Abstract
Bioluminescence resonance energy transfer (BRET) is a well-established method for investigating protein-protein interactions. Here we present a novel BRET approach to monitor ligand binding to G protein-coupled receptors (GPCRs) on the surface of living cells made possible by the use of fluorescent ligands in combination with a novel bioluminescent protein (NanoLuc) that can be readily expressed on the N-terminus of GPCRs.
The ability to monitor protein-protein or drug-protein interactions with ease and sensitivity is the cornerstone of cell biology and pharmacology. Bioluminescence resonance energy transfer (BRET) has become the proximity assay of choice for many researchers due to its ease of use and capacity for real time monitoring in live cells1,2,3. Renilla luciferase variant Rluc8 and green fluorescent protein variant Venus are a notable example BRET combination4, however various donor-acceptor pairs have been utilised successfully for different applications1. To date, drug-protein interactions have not been directly studied using BRET, although it has recently been shown that BRET can be used to detect drug concentration using bioluminescent sensor proteins5. However, the successful development of many different fluorescent agonists and antagonists for G protein-coupled receptors (GPCRs)6 provides an opportunity for them to be used as energy acceptors to measure BRET between fluorescently-labeled ligand and luciferase-tagged receptor.
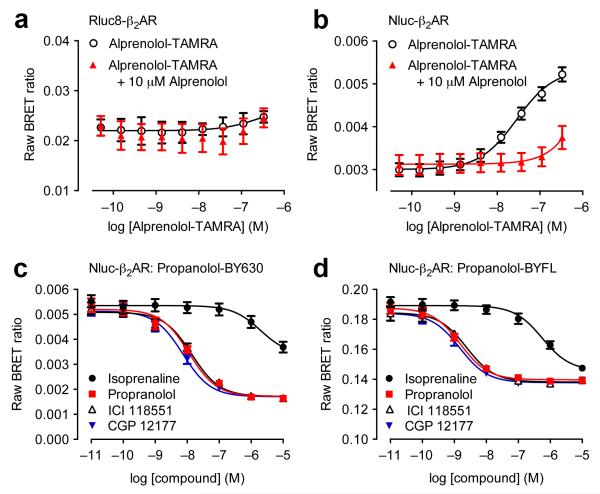
We have developed an assay that can measure ligand binding to GPCRs using BRET in living cells. We initially investigated whether β2-adrenoceptors (β2ARs) tagged on their extracellular N-terminus with a luminescent protein can be expressed in living cells. We assessed the aforementioned Rluc8 and recently described NanoLuc (Nluc)7 luciferase engineered from the luciferase found in deep sea shrimp, Oplophorus. In HEK293 cells, increasing levels of transiently-transfected cDNA caused concurrent increases in luminescence with both Rluc8- and Nluc-tagged β2AR. Nluc-β2AR produced substantially greater luminescence signals than Rluc8-β2AR, and with a spectrum left-shifted by circa 20 nm (Supplementary Fig. 1). BRET signals are dependent upon energy transfer between a bioluminescent donor (Rluc8 or Nluc) and fluorescent acceptor. We initially selected a TAMRA-labeled (carboxytetramethylrhodamine) β2AR antagonist (alprenolol-TAMRA) as a fluorescent acceptor ligand since it has spectral characteristics theoretically amenable both for accepting energy from either bioluminescent protein donors (peak excitation at 565 nm), and for emitting light at wavelengths that can be clearly distinguished from luciferase light emission (peak emission at 580 nm). We added increasing concentrations of TAMRA-labeled alprenolol to HEK293 cells transiently-transfected with luciferase tagged-β2AR prior to direct addition of luciferase substrate (coelenterazine h for Rluc8 and furimazine for Nluc) in continued presence of fluorescent ligand. We could readily detect receptor specific binding using Nluc as the BRET donor but not when Rluc8 was used (Fig. 1a,b). The main reason for this is likely to be the capacity of these N-terminally Nluc-tagged receptors to traffic appropriately to the plasma membrane (Supplementary Fig. 2). Thus, we observed a clear concentration-dependent ligand-binding BRET signal in cells expressing Nluc-β2AR, which was completely prevented by competition with a high concentration (10 μM) of unlabeled alprenolol (Fig. 1b).
Figure 1.
