resistant to NAL and CIP in Italy, prudent use of quinolones as empirical therapy for typhoid fever is required to prevent emergence of resistant strains that cause treatment failures. Good surveillance is required to target CIP-resistant S. enterica ser. Typhi that may cause treatment failure. Moreover, improving vaccination of the inhabitants of endemic areas and publicizing the S. enterica ser. Typhi immunization program to people who plan to travel to these regions may serve as effective measures to prevent the dissemination of enteric fever.
Acknowledgments
We thank Dr. Alberto Bellio and Dr. Amaranta Traversa for their collaboration in collecting the data for the Piedmontese isolates.
Data Availability
All relevant data are within the paper.
Funding Statement
Funding was provided by the Italian Ministry of Health, Centro Nazionale per la Prevenzione ed il Controllo delle Malattie (CCM) [project “Sorveglianza delle malattie trasmesse da alimenti e acqua (EnterNet): adeguamento del sistema italiano.al quadro normativo europeo” 5M10-1616-2014]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Parry CM, Threlfall EJ. Antimicrobial resistance in typhoidal and nontyphoidal salmonellae . Curr Opin Infect Dis. 2008; 21:531–538. 10.1097/QCO.0b013e32830f453a [DOI] [PubMed] [Google Scholar]
- 2. Parry CM. Antimicrobial drug resistance in Salmonella enterica . Curr Opin Infect Dis. 2003; 16:467–472. [DOI] [PubMed] [Google Scholar]
- 3. Chandel DS, Chaudhry R. Enteric fever treatment failures: A global concern. Emerg Infect Dis. 2001; 7:762–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Casin I, Breuil J, Darchis JP, Guelpa C, Collatz E. Fluoroquinolone resistance linked to GyrA, GyrB, and ParC mutations in Salmonella enterica typhimurium isolates in humans. Emerg Infect Dis. 2003; 9(11):1455–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eaves DJ, Randall L, Gray DT, Buckley A, Woodward MJ, White AP et al. Prevalence of mutations within the quinolone resistance-determining region of gyrA, gyrB, parC, and parE and association with antibiotic resistance in quinolone-resistant Salmonella enterica . Antimicrob Agents Chemother. 2004; 48:4012–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pfeifer Y, Matten J, Rabsch W. Salmonella enterica serovar Typhi with CTX-M beta-lactamase, Germany. Emerg Infect Dis. 2009; 15:1533 10.3201/eid1509.090567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Keddy KH, Smith AM, Sooka A, Ismail H, Oliver S. Fluoroquinolone-Resistant Typhoid, South Africa. Emerg Infect Dis. 2010; 16:879–880. 10.3201/eid1605.091917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Geetha VK, Yugendran T, Srinivasan R, Harish BN. Plasmid-mediated quinolone resistance in typhoidal Salmonellae: a preliminary report from South India. Indian J Med Microbiol 2014; 32: 31–34. 10.4103/0255-0857.124292 [DOI] [PubMed] [Google Scholar]
- 9. Miró E, Vergés C, García I, Mirelis B, Navarro F, Coll P et al. Resistance to quinolones and beta-lactams in Salmonella enterica due to mutations in topoisomerase-encoding genes, altered cell permeability and expression of an active efflux system. Enferm Infecc Microbiol Clin. 2004; 22:204–211. [DOI] [PubMed] [Google Scholar]
- 10. Hume S, Schulz T, Vinton P, Korman T, Torresi J. Increasing rates and clinical consequences of nalidixic acid-resistant isolates causing enteric fever in returned travellers: an 18-year experience. Eur J Clin Microbiol Infect Dis. 2009; 28:963–970. 10.1007/s10096-009-0732-6 [DOI] [PubMed] [Google Scholar]
- 11. Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemühl J, Grimont PA et al. Supplement 2003–2007 (No. 47) to the White-Kauffmann-Le Minor scheme. Res Microbiol. 2010; 161:26–29. 10.1016/j.resmic.2009.10.002 [DOI] [PubMed] [Google Scholar]
- 12. Threlfall EJ, Fisher IS, Berghold C, Gerner-Smidt P, Tschäpe H, Cormican M et al. Antimicrobial drug resistance in isolates of Salmonella enterica from cases of salmonellosis in humans in Europe in 2000: results of international multi-centre surveillance. Euro Surveill. 2003; 8:41–45. [DOI] [PubMed] [Google Scholar]
