Abstract
Background
To improve the outcome of patients suffering from gastric cancer, a better understanding of underlying genetic and epigenetic events in this malignancy is required. Although CpG island methylator phenotype (CIMP) and microsatellite instability (MSI) have been shown to play pivotal roles in gastric cancer pathogenesis, the clinical significance of these events on survival outcomes in patients with gastric cancer remains unknown.
Methods
This study included a patient cohort with pathologically confirmed gastric cancer who had surgical resections. A cohort of 68 gastric cancers was analyzed. CIMP and MSI statuses were determined by analyzing promoter CpG island methylation status of 28 genes/loci, and genomic instability at 10 microsatellite markers, respectively. A Cox’s proportional hazards model was performed for multivariate analysis including age, stage, tumor differentiation, KRAS mutation status, and combined CIMP/MLH1 methylation status in relation to overall survival (OS).
Results
By multivariate analysis, longer OS was significantly correlated with lower pathologic stage (P = 0.0088), better tumor differentiation (P = 0.0267) and CIMP-high and MLH1 3' methylated status (P = 0.0312). Stratification of CIMP status with regards to MLH1 methylation status further enabled prediction of gastric cancer prognosis.
Conclusions
CIMP and/or MLH1 methylation status may have a potential to be prognostic biomarkers for patients with gastric cancer.
Introduction
Gastric cancer is the second leading cause of cancer-related deaths, with about 700,000 confirmed mortalities annually worldwide, although the incidence has gradually decreased [1–3]. Gastric cancer is generally diagnosed at an advanced stage, which is the primary cause of its poor prognosis [4]. To improve the outcome of gastric cancer, identification of genetic or epigenetic events in the progression of gastric cancer is required. The most important epigenetic event in the progression of cancer is methylation of promoter CpG regions of key tumor suppressor genes. CpG islands are almost 1-kb long sequences of DNA with high guanine—cytosine content in promoter regions of the genes [5]. In contrast to normal cells, CpG islands within tumor suppressor genes in cancer cells are often hypermethylated, leading to a CpG island methylator phenotype (CIMP) [6,7]. Epigenetic silencing of tumor-related genes due to CpG island methylation has recently been reported in gastric cancer. Aberrant CpG island methylation of >100 growth-regulatory genes in gastric cancer has thus far been reported [8–24], however, the clinical significance of CIMP in gastric cancer remains unexplored and poorly understood.
In contrast, mismatch repair (MMR) deficiency in gastric cancer is also an important genetic event. Genomic instability within the number of microsatellite repeats (or microsatellites) is termed microsatellite instability (MSI). MSI is a feature caused by a defective DNA MMR system. Functional inactivation of MMR genes, such as MLH1 or MSH2, by promoter methylation is responsible for the MSI-high (MSI-H) phenotype in gastric cancer. In a previous study, gastric cancer with MSI-H showed a higher frequency of antral location, intestinal subtype, lower incidence of lymph node metastasis, and improved survival, compared to microsatellite stable (MSS) or MSI-L gastric cancers [17,25–30]. However, the clinical significance of MMR deficiency in gastric cancer remains unknown.
In view of this gap in knowledge, in this study, we explored the significance of CIMP and MMR deficiency in gastric cancer and determined their contribution as prognostic markers in patients with gastric cancer.
Materials and Methods
Tissue specimens
This study included a cohort of patients with pathologically confirmed gastric cancer who had undergone surgical resection at Okayama University Hospital (Okayama, Japan) from 1998–2004. A total of 68 gastric cancer tissues and their matched normal gastric mucosa were analyzed. All normal gastric mucosa tissues were obtained from sites adjacent to, but at least 5 cm away from, the original tumor. All patients provided written informed consent and the study was approved by the ethical committee of the Okayama University Hospital. All patients provided written informed consent for usage of their data for future analyses. All gastric cancers and normal gastric mucosa were fresh-frozen tissue specimens, from which DNA was extracted using a QIAamp DNA Mini Kit (QIAGEN).
MSI analysis
MSI analysis was performed by examination of 2 mononucleotide repeats (BAT25 and BAT26), 12 dinucleotide repeats (D17S250, D18S35, D18S58, D18S69, D2S123, D4S1559, D4S2381, D4S470, D5S107, D5S346, and D8S87, TP53), and one tetranucleotide repeats, MYCL, as described previously [31]. Tumors showing allelic shifts in ≥5 of 15 markers were classified as MSI-H (hereon referred to as “MSI”), and the rest were classified as microsatellite stable (MSS).
Sodium bisulfite modification and CIMP analyses
Because frequent hypermethylation of several genes is one of the characteristic features of tumors with CIMP, we investigated the methylation status of 28 promoter CpG island-related loci (APC, CACNA1G, CHFR, COX2, DAPK, DCC, HPP1, MGMT-Mp region, MGMT-Eh region, MINT1, MINT2, MINT31, MLH1 5', MLH1 3', p14, p16, RASSF1A, RASSF2A-region1, RASSF2A-region2, RASSF3, RASSF5, RASSF6, RUNX3, SFRP2-region1, SFRP2-region2, UNC5C, 3OST2, FOXL2), and the corresponding primer sequences are listed in S1 Table. Genomic DNA was bisulfite-modified to convert all unmethylated cytosine residues to uracils. In brief, 0.5–2.0 μg of DNA were denatured in NaOH, treated with sodium bisulfite, and purified using the Wizard DNA Clean-up System (Promega). The methylation status of each CIMP-related locus was evaluated by combined bisulfite restriction analysis (COBRA). Polymerase chain reaction (PCR) for COBRA was performed on a bisulfite-modified template DNA in a 25-μL PCR mixture containing 12.5 μL of HotStarTaq Master Mix kit (Qiagen), 0.5 μmol/L of each PCR primer, and approximately 25 ng of bisulfite-modified DNA. PCR products were digested by addition of restriction enzyme at 37°C for 12 h. The digested DNA was separated on 3% agarose gels in 1× Tris—acetate—EDTA buffer and stained with ethidium bromide. Human normal colonic DNA treated with SssI methylase (New England Biolabs) was used as a positive control for methylated alleles, and DNA from normal lymphocytes was used as a control for unmethylated alleles. Water was used as a negative PCR control to monitor PCR contamination. CIMP-high was defined as not less than 10 of the methylation of these loci.
KRAS mutation analyses
Direct sequencing was performed to identify KRAS exon 2 (codon 12/13) mutations. PCR for KRAS gene was performed in a 25-μL PCR mixture containing 12.5 μL of HotStarTaq Master Mix kit with primers. The QIAquick PCR Purification kit was used to purify PCR products, and they were directly sequenced on an ABI 310 DNA sequencer [32].
Statistical analyses
JMP software (ver 10.0, SAS Institute Inc.) was used to perform statistical analysis. Student’s t-test was used to compare continuous variables, and Fisher’s exact test was used to analyze categorical variables. Overall survival (OS) was measured from the operation date to the date of death. The Kaplan—Meier method and log-rank statistics for differences between various prognostic factors were used to estimate OS distributions. Cox proportional hazard models were used to calculate the hazard ratio (HR) with corresponding 95% confidence interval (CI). Univariate or multivariate logistic regression analysis was performed to determine the differences in HR between each group. All reported P values are two-sided, and P < 0.05 was considered to indicate statistical significance.
Results
Study population
In this study, we investigated 68 patients with gastric cancer. In the promoter CpGs of the MLH1 gene, spread of CpG methylation within its 3' region was determined to be a critical for MLH1 expression [33]. A description of the patient cohort and various clinicopathological features based on sex, age, stage, tumor differentiation, MSI status, KRAS mutation, and MLH1 3' methylation is shown in Table 1. These statuses were compared between the CIMP-high and CIMP-low groups. Of these parameters, MSI status and MLH1 3' methylation status showed remarkable differences between the 2 groups (Table 1).