Suitability of NanoLuc for BRET binding studies. (a,b) BRET ligand binding assays for transiently-transfected Rluc8-β2AR (a) and Nluc-β2AR (b) treated with increasing concentrations of alprenolol-TAMRA in the absence or presence of 10 μM unlabeled alprenolol. Data are mean ± s.e.m. of three experiments performed in quadruplicate. (c,d) Inhibition of the BRET signal for HEK293 cells stably-expressing Nluc-β2AR treated with 10 nM propranolol-BY630 (c) or propranolol-BYFL (d) and increasing concentrations of unlabelled ligands as shown. Each data point represents mean ± s.e.m. of five (all curves in (c) and propranolol in (d)) or four (d) separate experiments. In each experiment we made triplicate determinations for each data point. We calculated KD values indicating the affinity of propranolol-BY630 and propranolol-BYFL (mean ± s.e.m.) from saturation binding assays as 18.9 ± 4.1 nM (n=6) and 42.8 ± 10.8 nM (n=8) respectively. Subsequently, we calculated the respective pKi values indicating the affinity of propranolol, ICI 118551 and CGP12177 from the corresponding IC50 values using the Cheng-Prusoff Equation: 8.13 ± 0.05, 8.04 ± 0.04, 8.32 ± 0.03 (competing with propranolol-BY630) and 8.89 ± 0.09, 8.69 ± 0.14, 8.92 ± 0.03 (competing with propranolol-BYFL). These values (particularly those obtained with propranolol-BYFL) are comparable to those obtained by Baker (2005)20. See text for further discussion of the small differences between values obtained with propranolol-BY630 and propranolol-BYFL.
To test compatibility with various fluorophores, we used fluorescent propranolol derivatives conjugated to either BODIPY-630/650 (excitation 630 nm, emission 650 nm; propranolol-BY630) or BODIPY-FL (excitation 503 nm, emission 512 nm; propranolol-BYFL) combined with Nluc-β2AR (Fig. 1c,d). We observed specific binding with both fluorescent ligands, which was inhibited by isoprenaline, propranolol, ICI 118551 and CGP 12177. Therefore Nluc has a substantial dynamic range compatible with excitation wavelengths of both BODIPY-630/650 and BODIPY-FL fluorophores. We observed a reduced signal-to-background ratio with propranolol-BYFL (Supplementary Fig. 3), which is due to the large degree of donor background present in the BRET acceptor channel. Consequently BODIPY-630/650 is generally a preferred choice of acceptor. However, despite suboptimal performance with the BODIPY-FL, our results support use of a variety of fluorescent dyes as potentially robust tracers for ligand binding applications for GPCRs. As with distinct radioligands acting on the same receptor, different fluorophores may result in a ligand exhibiting slightly different affinities8 and binding modes. This could potentially shift apparent affinities of competing ligands due to the probe-dependency of cooperative interactions between protomers within a receptor complex9. This factor may also influence fluorophore choice, opening up interesting avenues of potential research into cooperativity mechanisms by utilising multiple fluorescent ligands (see below).
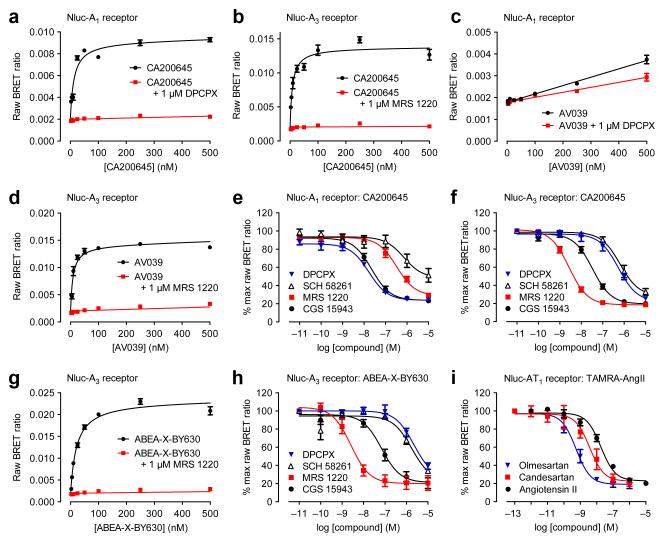