- 13. García-Fernández A, Fortini D, Veldman K, Mevius D, Carattoli A. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella . J Antimicrob Chemother 2009; 63: 274–281. 10.1093/jac/dkn470 [DOI] [PubMed] [Google Scholar]
- 14. Kim HB, Wang M, Park CH, Kim EC, Jacoby GA, Hooper DC. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae . Antimicrob Agents Chemother. 2009; 53: 3582–3584. 10.1128/AAC.01574-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hunter SB, Vauterin P, Lambert-Fair MA, Van Duyne MS, Kubota K, Graves L et al. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J Clin Microbiol. 2005; 43:1045–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Butt T, Ahmad RN, Mahmood A, Zaidi S. Ciprofloxacin treatment failure in typhoid fever case, Pakistan. Emerg Infect Dis. 2003; 9:1621–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harish BN, Menezes GA. Antimicrobial resistance in typhoidal salmonellae . Indian J Med Microbiol. 2011; 29:223–229 10.4103/0255-0857.83904 [DOI] [PubMed] [Google Scholar]
- 18. Crump JA, Mintz ED. Global trends in typhoid and paratyphoid fever. Clin Infect Dis. 2010; 50: 241–246. 10.1086/649541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Commons RJ, McBryde E, Valcanis M, Powling J, Street A, Hogg G. Twenty-six years of enteric fever in Australia: an epidemiological analysis of antibiotic resistance. Med J Aust. 2012; 196:332–336. [DOI] [PubMed] [Google Scholar]
- 20. Farmakiotis D, Varughese J, Sue P, Andrews P, Brimmage M, Dobroszycki J et al. Typhoid Fever in an inner city hospital: a 5-year retrospective review. Travel Med. 2013; 20:17–21. [DOI] [PubMed] [Google Scholar]
- 21. Singhal L, Gupta PK, Kale P, Gautam V, Ray P. Trends in antimicrobial susceptibility of Salmonella Typhi from North India (2001–2012). Indian J Med Microbiol. 2014; 32:149–152. 10.4103/0255-0857.129799 [DOI] [PubMed] [Google Scholar]
- 22. Hume S, Schulz T, Vinton P, Korman T, Torresi J. Increasing rates and clinical consequences of nalidixic acid-resistant isolates causing enteric fever in returned travelers: an 18-year experience. Eur J Clin Microbiol Infect Dis. 2009; 28: 963–970. 10.1007/s10096-009-0732-6 [DOI] [PubMed] [Google Scholar]
- 23. Ruiz J. Mechanisms of resistance to quinolones: Target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother 2003; 51:1109–1117. [DOI] [PubMed] [Google Scholar]
- 24. Turner AK, Nair S, Wain J. The acquisition of full fluoroquinolone resistance in Salmonella Typhi by accumulation of point mutations in the topoisomerase targets. J Antimicrob Chemother. 2006; 58:733–740. [DOI] [PubMed] [Google Scholar]
- 25. Acharya D, Trakulsomboon S, Madhup SK, Korbsrisate S. Antibiotic susceptibility pattern and the indicator of decreased ciprofloxacin susceptibility of Salmonella enterica serovar Typhi isolated from Dhulikhel Hospital, Nepal. Jpn J Infect Dis. 2012; 65:264–267. [DOI] [PubMed] [Google Scholar]
- 26. Lee CJ, Su LH, Huang YC, Chiu CH. First isolation of ciprofloxacin-resistant Salmonella enterica serovar Typhi in Taiwan. J Microbiol Immunol Infect. 2013; 46:469–473. 10.1016/j.jmii.2013.01.002 [DOI] [PubMed] [Google Scholar]
- 27. Chau TT, Campbell JI, Galindo CM, Van Minh Hoang N, Diep TS, Nga TT et al. Antimicrobial drug resistance of Salmonella enterica serovar Typhi in Asia and molecular mechanism of reduced susceptibility to the fluoroquinolones. Antimicrob Agents Chemother. 2007; 51:4315–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roumagnac P, Weill FX, Dolecek C, Baker S, Brisse S, Chinh NT et al. Evolutionary history of Salmonella Typhi. Science. 2006; 31:1301–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Menezes GA, Harish BN, Khan MA, Goessens WH, Hays JP. Antimicrobial resistance trends in blood culture positive Salmonella Typhi isolates from Pondicherry, India, 2005–2009. Clin Microbiol Infect. 2012; 18:239–245. 10.1111/j.1469-0691.2011.03546.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.