Table 1. Clinicopathological features of the patients according to CIMP status.
| Characteristics | Total | CIMP-high (%) | CIMP-low (%) | P |
|---|---|---|---|---|
| No. of patients | 68 | 30 (44.1) | 38 (55.9) | |
| Sex | 1.0000 | |||
| Male | 46 | 20 (43.5) | 26 (56.5) | |
| Female | 22 | 10 (45.4) | 12 (54.6) | |
| Age at surgery | 0.1975 | |||
| <70 | 45 | 17 (37.8) | 28 (62.2) | |
| ≥70 | 23 | 13 (56.5) | 10 (43.5) | |
| Stage | 0.6311 | |||
| I/II | 32 | 13 (40.6) | 19 (59.4) | |
| III/IV | 36 | 17 (47.2) | 19 (52.8) | |
| Tumor Differentiation | 0.8073 | |||
| Well/moderate | 32 | 15 (46.9) | 17 (53.1) | |
| Poor | 36 | 15 (41.7) | 21 (58.3) | |
| MSI status | 0.0178* | |||
| MSS | 58 | 22 (37.9) | 36 (62.1) | |
| MSI | 10 | 8 (80.0) | 2 (20.0) | |
| KRAS | 1.0000 | |||
| Wild-type | 65 | 29 (44.6) | 36 (55.4) | |
| Mutated | 3 | 1 (33.3) | 2 (66.7) | |
| MLH1 3′ Methylation | 0.0008*** | |||
| U | 57 | 20 (35.1) | 37 (64.9) | |
| M | 11 | 10 (90.9) | 1 (9.1) |
* P < 0.05,
*** P < 0.001
Methylation status
28 CpG loci including APC, CACNA1G, CHFR, COX2, DAPK, DCC, HPP1, MGMT-Mp region, MGMT-Eh region, MINT1, MINT2, MINT31, MLH1 5', MLH1 3', p14, p16, RASSF1A, RASSF2A-region1, RASSF2A-region2, RASSF3, RASSF5, RASSF6, RUNX3, SFRP2-region1, SFRP2-region2, UNC5C, 3OST2 and FOXL2 were analyzed for determining the CIMP status of each gastric cancer. Methylation spectrum of these loci is shown in tile map (Fig 1a). In these loci, CACNA1G, CHFR, DCC, HPP1, MINT1, MINT2, MINT31, MLH1 5', MLH1 3', p16, RASSF2A-region1, RASSF2A-region2, RUNX3, SFRP2-region2, UNC5C, 3OST2, and FOXL2 were significantly methylated in the CIMP-high group (Table 2).
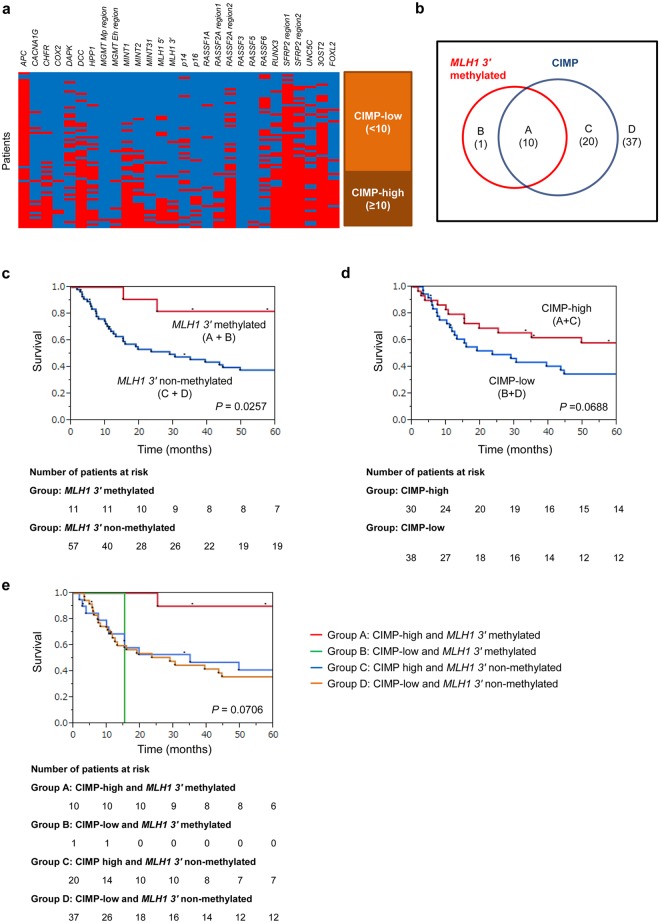
Fig 1. Relationship between CIMP and MLH1 methylation.
a) Tile map showing methylation pattern. Twenty eight loci (APC, CACNA1G, CHFR, COX2, DAPK, DCC, HPP1, MGMT-Mp region, MGMT-Eh region, MINT1, MINT2, MINT31, MLH1 5', MLH1 3', p14, p16, RASSF1A, RASSF2A-region1, RASSF2A-region2, RASSF3, RASSF5, RASSF6, RUNX3, SFRP2-region1, SFRP2-region2, UNC5C, 3OST2, FOXL2) were analyzed to determine CIMP status. Thirty patients with not less than ten methylated loci were identified as the CIMP-high group. b) Venn diagram showing the overlap of MLH1 3' methylation and CIMP status. The overlapping relationship between MLH1 3' methylation and CIMP status was analyzed. 10 patients were in the combined in the CIMP-high/MLH1 3' methylated (A), 1 patients were in the CIMP-low/MLH1 3' methylated (B), 20 patients were in the CIMP high/MLH1 3' non-methylated (C), and 37 patients were in the CIMP-low/MLH1 3' non-methylated groups (D). c) Kaplan–Meier estimate of OS in patients with MLH1 3' methylated or non-methylated gastric cancers. Kaplan–Meier survival curves were generated according to MLH1 3' methylation status. The 5-year survival rates were analyzed for the MLH1 3' methylated and non-methylated groups. The survival rate was significantly higher in the MLH1 3' methylated group than in the non-methylated group (log-rank P = 0.0257). d) Kaplan–Meier estimate of OS in patients with CIMP-high or CIMP-low gastric cancers. Kaplan–Meier survival curves were generated according to CIMP status. The 5-year survival rate was analyzed for the CIMP-high group and CIMP-low group. The survival rate was slightly higher in the CIMP-high group than in the CIMP-low group, but the difference was not significant (log-rank P = 0.0688). e) Contribution of CIMP and mismatch repair deficiency status to survival rate. Kaplan–Meier survival curves were generated according to CIMP and MLH1 methylation status. The patients were classified on the basis of combined CIMP status and MLH1 methylation status into the CIMP-high/MLH1 3' methylated (A), CIMP-low/MLH1 3' methylated (B), CIMP high/MLH1 3' non-methylated (C), and CIMP-low/MLH1 3' non-methylated groups (D). Overall survival rates were higher in the combined CIMP-high/MLH1 3' methylated group, compared to the other groups where the differences were not statistically significant (log-rank P = 0.0706).