To further exemplify BRET ligand binding with multiple fluorescent ligands, we used HEK293 cells stably-transfected with N-terminally Nluc-labeled adenosine A1 or A3 receptors and treated them with increasing concentrations of the BODIPY-630/650-labeled antagonist CA20064510. We determined non-specific binding using a high concentration of unlabelled antagonist (DPCPX for Nluc-A1 or MRS 1220 for Nluc-A3) and measured BRET after direct furimazine addition. We observed a saturable signal for both receptors with low non-specific binding (Fig. 2a,b) across the full concentration range of fluorescent ligand. KD values for specific binding were 7.5 ± 2.4 nM for Nluc-A1 and 7.6 ± 3.7 nM for Nluc-A3 (mean ± s.e.m. of n=4), consistent with unmodified receptors11. In addition, we examined kinetics of CA200645 binding to Nluc-A1 (Supplementary Fig. 4), yielding a similar KD to that obtained with saturation binding (20.4 ± 6.9 nM, n=3, p>0.05 unpaired t-test vs saturation KD). As affinity values for CA200645 at both adenosine receptors were very similar, it was important to confirm that the specific ligand binding BRET signals generated had the appropriate pharmacology for the specific receptor under study, and were not simply a consequence of non-specific membrane interactions due to the lipophilicity of BODIPY. We used an A3-selective fluorescent ligand AV039 (compound 19 in ref. 12) containing the same BODIPY fluorophore and tested its ability to bind to Nluc-A1 (Fig. 2c) and Nluc-A3 (Fig. 2d) expressing cells. We did not detect a saturable specific BRET signal at Nluc-A1 at concentrations of AV039 up to 500 nM (Fig. 2c). In contrast, we detected clear specific binding at Nluc-A3 (Fig. 2d) yielding a KD for AV039 of 24.6 ± 8.3 nM.
Figure 2.
Extending use of NanoBRET. (a,b) We treated Nluc-A1 (a) and Nluc-A3 (b) with increasing CA200645 concentrations with non-specific binding established with 1 μM DPCPX for Nluc-A1 (a) and 1 μM MRS 1220 for Nluc-A3 (b). (c,d) We observed a specific saturable signal in Nluc-A3 (d) cells but not in Nluc-A1 (c) cells treated with increasing AV039 concentrations in the presence of 1 μM MRS 1220 (d) or 1 μM DPCPX (c). In (c), the only statistically significant difference was at 250 nM AV039 (two-way ANOVA; p<0.05). (e,f) We treated Nluc-A1 (e) and Nluc-A3 (f) cells with 25 nM CA200645 and increasing unlabeled ligand concentrations. (g) We generated saturation BRET binding curves for binding fluorescent agonist ABEA-X-BY630 to Nluc-A3 in absence or presence of 1μM MRS 1220. (h) We monitored the ability of increasing concentrations of DPCPX, SCH 58261, MRS 1220 and CGS 15943 to decrease Nluc-A3 to ABEA-X-BY630 BRET. (i) We treated Nluc-AT1 with 1 μM TAMRA-AngII and increasing concentrations of angiotensin II, candesartan and olmesartan. We measured BRET after furimazine addition. Panels a, b, c, d and g are representative of four experiments (in triplicate; error bars are s.e.m. of triplicate points). Data in e, f, h and i represent mean ± s.e.m. of three experiments (in duplicate; i) or four experiments (in triplicate; e, f, h). Exceptions are DPCPX in (e) which is mean ± s.e.m. of five experiments (in triplicate) and MRS1220 in (f) which is mean ± s.e.m. of three experiments (in triplicate).
We then investigated the ability of a panel of ligands to inhibit specific binding of CA200645 to Nluc-A1 and Nluc-A3 (Fig. 2e,f and Supplementary Fig. 5) and calculated affinity (pKi) values (Supplementary Table 1). Importantly the A1-selective antagonist DPCPX showed high affinity at Nluc-A1 and low affinity at Nluc-A3 and conversely the A3-selective antagonist MRS1220 showed high affinity at Nluc-A3 and lower affinity at Nluc-A1. Furthermore, affinities were comparable to those obtained using radioligand binding assays13 (Supplementary Table 1). We also obtained comparable results with the A3-selective ligand AV039 in Nluc-A3 cells (Supplementary Fig. 6 and Supplementary Table 2).