Table 2. Methylation features according to CIMP status.
| CIMP-high | CIMP-low | |||||
|---|---|---|---|---|---|---|
| Gene | Positive (%) | Negative (%) | Positive (%) | Negative (%) | Total | P |
| APC | 28 (93.3) | 2 (6.7) | 36 (94.7) | 2 (5.3) | 68 | 1.0000 |
| CACNA1G | 10 (33.3) | 20 (66.7) | 3 (7.9) | 35 (92.1) | 68 | 0.0122 * |
| CHFR | 22 (73.3) | 8 (26.7) | 1 (2.6) | 37 (97.4) | 68 | <0.0001 *** |
| COX2 | 3 (10.0) | 27 (90.0) | 0 (0.0) | 38 (100.0) | 68 | 0.0810 |
| DAPK | 7 (23.3) | 23 (76.7) | 15 (39.5) | 23 (60.5) | 68 | 0.1969 |
| DCC | 29 (96.7) | 1 (3.3) | 11 (29.0) | 27 (71.0) | 68 | <0.0001 *** |
| HPP1 | 17 (56.7) | 13 (43.3) | 4 (10.5) | 34 (89.5) | 68 | <0.0001 *** |
| MGMT-Mp region | 1 (3.3) | 29 (96.7) | 0 (0.0) | 38 (100.0) | 68 | 0.4412 |
| MGMT-Eh region | 5 (16.7) | 25 (83.3) | 1 (2.6) | 37 (97.4) | 68 | 0.0801 |
| MINT1 | 25 (83.3) | 5 (16.7) | 7 (18.4) | 31 (81.6) | 68 | <0.0001 *** |
| MINT2 | 21 (70.0) | 9 (30.0) | 4 (10.5) | 34 (89.5) | 68 | <0.0001 *** |
| MINT31 | 8 (26.7) | 22 (73.3) | 0 (0.0) | 38 (100.0) | 68 | 0.0008 *** |
| MLH1 5′ | 14 (46.7) | 16 (53.3) | 4 (10.5) | 34 (89.5) | 68 | 0.0018 ** |
| MLH1 3′ | 10 (33.3) | 20 (66.7) | 1 (2.6) | 37 (97.4) | 68 | 0.0008 *** |
| p14 | 6 (20.0) | 24 (80.0) | 8 (21.1) | 30 (78.9) | 68 | 1.0000 |
| p16 | 10 (33.3) | 20 (66.7) | 0 (0.0) | 38 (100.0) | 68 | 0.0001 *** |
| RASSF1A | 2 (6.67) | 28 (93.3) | 2 (5.3) | 36 (94.7) | 68 | 1.0000 |
| RASSF2A-region1 | 12 (40.0) | 18 (60.0) | 0 (0.0) | 38 (100.0) | 68 | <0.0001 *** |
| RASSF2A-region2 | 26 (86.7) | 4 (13.3) | 11 (29.0) | 27 (71.0) | 68 | <0.0001 *** |
| RASSF3 | 0 (0.0) | 30 (100.0) | 0 (0.0) | 38 (100.0) | 68 | 1.0000 |
| RASSF5 | 2 (6.7) | 28 (93.3) | 0 (0.0) | 38 (100.0) | 68 | 0.191 |
| RASSF6 | 10 (33.3) | 20 (66.7) | 15 (39.5) | 23 (60.5) | 68 | 0.6234 |
| RUNX3 | 24 (80.0) | 6 (20.0) | 8 (21.1) | 30 (78.9) | 68 | <0.0001 *** |
| SFRP2-region1 | 30 (100.0) | 0 (0.0) | 33 (86.8) | 5 (13.2) | 68 | 0.0618 |
| SFRP2-region2 | 30 (100.0) | 0 (0.0) | 19 (50.0) | 19 (50.0) | 68 | <0.0001 *** |
| UNC5C | 21 (70.0) | 9 (30.0) | 8 (21.1) | 30 (78.9) | 68 | <0.0001 *** |
| 3OST2 | 30 (100.0) | 0 (0.0) | 22 (57.9) | 16 (42.1) | 68 | <0.0001 *** |
| FOXL2 | 22 (73.3) | 8 (26.7) | 2 (5.3) | 36 (94.7) | 68 | <0.0001 *** |
* P < 0.05,
** P < 0.01,
*** P < 0.001
Survival outcomes in patients based upon MLH1 3' methylation and CIMP status in gastric cancers
The overlapping relationship between CIMP and MLH1 3' methylation status was analyzed. 10 patients were in the CIMP-high/MLH1 3' methylated, 1 patient was in the CIMP-low/MLH1 3' methylated, 20 patients were in the CIMP high/MLH1 3' non-methylated and 37 patients were in the CIMP-low/MLH1 3' non-methylated groups (Fig 1b). Kaplan–Meier survival curves were generated according to the MLH1 3' methylation status. The 5-year OS rates were determined for the MLH1 3' methylated and non-methylated groups. The 5-year OS rates were significantly higher in the MLH1 3' methylated group compared to the non-methylated group (log-rank P = 0.0257; Fig 1c).
Likewise, the 5-year OS rates were analyzed for the CIMP-high and CIMP-low groups, and the rate was slightly higher in the CIMP-high group than in the CIMP-low group but the difference was not significant (log-rank P = 0.0688; Fig 1d).
Contribution of mismatch repair deficiency and CIMP status to survival rate
The 5-year survival rates were analyzed and compared among these groups; CIMP-high/MLH1 3' methylated, CIMP-low/MLH1 3' methylated, CIMP high/MLH1 3' non-methylated and CIMP-low/MLH1 3' non-methylated groups. We noted that the overall survival rates were higher in the combined CIMP-high/MLH1 3' methylated group, compared to the other groups where the differences were not statistically significant (log-rank P = 0.0706; Fig 1e).
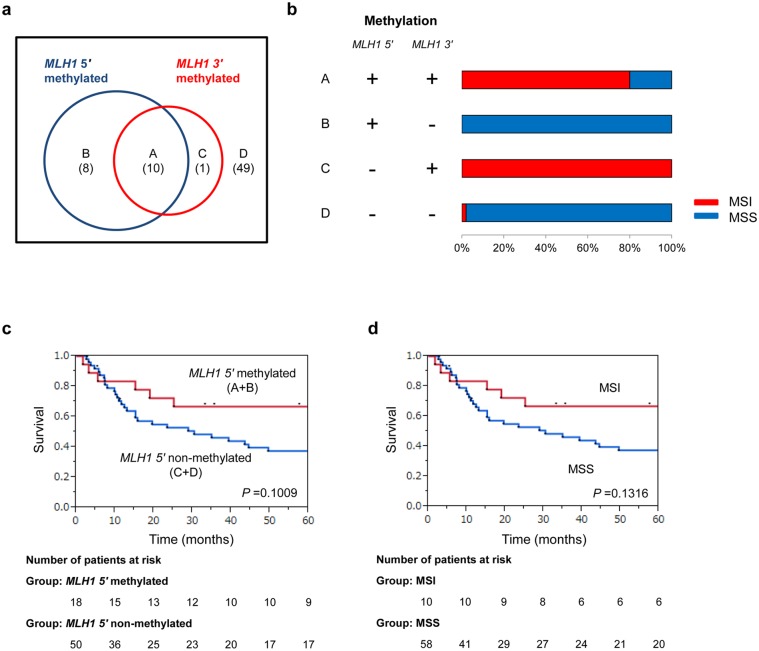
Relationship between MLH1 methylation and MSI
The overlapping relationship between MLH1 5' methylation and 3' methylation status was analyzed. 10 patients were classified as MLH1 5' methylated/3' methylated, 8 patients as MLH1 5' methylated/3' non-methylated, 1 patient as MLH1 5' non-methylated/3' methylated, and 49 patients as 5' non-methylated/3' non-methylated (Fig 2a). In the MLH1 3' methylated group, more than 80% patients showed MSI. In contrast, in the MLH1 3' non-methylated group, almost all cases showed MSS (Fig 2b). Kaplan–Meier survival curves were generated according to MLH1 5' methylation status. The 5-year survival rates were analyzed for the MLH1 5' methylated and non-methylated groups, and the rate was slightly higher in the MLH1 5' methylated group than in the non-methylated group but the differences were not significant (log-rank P = 0.1009; Fig 2c). Kaplan–Meier survival curves were generated according to MSI status. The 5-year survival rates were analyzed for the MSI and MSS groups, and the rate was slightly higher in the MSI group than in the MSS group but the differences were not significant (log-rank P = 0.1316; Fig 2d).
Fig 2. The relationship between MLH1 methylation and MSI.
a) Venn diagram showing the overlap of MLH1 5' methylation and 3' methylation status. The overlapping relationship between MLH1 5'—and 3' methylation status was analyzed. 10 patients were in the MLH1 5' methylated/3' methylated (A), 8 patients were in the MLH1 5' methylated/3' non-methylated (B), 1 patient was in the MLH1 5' non-methylated/3' methylated (C), and 49 patients were in the 5' non-methylated/3' non-methylated groups (D). b) The relationship between MLH1 methylation and MSI. In the MLH1 3' methylated group, >80% cases showed MSI. In contrast, in the MLH1 3' non-methylated group, almost all cases showed MSS. c) Kaplan–Meier estimate of OS in patients with MLH1 5' methylated or non-methylated gastric cancer. Kaplan–Meier survival curves were generated according to MLH1 5' methylation status. The 5-year survival rate was analyzed for the MLH1 5' group and non-methylated groups. The survival rate was slightly higher in the MLH1 5' methylated group than in the non-methylated group but the difference was not significant (log-rank P = 0.1009). d) Kaplan–Meier estimate of OS in patients with MSI or MSS gastric cancer. Kaplan–Meier survival curves were generated according to MSI status. The 5-year survival rate was analyzed for the MSI group and MSS group. The survival rate was slightly higher in the MSI group than in the non-methylated group but the difference was not significant (log-rank P = 0.1316).
Multivariate analysis for survival outcome predictors
A Cox proportional hazards model, including age, stage, differentiation, KRAS mutation status, and CIMP/MLH1 3' methylation status in relation to OS was used to perform multivariate analysis (Table 3). Only stage (P = 0.0088), differentiation (P = 0.0267), and CIMP/MLH1 methylation status (P = 0.0312) were statistically significant predictors of OS. Hazard ratio was significantly lower in the CIMP-high/MLH1 3' methylated group.
Table 3. Multivariate analysis of outcome predictors.
| Univariate | Multivariate | ||||
|---|---|---|---|---|---|
| Characteristic | Total | HR (95% CI) | P | HR (95% CI) | P |
| Age | 0.4476 | 0.4437 | |||
| <70 | 45 | 1.00 (Referent) | 1.00 (Referent) | ||
| ≥70 | 23 | 1.30 (0.65–2.48) | 1.37 (0.60–3.03) | ||
| Stage | 0.0003 *** | 0.0088 ** | |||
| Stage I and II | 32 | 1.00 (Referent) | 1.00 (Referent) | ||
| Stage III and IV | 36 | 3.50 (1.75–7.49) | 2.94 (1.31–7.09) | ||
| Differentiation | 0.0032 ** | 0.0267 * | |||
| Well/Moderate | 32 | 1.00 (Referent) | 1.00 (Referent) | ||
| Poorly | 36 | 2.76 (1.39–5.83) | 2.22 (1.09–4.80) | ||
| KRAS Mutation | 0.1690 | 0.5984 | |||
| Absent | 65 | 1.00 (Referent) | 1.00 (Referent) | ||
| Present | 3 | 0.31 (0.02–1.47) | 0.59 (0.03–3.29) | ||
| CIMP/MLH1 Methylation | 0.0311 * | 0.0312 * | |||
| CIMP−/MLH1-U | 37 | 1.00 (Referent) | 1.00 (Referent) | ||
| CIMP+/MLH1-U | 20 | 0.85 (0.40–1.70) | 0.6535 | 0.67 (0.29–1.47) | 0.3286 |
| CIMP+/MLH1-M | 10 | 0.19 (0.03–0.63) | 0.0042 ** | 0.18 (0.03–0.64) | 0.0048 ** |
| CIMP−/MLH1-M | 1 | 1.89 (0.10–9.24) | 0.5722 | 2.11 (0.10–15.61) | 0.5497 |
* P < 0.05,
** P < 0.01,
*** P < 0.001
Discussion
DNA methylation in cancer cells has become a topic of intense investigation. Inactivation of tumor suppressor genes by CpG methylation of promoter regions accelerates carcinogenesis because of aberrant cell cycle regulation and proliferation [15]. Some candidate aberrantly methylated genes in gastric cancer have been reported. RASSF1A, p14ARF, and MGMT [34–38]; CHRNA3, DOK1, and GNMT [39]; p16, hMLH1, MINT1, MINT2, MINT12, MINT25, and MINT31 [40]; APC, CDH1, MHL1, CDKN2A, CDKN2B, and RUNX3 [17]; CDH1 [41]; DKK3 [42]; PTEN [43]; MGMT[44]; TFPI2 [22]; CACNA2D3 [45]; PCDH10 [46]; SOX2 [47]; MAL [48]; and COX2 [49] were previously reported to be hypermethylated in gastric cancer. Methylation of p16 promoter CpG islands is a marker for malignant potential of dysplasia in the stomach [50]. Methylation of MGMT is associated with advanced stage and poor prognosis [44]. Aberrant DNA methylation in these genes may promote development of gastric cancer. However, precise gene targets of hypermethylation for carcinogenesis remain unknown [51,52].
Concurrent CpG methylation in multiple genes has been defined as CIMP in colorectal cancer (CRC) and gastric cancer [15,40,53–57], and has been shown to correlate with hypermethylation of tumor suppressor genes. However, the evidence for CIMP in gastric cancer is not as convincing as is the case for CRC [58,59]. In gastric cancer, CIMP-high has been described in 41% [40] and 31% [54] tumors. Patients with CIMP-high gastric cancer have significantly shorter survival than those with CIMP-low gastric cancer [54,60]. Another report showed that CIMP was associated with better survival in gastric cancer [54]. The recent meta-analysis has focused on the strong relation of CIMP with H. pylori, EBV, and MSI, but CIMP could not show a prognostic potential for gastric cancer [61].
In contrast, instability at the microsatellites repeats within various growth-regulatory genes is defined as MSI. A standard panel, such as the NCI panel, is recommended, including mononucleotide (BAT26 and BAT25) and dinucleotide (D2S123, D5S346, and D17S250) repeats [62]. Three levels of MSI can be identified: high-level MSI (MSI-H), low-level MSI (MSI-L), and MSS. The MSI-H phenotype in gastric cancer was reported to account for 5%–50% MSI positive neoplasms [17]. MSI is a feature caused by a defective DNA MMR system. Functional inactivation of MMR genes, such as MLH1 or MSH2, by mutational inactivation and promoter methylation is responsible for the MSI-H phenotype in gastric cancer. In particular, similar to CRC, methylation of MLH1 is associated with the MSI-H phenotype [15,40,63,64] because MLH1 methylation precedes the loss of protein expression. Leite M et al. reported that MLH1 promoter hypermethylation was observed in 78.7% (70/89) of the analyzed MSI cases [65]. Methylation of the 3' region of the MLH1 promoter, which is close to its transcriptional start site (TSS), is required for gene silencing. The 5' end of the promoter is also prone to methylation, but this is not functionally important unless the methylation extends to the critical 3' region [66,67]. MSI status is responsible for the mutation of genes regulating cell-cycle and apoptotic signaling, including TGFβRII, IGFIIR, TCF4, RIZ, BAX, CASPASE5, FAS, BCL10, and APAF1 [17,25] and genes maintaining genomic integrity, including MSH6, MSH3, MED1, RAD50, BLM, ATR, and MRE11 [17,68]. Gastric cancers with MSI-H show a higher frequency of antral location, intestinal subtype, lower incidence of lymph node metastasis, and improved survival relative to those of gastric cancer with MSS or MSI-L [17,25–30].