To determine whether the BRET ligand-binding assay was applicable to fluorescent agonists, we also undertook experiments with a fluorescent adenosine receptor agonist, ABEA-X-BY63014. In saturation binding experiments, we observed a clear saturable BRET signal at both Nluc-A3 (Fig. 2g) and Nluc-A1 (Supplementary Fig. 7a). ABEA-X-BY630 had a higher affinity for Nluc-A3 (KD = 38.4 ± 13.7 nM) than Nluc-A1 (KD = 167.0 ± 74.4 nM). Competition binding assays with ABEA-X-BY630 and the panel of eight ligands used above yielded comparable affinities to those obtained with the antagonist fluorescent ligands (Fig. 2h, Supplementary Fig. 7 and Supplementary Table 2). Again, affinities were comparable to those obtained using radioligand binding assays13 (Supplementary Table 2). However, we obtained subtle differences in pKi values for non-fluorescent competing ligands with different fluorescent ligands, particularly in the case of Nluc-A3 (Supplementary Fig. 8). This would be in keeping with known allosterism across the A3 homodimer interface9. Interestingly, we observed a similar phenomenon with the β2-adrenoceptor (Fig. 1) that is also known to form homodimers15.
We used a similar fluorescent agonist strategy to investigate ligand binding to a peptide receptor (angiotensin II receptor type 1; AT1) using a TAMRA-labelled angiotensin II (TAMRA-AngII). We performed competition BRET binding assays to investigate the ability of three AT1 ligands (angiotensin II, candesartan and olmesartan) to reduce the BRET signal. We observed a clear, concentration-dependent decrease in BRET signal in the presence of competing compounds (Fig. 2i), with olmesartan exhibiting highest affinity. To demonstrate assay sensitivity, we carried out competition binding assays at Nluc-AT1 using varying concentrations of TAMRA-AngII and found at concentrations down to 100 nM TAMRA-AngII a specific signal was still observed that could be reduced by all three competing compounds (Supplementary Fig. 9).
Traditionally, binding assays have used radioactive ligands to probe targets; however, this has become increasingly costly and undesirable for practical reasons16,17. Furthermore, for technical reasons, many of the ligand-receptor affinities published in the literature have consequently been derived using cell membranes assayed at 4°C, with assumptions made about applicability to receptors in live cells at 37°C, as they are in the body. More recently, FRET ligand binding assays have been developed and successfully utilized16,18, setting a precedent for applicability of resonance energy transfer approaches. However, there are a number of reasons why BRET is distinct from FRET, and indeed, is often used in preference1. This study has demonstrated that BRET ligand binding assays provide an exciting alternative to radioligand binding assays, with the important advantage of being able to monitor ligand-receptor interactions in live cells, at 37°C and in real-time. Separation of free and bound fluorescent ligand is not required due to the exquisite distance-dependence of BRET19, nor is an additional step of conjugating a fluorophore to the N-terminal domain of the receptor of interest, as required with SNAP/CLIP technology commonly used with time-resolved FRET16. Indeed, as illustrated by our data, no wash steps or lysis are required, making this approach truly homogenous. Consequently BRET ligand binding has considerable potential for future drug discovery and profiling applications.
ONLINE METHODS
cDNA constructs
We cloned β2AR and AT1 receptor cDNAs into a pF-sNnK vector (Promega), encoding a fusion of the secretory signal peptide sequence of IL6 on the N-terminus of NanoLuc (Nluc). The resulting open reading frame (ORF) therefore encoded a fusion of secreted Nluc at the N-terminus of β2AR or AT1 receptor, with Gly-Ser-Ser-Gly linkers between the Nluc ORF and the GPCR ORF. N-terminally Rluc8-labeled β2AR was generated by substituting the Nluc ORF for the Rluc8 ORF. We generated Nluc-labeled adenosine receptor constructs by amplifying the full length sequence of Nluc luciferase (as provided by Promega in the pNL1.1 vector) and fusing it in frame with the membrane signal sequence of the 5HT3A receptor within pcDNA3.1 to yield sig-Nluc. We then fused the full-length human sequence of the adenosine receptor of choice (with the methionine start signal removed) to the 3′ end of the sig-Nluc in pcDNA3.1. This gave the constructs designated as Nluc-A1 receptor and Nluc-A3 receptor, both of which include the signal sequence.
Ligands
CA200645, propranolol-BY630 (propranolol-βalanine-βalanine-X-BODIPY-630/650) and propranolol-BYFL (propranolol-βalanine-βalanine-X-BODIPY-FL) were from CellAura. AV039 and ABEA-X-BY630 (ABEA-X-BODIPY-630/650) were synthesised by the University of Nottingham as described by Vernall et al.12 and Middleton et al.14. Alprenolol-TAMRA was synthesised by Promega. TAMRA-AngII was from AnaSpec. Alprenolol and angiotensin II were from Sigma. Candesartan and olmesartan were from Zhou Fang Pharm Chemical. DPCPX, SCH 58261, MRS 1220, CGS 15943, ZM 241385, XAC, PSB 603, isoprenaline, propranolol, ICI 118551 and CGP 12177 were from Tocris.
Stable cell line generation
We maintained HEK293 cells (for Nluc-A1; from ATCC) or HEK293G cells (Glosensor™ cAMP HEK293 for Nluc-A3; from Promega) in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum (FCS) and 2 mM L-glutamine at 37°C, 5% CO2. We generated mixed population cell lines by transfecting the required Nluc-adenosine receptor construct using Lipofectoamine (Life Technologies) according to the manufacturer’s instructions and then subjecting the cells to selective pressure (1 mg/ml G418) for 2–3 weeks. We then dilution cloned the Nluc-A1 and A3 receptor cell lines to obtain cell lines originating from a single cell. The Nluc-β2AR stable cell line was from Promega. We confirm that all cell lines used were mycoplasma free.
BRET β2AR ligand binding assays
We carried out transient transfections of HEK293 cells (ATCC) using FuGENE (Promega) at a 3:1 lipid:DNA ratio. We then seeded cells and lipid-DNA complexes into 96-well plates at a density of 20,000 cells per well (in DMEM supplemented with 10% FCS (Gibco)), with 50 ng of DNA per well. 24 hours post-transfection, we removed the media from each well and replaced it with OptiMEM without phenol red (Gibco). For experiments using alprenolol-TAMRA, we then incubated for 180 min in OptiMEM (without phenol red). We then added serially-diluted alprenolol-TAMRA in the absence or presence of 10 μM alprenolol and incubated for 120 min at room temperature. We then added the required substrate (furimazine for Nluc-β2AR and coelenterazine h for Rluc8-β2AR) to a final concentration of 10 μM. We then measured BRET using the CLARIOstar plate reader (BMG Labtech) at room temperature. We sequentially measured filtered light emissions at 450 nm (80 nm bandpass) and >610 nm (longpass), and calculated the raw BRET ratio by dividing the >610 nm emission by the 450 nm emission. For competition experiments using fluorescent-propranolol derivatives, we incubated Nluc-β2AR stably-transfected HEK293 cells with 10 nM propranolol-BY630 or propranolol-BYFL and the required concentration of competing ligand diluted in HEPES buffered saline solution (HBSS, 25 mM HEPES, 10 mM glucose, 146 mM NaCl, 5 mM KCl, 1 mM MgSO4, 2 mM sodium pyruvate, 1.3 mM CaCl2) for 1 h at 37°C. For saturation experiments, we incubated β2AR cells with increasing concentrations of propranolol-BY630 or propranolol-BYFL in the presence or absence of 1 μM propranolol for 1 h at 37°C. We then measured the luminescence and fluorescence using the PHERAstar FS plate reader (BMG Labtech) at room temperature. We measured filtered light emissions at 460 nm (80 nm bandpass) and 535 nm (60 nm bandpass) for propranolol-BYFL and at 460 nm (80 nm bandpass) and >610 nm (longpass) for propranolol-BY630. We calculated the raw BRET ratio by dividing the >610 nm emission or 535 nm emission by the 460 nm emission. We have adopted the term ‘raw BRET ratio’ as no background ratio has been subtracted.
BRET A1 and A3 receptor ligand binding assays
We performed saturation and competition binding assays on stably-transfected cells that we seeded 24 h prior to experimentation in white Thermo Scientific Matrix 96-well microplates. We removed the media from each well and replaced it with HBSS with the required concentration of fluorescent ligand and competing ligand. For pre-incubation experiments with ABEA-X-BY630, we incubated competing unlabeled ligand for 30 min prior to the addition of 250 nM ABEA-X-BY630 for Nluc-A1 expressing cells and 50 nM ABEA-X-BY630 for Nluc-A3 expressing cells. For saturation and competition experiments, upon the addition of fluorescent ligand we incubated cells for 1 h at 37°C (no CO2) and then added the Nluc substrate furimazine (Promega) to a final concentration of 10 μM. For association kinetic experiments on Nluc-A1 expressing cells, we removed media from each well, replaced it with HBSS containing 10 μM furimazine and incubated for 15 min at room temperature in the PHERAstar FS plate reader (BMG Labtech) to allow the signal to reach equilibrium. We then added the required concentration of CA200645, immediately reinserted the plate and read every well once per min for 60 min. For all experiments, we measured the luminescence and resulting BRET using the PHERAstar FS plate reader (BMG Labtech) at room temperature. We again sequentially measured filtered light emissions at 460 nm (80 nm bandpass) and >610 nm (longpass), and calculated the raw BRET ratio by dividing the >610 nm emission by the 460 nm emission.