As described above, CIMP and MMR-deficiency status are key features of gastric cancer and may reflect survival differences in patients suffering from this malignancy. Significant correlation between CIMP and MSI has been reported in GC [61]. However, data regarding the synergistic effects of these parameters are scarce, and multivariate analysis of these genetic and epigenetic parameters is required. In our study, CACNA1G, CHFR, DCC, HPP1, MINT1, MINT2, MINT31, MLH1 5', MLH1 3', p16, RASSF2A-region1, RASSF2A-region2, RUNX3, SFRP2-region2, UNC5C, 3OST2, and FOXL2 were significantly methylated in the CIMP-high group. In particular, we reported promoter methylation of FOXL2 in gastric cancer. FOXL2 is a gene encoding a forkhead transcription factor and is essential for ovarian function [69]. FOXL2 regulates the cell cycle by inducing G1 arrest and protects cells from oxidative damage by promoting oxidized DNA repair and by increasing the amount of the anti-oxidant agent glutathione [69]. FOXL2 suppresses proliferation, invasion and promotes apoptosis of cervical cancer cells [70]. The promoter methylation of FOXL2 may have a significant role in tumorigenesis in gastric cancer. In contrast, MLH1 3' methylation was required for MMR deficiency and showed MSI. The MLH1 3' methylation group had a tendency toward a good prognosis in the Kaplan–Meier survival estimate.
However, CIMP and MMR deficiency are dependent on each other. MSI-associated sporadic CRCs arise through a process that involves CIMP [66,71]; therefore, integrated statistical analysis of CIMP and MMR deficiency should be performed. For example, in duodenal adenocarcinomas, CIMP/MLH1 methylation status showed a significant prognostic value in both OS and time-to-recurrence (TTR) in multivariate analysis [72]. Patients with CIMP-high/MLH1-unmethylated tumors had the worst OS and TTR [72]. In our multivariate analysis of patients with gastric cancer, only the CIMP-high/MLH1 3' methylated group had a good prognosis. The reason for good prognosis in the CIMP-high/MLH1 3' methylated group remains unknown. This phenomenon may be because of synergistic inactivation of vital genes due to mutation and promoter methylation. To further confirm our findings, we performed an independent validation of our results from in patient data submitted to The Cancer Genome Atlas database (TCGA). [73–75] CIMP-high/MLH1 hyper-methylated group showed more frequent lymph node metastasis (p = 0.0009), advanced disease stage (p = 0.0078; S1 File), and slightly better Disease free survival (S2 File). On the other hand, CIMP/MSI status can’t show the significant value as prognostic marker (S2 Table). This result shows that MLH1 has the most important role among MMR genes in the carcinogenesis of GC. Further investigation is required to elucidate the relationship between CIMP status and MMR deficiency. This approach will lead to a new strategy for the treatment of gastric cancer.
In conclusion, our data suggests that stratification of patients with CIMP based on MLH1 methylation status may enable prediction of gastric cancer prognosis. The CIMP-high/MLH1 3' methylated group had good prognosis, but other groups may require intensive treatment for improvement of survival, which needs to be validated in future studies.
Supporting Information
We investigated the methylation status of 17 promoter CpG island-related loci (APC, CACNA1G, CHFR, DAPK, DCC, MGMT, MINT, MLH1, p16, RASSF1, RASSF2, RASSF3, RASSF5, RASSF6, RUNX3, SFRP2, and UNC5C) in TCGA database (TCGA provisional). Upper 25% of each locus was determined as hyper-methylated. CIMP-high was defined as not less than 5 of the hyper-methylation of these loci (Figure A). The overlapping relationship between CIMP and MLH1 methylation status was analyzed. 55 patients were in the CIMP-high/MLH1 methylated, 29 patients were in the CIMP-low/MLH1 methylated, 77 patients were in the CIMP high/MLH1 non-methylated and 177 patients were in the CIMP-low/MLH1 non-methylated groups (Figure B). Correlation between tumor depth, lymph node metastasis, distant metastasis, Stage and CIMP/MLH1 methylation status were analyzed using Fisher’s exact test. Positive lymph node metastasis (p = 0.0009) and higher Stage (p = 0.0078) were positively correlated with CIMP-high/MLH1 methylated group (Figure C).
(DOCX)
Kaplan—Meier survival curves were generated according to the MLH1 methylation status. The disease free survival rates were determined for the MLH1 methylated and non-methylated groups. Disease free survival rates were slightly higher in the MLH1 methylated group compared to the non-methylated group but the difference was not significant (log-rank P = 0.1173) (Figure A). Disease free survival rates were analyzed for the CIMP-high and CIMP-low groups, and the rate was slightly higher in the CIMP-high group than in the CIMP-low group but the difference was not significant (log-rank P = 0.1847) (Figure B). Disease free survival rates were analyzed and compared between CIMP-high/MLH1 methylated and other groups. We noted that the disease free survival rates were slightly higher in the combined CIMP-high/MLH1 methylated group, compared to the other groups where the differences were not statistically significant (log-rank P = 0.1073) (Figure C).
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
The authors would like to thank Mr. Toru Nakai, Mrs. Tae Yamanishi, and Mr. Akihiro Nyuya for technical assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by KAKENHI grants 19390351 and 20590572 to TN, and by grants R01 CA72851 and 181572 from the National Cancer Institute, National Institutes of Health, and funds from the Baylor Research Institute to AG.
References
- 1. Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2006;24(14):2137–50. Epub 2006/05/10. 10.1200/JCO.2005.05.2308 . [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2010;19(8):1893–907. Epub 2010/07/22. 10.1158/1055-9965.EPI-10-0437 . [DOI] [PubMed] [Google Scholar]
- 3. Nardone G. Review article: molecular basis of gastric carcinogenesis. Alimentary pharmacology & therapeutics. 2003;17 Suppl 2:75–81. Epub 2003/06/06. . [DOI] [PubMed] [Google Scholar]
- 4. Heemskerk VH, Lentze F, Hulsewe KW, Hoofwijk TG. Gastric carcinoma: review of the results of treatment in a community teaching hospital. World journal of surgical oncology. 2007;5:81 Epub 2007/07/31. 10.1186/1477-7819-5-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rollins RA, Haghighi F, Edwards JR, Das R, Zhang MQ, Ju J, et al. Large-scale structure of genomic methylation patterns. Genome research. 2006;16(2):157–63. Epub 2005/12/21. 10.1101/gr.4362006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shi H, Wang MX, Caldwell CW. CpG islands: their potential as biomarkers for cancer. Expert review of molecular diagnostics. 2007;7(5):519–31. Epub 2007/09/26. 10.1586/14737159.7.5.519 . [DOI] [PubMed] [Google Scholar]
- 7. Laird PW. The power and the promise of DNA methylation markers. Nature reviews Cancer. 2003;3(4):253–66. Epub 2003/04/03. 10.1038/nrc1045 . [DOI] [PubMed] [Google Scholar]
- 8. Tamura G. Alterations of tumor suppressor and tumor-related genes in the development and progression of gastric cancer. World journal of gastroenterology: WJG. 2006;12(2):192–8. Epub 2006/02/17. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tamura G. Promoter methylation status of tumor suppressor and tumor-related genes in neoplastic and non-neoplastic gastric epithelia. Histology and histopathology. 2004;19(1):221–8. Epub 2004/01/01. . [DOI] [PubMed] [Google Scholar]
- 10. Tamura G. Genetic and epigenetic alterations of tumor suppressor and tumor-related genes in gastric cancer. Histology and histopathology. 2002;17(1):323–9. Epub 2002/01/30. . [DOI] [PubMed] [Google Scholar]
- 11. Rashid A, Issa JP. CpG island methylation in gastroenterologic neoplasia: a maturing field. Gastroenterology. 2004;127(5):1578–88. Epub 2004/11/03. . [DOI] [PubMed] [Google Scholar]
- 12. Sato F, Meltzer SJ. CpG island hypermethylation in progression of esophageal and gastric cancer. Cancer. 2006;106(3):483–93. Epub 2005/12/20. 10.1002/cncr.21657 . [DOI] [PubMed] [Google Scholar]
- 13. Vogiatzi P, Vindigni C, Roviello F, Renieri A, Giordano A. Deciphering the underlying genetic and epigenetic events leading to gastric carcinogenesis. Journal of cellular physiology. 2007;211(2):287–95. Epub 2007/01/24. 10.1002/jcp.20982 . [DOI] [PubMed] [Google Scholar]
- 14. Ottini L, Falchetti M, Lupi R, Rizzolo P, Agnese V, Colucci G, et al. Patterns of genomic instability in gastric cancer: clinical implications and perspectives. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2006;17 Suppl 7:vii97–102. Epub 2006/06/09. 10.1093/annonc/mdl960 . [DOI] [PubMed] [Google Scholar]
- 15. Choi IS, Wu TT. Epigenetic alterations in gastric carcinogenesis. Cell research. 2005;15(4):247–54. Epub 2005/04/29. 10.1038/sj.cr.7290293 . [DOI] [PubMed] [Google Scholar]
- 16. Yasui W, Oue N, Aung PP, Matsumura S, Shutoh M, Nakayama H. Molecular-pathological prognostic factors of gastric cancer: a review. Gastric cancer: official journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association. 2005;8(2):86–94. Epub 2005/05/03. 10.1007/s10120-005-0320-0 . [DOI] [PubMed] [Google Scholar]
- 17. Nobili S, Bruno L, Landini I, Napoli C, Bechi P, Tonelli F, et al. Genomic and genetic alterations influence the progression of gastric cancer. World journal of gastroenterology: WJG. 2011;17(3):290–9. Epub 2011/01/22. 10.3748/wjg.v17.i3.290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yamashita K, Sakuramoto S, Watanabe M. Genomic and epigenetic profiles of gastric cancer: potential diagnostic and therapeutic applications. Surgery today. 2011;41(1):24–38. Epub 2010/12/31. 10.1007/s00595-010-4370-5 . [DOI] [PubMed] [Google Scholar]
- 19. Yasui W, Sentani K, Motoshita J, Nakayama H. Molecular pathobiology of gastric cancer. Scandinavian journal of surgery: SJS: official organ for the Finnish Surgical Society and the Scandinavian Surgical Society. 2006;95(4):225–31. Epub 2007/01/26. . [DOI] [PubMed] [Google Scholar]
- 20. Fan XY, Hu XL, Han TM, Wang NN, Zhu YM, Hu W, et al. Association between RUNX3 promoter methylation and gastric cancer: a meta-analysis. BMC gastroenterology. 2011;11:92 Epub 2011/08/27. 10.1186/1471-230X-11-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yuasa Y, Nagasaki H, Akiyama Y, Sakai H, Nakajima T, Ohkura Y, et al. Relationship between CDX2 gene methylation and dietary factors in gastric cancer patients. Carcinogenesis. 2005;26(1):193–200. Epub 2004/10/23. 10.1093/carcin/bgh304 . [DOI] [PubMed] [Google Scholar]
- 22. Jee CD, Kim MA, Jung EJ, Kim J, Kim WH. Identification of genes epigenetically silenced by CpG methylation in human gastric carcinoma. Eur J Cancer. 2009;45(7):1282–93. Epub 2009/02/07. 10.1016/j.ejca.2008.12.027 . [DOI] [PubMed] [Google Scholar]
- 23. Niwa T, Tsukamoto T, Toyoda T, Mori A, Tanaka H, Maekita T, et al. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer research. 2010;70(4):1430–40. Epub 2010/02/04. 10.1158/0008-5472.CAN-09-2755 . [DOI] [PubMed] [Google Scholar]
- 24. Ushijima T, Nakajima T, Maekita T. DNA methylation as a marker for the past and future. Journal of gastroenterology. 2006;41(5):401–7. Epub 2006/06/27. 10.1007/s00535-006-1846-6 . [DOI] [PubMed] [Google Scholar]
- 25. Iacopetta BJ, Soong R, House AK, Hamelin R. Gastric carcinomas with microsatellite instability: clinical features and mutations to the TGF-beta type II receptor, IGFII receptor, and BAX genes. The Journal of pathology. 1999;187(4):428–32. Epub 1999/07/09. . [DOI] [PubMed] [Google Scholar]
- 26. Falchetti M, Saieva C, Lupi R, Masala G, Rizzolo P, Zanna I, et al. Gastric cancer with high-level microsatellite instability: target gene mutations, clinicopathologic features, and long-term survival. Human pathology. 2008;39(6):925–32. Epub 2008/04/29. 10.1016/j.humpath.2007.10.024 . [DOI] [PubMed] [Google Scholar]
- 27. Hayden JD, Cawkwell L, Quirke P, Dixon MF, Goldstone AR, Sue-Ling H, et al. Prognostic significance of microsatellite instability in patients with gastric carcinoma. Eur J Cancer. 1997;33(14):2342–6. Epub 1998/06/09. . [DOI] [PubMed] [Google Scholar]
- 28. Yamamoto H, Perez-Piteira J, Yoshida T, Terada M, Itoh F, Imai K, et al. Gastric cancers of the microsatellite mutator phenotype display characteristic genetic and clinical features. Gastroenterology. 1999;116(6):1348–57. Epub 1999/05/29. . [DOI] [PubMed] [Google Scholar]
- 29. Choi SW, Choi JR, Chung YJ, Kim KM, Rhyu MG. Prognostic implications of microsatellite genotypes in gastric carcinoma. International journal of cancer Journal international du cancer. 2000;89(4):378–83. Epub 2000/08/24. . [DOI] [PubMed] [Google Scholar]
- 30. Corso G, Pedrazzani C, Marrelli D, Pascale V, Pinto E, Roviello F. Correlation of microsatellite instability at multiple loci with long-term survival in advanced gastric carcinoma. Arch Surg. 2009;144(8):722–7. Epub 2009/08/19. 10.1001/archsurg.2009.42 . [DOI] [PubMed] [Google Scholar]
- 31. Kambara T, Matsubara N, Nakagawa H, Notohara K, Nagasaka T, Yoshino T, et al. High frequency of low-level microsatellite instability in early colorectal cancer. Cancer research. 2001;61(21):7743–6. Epub 2001/11/03. . [PubMed] [Google Scholar]
- 32. Nagasaka T, Koi M, Kloor M, Gebert J, Vilkin A, Nishida N, et al. Mutations in both KRAS and BRAF may contribute to the methylator phenotype in colon cancer. Gastroenterology. 2008;134(7):1950–60, 60 e1. Epub 2008/04/26. 10.1053/j.gastro.2008.02.094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nagasaka T, Sasamoto H, Notohara K, Cullings HM, Takeda M, Kimura K, et al. Colorectal cancer with mutation in BRAF, KRAS, and wild-type with respect to both oncogenes showing different patterns of DNA methylation. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2004;22(22):4584–94. Epub 2004/11/16. 10.1200/JCO.2004.02.154 . [DOI] [PubMed] [Google Scholar]
- 34. Hibi K, Sakata M, Yokomizo K, Kitamura YH, Sakuraba K, Shirahata A, et al. Methylation of the MGMT gene is frequently detected in advanced gastric carcinoma. Anticancer research. 2009;29(12):5053–5. Epub 2010/01/02. . [PubMed] [Google Scholar]
- 35. Kang GH, Lee S, Kim JS, Jung HY. Profile of aberrant CpG island methylation along multistep gastric carcinogenesis. Laboratory investigation; a journal of technical methods and pathology. 2003;83(4):519–26. Epub 2003/04/16. . [DOI] [PubMed] [Google Scholar]
- 36. Sarbia M, Geddert H, Klump B, Kiel S, Iskender E, Gabbert HE. Hypermethylation of tumor suppressor genes (p16INK4A, p14ARF and APC) in adenocarcinomas of the upper gastrointestinal tract. International journal of cancer Journal international du cancer. 2004;111(2):224–8. Epub 2004/06/16. 10.1002/ijc.20212 . [DOI] [PubMed] [Google Scholar]
- 37. Ye M, Xia B, Guo Q, Zhou F, Zhang X. Association of diminished expression of RASSF1A with promoter methylation in primary gastric cancer from patients of central China. BMC cancer. 2007;7:120 Epub 2007/07/05. 10.1186/1471-2407-7-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guo W, Dong Z, Chen Z, Yang Z, Wen D, Kuang G, et al. Aberrant CpG island hypermethylation of RASSF1A in gastric cardia adenocarcinoma. Cancer investigation. 2009;27(4):459–65. Epub 2009/01/23. 10.1080/07357900802620828 . [DOI] [PubMed] [Google Scholar]
- 39. Balassiano K, Lima S, Jenab M, Overvad K, Tjonneland A, Boutron-Ruault MC, et al. Aberrant DNA methylation of cancer-associated genes in gastric cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC-EURGAST). Cancer letters. 2011;311(1):85–95. Epub 2011/08/13. 10.1016/j.canlet.2011.06.038 . [DOI] [PubMed] [Google Scholar]
- 40. Toyota M, Ahuja N, Suzuki H, Itoh F, Ohe-Toyota M, Imai K, et al. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer research. 1999;59(21):5438–42. Epub 1999/12/20. . [PubMed] [Google Scholar]
- 41. Graziano F, Arduini F, Ruzzo A, Bearzi I, Humar B, More H, et al. Prognostic analysis of E-cadherin gene promoter hypermethylation in patients with surgically resected, node-positive, diffuse gastric cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2004;10(8):2784–9. Epub 2004/04/23. . [DOI] [PubMed] [Google Scholar]
- 42. Yu J, Tao Q, Cheng YY, Lee KY, Ng SS, Cheung KF, et al. Promoter methylation of the Wnt/beta-catenin signaling antagonist Dkk-3 is associated with poor survival in gastric cancer. Cancer. 2009;115(1):49–60. Epub 2008/12/04. 10.1002/cncr.23989 . [DOI] [PubMed] [Google Scholar]
- 43. Kang YH, Lee HS, Kim WH. Promoter methylation and silencing of PTEN in gastric carcinoma. Laboratory investigation; a journal of technical methods and pathology. 2002;82(3):285–91. Epub 2002/03/16. . [DOI] [PubMed] [Google Scholar]
- 44. Park TJ, Han SU, Cho YK, Paik WK, Kim YB, Lim IK. Methylation of O(6)-methylguanine-DNA methyltransferase gene is associated significantly with K-ras mutation, lymph node invasion, tumor staging, and disease free survival in patients with gastric carcinoma. Cancer. 2001;92(11):2760–8. Epub 2001/12/26. . [DOI] [PubMed] [Google Scholar]
- 45. Wanajo A, Sasaki A, Nagasaki H, Shimada S, Otsubo T, Owaki S, et al. Methylation of the calcium channel-related gene, CACNA2D3, is frequent and a poor prognostic factor in gastric cancer. Gastroenterology. 2008;135(2):580–90. Epub 2008/07/01. 10.1053/j.gastro.2008.05.041 . [DOI] [PubMed] [Google Scholar]
- 46. Yu J, Cheng YY, Tao Q, Cheung KF, Lam CN, Geng H, et al. Methylation of protocadherin 10, a novel tumor suppressor, is associated with poor prognosis in patients with gastric cancer. Gastroenterology. 2009;136(2):640–51e1. Epub 2008/12/17. 10.1053/j.gastro.2008.10.050 . [DOI] [PubMed] [Google Scholar]
- 47. Otsubo T, Akiyama Y, Yanagihara K, Yuasa Y. SOX2 is frequently downregulated in gastric cancers and inhibits cell growth through cell-cycle arrest and apoptosis. British journal of cancer. 2008;98(4):824–31. Epub 2008/02/13. 10.1038/sj.bjc.6604193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Buffart TE, Overmeer RM, Steenbergen RD, Tijssen M, van Grieken NC, Snijders PJ, et al. MAL promoter hypermethylation as a novel prognostic marker in gastric cancer. British journal of cancer. 2008;99(11):1802–7. Epub 2008/11/13. 10.1038/sj.bjc.6604777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. de Maat MF, van de Velde CJ, Umetani N, de Heer P, Putter H, van Hoesel AQ, et al. Epigenetic silencing of cyclooxygenase-2 affects clinical outcome in gastric cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2007;25(31):4887–94. Epub 2007/11/01. 10.1200/JCO.2006.09.8921 . [DOI] [PubMed] [Google Scholar]
- 50. Sun Y, Deng D, You WC, Bai H, Zhang L, Zhou J, et al. Methylation of p16 CpG islands associated with malignant transformation of gastric dysplasia in a population-based study. Clinical cancer research: an official journal of the American Association for Cancer Research. 2004;10(15):5087–93. Epub 2004/08/07. 10.1158/1078-0432.CCR-03-0622 . [DOI] [PubMed] [Google Scholar]
- 51. Johnson IT. New approaches to the role of diet in the prevention of cancers of the alimentary tract. Mutation research. 2004;551(1–2):9–28. Epub 2004/07/01. 10.1016/j.mrfmmm.2004.02.017 . [DOI] [PubMed] [Google Scholar]
- 52. Johnson IT, Belshaw NJ. Environment, diet and CpG island methylation: epigenetic signals in gastrointestinal neoplasia. Food and chemical toxicology: an international journal published for the British Industrial Biological Research Association. 2008;46(4):1346–59. Epub 2007/11/03. 10.1016/j.fct.2007.09.101 . [DOI] [PubMed] [Google Scholar]
- 53. Issa JP. CpG island methylator phenotype in cancer. Nature reviews Cancer. 2004;4(12):988–93. Epub 2004/12/02. 10.1038/nrc1507 . [DOI] [PubMed] [Google Scholar]
- 54. An C, Choi IS, Yao JC, Worah S, Xie K, Mansfield PF, et al. Prognostic significance of CpG island methylator phenotype and microsatellite instability in gastric carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11(2 Pt 1):656–63. Epub 2005/02/11. . [PubMed] [Google Scholar]
- 55. Oue N, Mitani Y, Motoshita J, Matsumura S, Yoshida K, Kuniyasu H, et al. Accumulation of DNA methylation is associated with tumor stage in gastric cancer. Cancer. 2006;106(6):1250–9. Epub 2006/02/14. 10.1002/cncr.21754 . [DOI] [PubMed] [Google Scholar]
- 56. Park SY, Kook MC, Kim YW, Cho NY, Jung N, Kwon HJ, et al. CpG island hypermethylator phenotype in gastric carcinoma and its clinicopathological features. Virchows Archiv: an international journal of pathology. 2010;457(4):415–22. Epub 2010/08/26. 10.1007/s00428-010-0962-0 . [DOI] [PubMed] [Google Scholar]
- 57. Oue N, Oshimo Y, Nakayama H, Ito R, Yoshida K, Matsusaki K, et al. DNA methylation of multiple genes in gastric carcinoma: association with histological type and CpG island methylator phenotype. Cancer science. 2003;94(10):901–5. Epub 2003/10/15. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kang GH, Lee S, Cho NY, Gandamihardja T, Long TI, Weisenberger DJ, et al. DNA methylation profiles of gastric carcinoma characterized by quantitative DNA methylation analysis. Laboratory investigation; a journal of technical methods and pathology. 2008;88(2):161–70. Epub 2007/12/26. 10.1038/labinvest.3700707 . [DOI] [PubMed] [Google Scholar]
- 59. Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nature genetics. 2006;38(7):787–93. Epub 2006/06/29. 10.1038/ng1834 . [DOI] [PubMed] [Google Scholar]
- 60. Kusano M, Toyota M, Suzuki H, Akino K, Aoki F, Fujita M, et al. Genetic, epigenetic, and clinicopathologic features of gastric carcinomas with the CpG island methylator phenotype and an association with Epstein-Barr virus. Cancer. 2006;106(7):1467–79. Epub 2006/03/07. 10.1002/cncr.21789 . [DOI] [PubMed] [Google Scholar]
- 61. Zong L, Seto Y. CpG island methylator phenotype, Helicobacter pylori, Epstein-Barr virus, and microsatellite instability and prognosis in gastric cancer: a systematic review and meta-analysis. PloS one. 2014;9(1):e86097 Epub 2014/01/30. 10.1371/journal.pone.0086097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer research. 1998;58(22):5248–57. Epub 1998/11/21. . [PubMed] [Google Scholar]
- 63. Lee JH, Park SJ, Abraham SC, Seo JS, Nam JH, Choi C, et al. Frequent CpG island methylation in precursor lesions and early gastric adenocarcinomas. Oncogene. 2004;23(26):4646–54. Epub 2004/04/06. 10.1038/sj.onc.1207588 . [DOI] [PubMed] [Google Scholar]
- 64. Leung WK, Yu J, Ng EK, To KF, Ma PK, Lee TL, et al. Concurrent hypermethylation of multiple tumor-related genes in gastric carcinoma and adjacent normal tissues. Cancer. 2001;91(12):2294–301. Epub 2001/06/20. . [PubMed] [Google Scholar]
- 65. Leite M, Corso G, Sousa S, Milanezi F, Afonso LP, Henrique R, et al. MSI phenotype and MMR alterations in familial and sporadic gastric cancer. International journal of cancer Journal international du cancer. 2011;128(7):1606–13. Epub 2010/06/10. 10.1002/ijc.25495 . [DOI] [PubMed] [Google Scholar]
- 66. Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138(6):2073–87e3. Epub 2010/04/28. 10.1053/j.gastro.2009.12.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Deng G, Nguyen A, Tanaka H, Matsuzaki K, Bell I, Mehta KR, et al. Regional hypermethylation and global hypomethylation are associated with altered chromatin conformation and histone acetylation in colorectal cancer. International journal of cancer Journal international du cancer. 2006;118(12):2999–3005. Epub 2006/01/21. 10.1002/ijc.21740 . [DOI] [PubMed] [Google Scholar]
- 68. Ottini L, Falchetti M, Saieva C, De Marco M, Masala G, Zanna I, et al. MRE11 expression is impaired in gastric cancer with microsatellite instability. Carcinogenesis. 2004;25(12):2337–43. Epub 2004/08/21. 10.1093/carcin/bgh257 . [DOI] [PubMed] [Google Scholar]
- 69. Benayoun BA, Georges AB, L'Hote D, Andersson N, Dipietromaria A, Todeschini AL, et al. Transcription factor FOXL2 protects granulosa cells from stress and delays cell cycle: role of its regulation by the SIRT1 deacetylase. Human molecular genetics. 2011;20(9):1673–86. Epub 2011/02/04. 10.1093/hmg/ddr042 . [DOI] [PubMed] [Google Scholar]
- 70. Liu XL, Meng YH, Wang JL, Yang BB, Zhang F, Tang SJ. FOXL2 suppresses proliferation, invasion and promotes apoptosis of cervical cancer cells. International journal of clinical and experimental pathology. 2014;7(4):1534–43. Epub 2014/05/13. [PMC free article] [PubMed] [Google Scholar]
- 71. Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(15):8681–6. Epub 1999/07/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fu T, Pappou EP, Guzzetta AA, Jeschke J, Kwak R, Dave P, et al. CpG island methylator phenotype-positive tumors in the absence of MLH1 methylation constitute a distinct subset of duodenal adenocarcinomas and are associated with poor prognosis. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18(17):4743–52. Epub 2012/07/25. 10.1158/1078-0432.CCR-12-0707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.The Cancer Genome Atlas. https://tcga-datancinihgov/tcga/.
- 74. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6(269):pl1 Epub 2013/04/04. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2(5):401–4. Epub 2012/05/17. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
We investigated the methylation status of 17 promoter CpG island-related loci (APC, CACNA1G, CHFR, DAPK, DCC, MGMT, MINT, MLH1, p16, RASSF1, RASSF2, RASSF3, RASSF5, RASSF6, RUNX3, SFRP2, and UNC5C) in TCGA database (TCGA provisional). Upper 25% of each locus was determined as hyper-methylated. CIMP-high was defined as not less than 5 of the hyper-methylation of these loci (Figure A). The overlapping relationship between CIMP and MLH1 methylation status was analyzed. 55 patients were in the CIMP-high/MLH1 methylated, 29 patients were in the CIMP-low/MLH1 methylated, 77 patients were in the CIMP high/MLH1 non-methylated and 177 patients were in the CIMP-low/MLH1 non-methylated groups (Figure B). Correlation between tumor depth, lymph node metastasis, distant metastasis, Stage and CIMP/MLH1 methylation status were analyzed using Fisher’s exact test. Positive lymph node metastasis (p = 0.0009) and higher Stage (p = 0.0078) were positively correlated with CIMP-high/MLH1 methylated group (Figure C).
(DOCX)
Kaplan—Meier survival curves were generated according to the MLH1 methylation status. The disease free survival rates were determined for the MLH1 methylated and non-methylated groups. Disease free survival rates were slightly higher in the MLH1 methylated group compared to the non-methylated group but the difference was not significant (log-rank P = 0.1173) (Figure A). Disease free survival rates were analyzed for the CIMP-high and CIMP-low groups, and the rate was slightly higher in the CIMP-high group than in the CIMP-low group but the difference was not significant (log-rank P = 0.1847) (Figure B). Disease free survival rates were analyzed and compared between CIMP-high/MLH1 methylated and other groups. We noted that the disease free survival rates were slightly higher in the combined CIMP-high/MLH1 methylated group, compared to the other groups where the differences were not statistically significant (log-rank P = 0.1073) (Figure C).
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.