BRET AT1 receptor ligand binding assays
We maintained HEK293FT cells (Life Technologies) at 37°C, 5% CO2 in complete medium (Dulbecco’s modified Eagle’s medium (DMEM) containing 0.3 mg/ml glutamine, 100 IU/ml penicillin, and 100 μg/ml streptomycin) supplemented with 10% FCS (Gibco). We carried out transient transfections in a 96-well plate using FuGENE (Promega). For each well we incubated 1 ng of Nluc-AT1 receptor cDNA and 49 ng of pcDNA3 (Life Technologies) for 10 min at room temperature with a mix of 0.5 μl of FuGENE and 49.5 μl of serum-free DMEM (pre-incubated at room temperature for 5 min). We then incubated cells (105 in 150 μl/well) in DMEM supplemented with 10% FCS with the final DNA-FuGENE mix (50 μl/well). We carried out assays 48 h post transfection after removing medium. We treated cells with competitor ligand for 30 min followed by addition of TAMRA-AngII for a further 30 min. We carried out ligand incubations at 37°C, 5% CO2. We then added the Nluc substrate furimazine (Promega) to a final concentration of 10 μM and measured luminescence immediately. We measured BRET at 37°C using the PHERAStar FS plate reader (BMG Labtech). We sequentially measured filtered light emissions at 460 nm (80 nm bandpass) and >610 nm (longpass), and calculated the raw BRET ratio by dividing the >610 nm emission by the 460 nm emission.
Measurement of Nluc and Rluc8 emission spectra
To determine the emission spectra of Nluc and Rluc8, we transiently transfected HEK293 cells (ATCC) with the expression constructs for Nluc-β2AR and Rluc8-β2AR as described above for the β2AR ligand binding assays. 24 hours post-transfection, we removed the media from each well and replaced it with OptiMEM without phenol red (Gibco), followed by incubation for 180 min at 37°C. Immediately prior to measurement we added the luciferase substrates furimazine (Nluc) or coelenterazine h (Rluc8) at a final concentration of 10 μM. We then determined the emission spectra with a CLARIOstar plate reader (BMG Labtech) using the luminescent scanning option (20 nm bandwidth, 1 nm resolution, integration time 500 msec, gain 3000).
Bioluminescent imaging of Nluc-β2AR and Rluc8-β2AR
We performed bioluminescent imaging experiments to determine the localization of the Nluc-β2AR and Rluc8-β2AR fusion proteins. We performed all imaging experiments using the Olympus LV200 bioluminescence microscope equipped with a Hamamatsu ImagEM EMCCD camera and a 100x/1.4 UPLanSApo. We transfected HEK293 cells with expression constructs for Nluc-β2AR and Rluc8-β2AR, plated in 35 mm optically clear dishes (Ibidi) at a density of 200,000 cells per dish in 2 ml growth medium (DMEM supplemented with 10% FCS) and incubated for 24 h in a tissue culture incubator. We then replaced the growth medium with 1 ml OptiMEM followed by incubation for 180 min at 37°C. Immediately prior to image acquisition, we replaced the media with OptiMem including the luciferase substrates furimazine (Nluc) or coelenterazine h (Rluc8) at a final concentration of 10 μM. We identified suitable fields of view based on imaging of total luminescent signal of the donor fusion. We acquired images using an EM gain of 200 and exposure times of 1 sec (Nluc-β2AR) and 20 sec (Rluc8-β2AR). We acquired all images using Olympus cellSens software and performed image processing with ImageJ software.
Data presentation and statistical analysis
We presented and analysed data using Prism software (GraphPad).
We simultaneously fitted the total and non-specific saturation binding curves using the following equation:
where Bmax is the maximal response, [B] is the concentration of fluorescent ligand in nM, KD is the equilibrium dissociation constant in nM, M is the slope of the non-specific binding component and C is the intercept with the Y-axis.
We fitted the competition binding curves to calculate the Ki of the unlabeled ligands using the Cheng-Prusoff equation:
where [L] is the concentration of fluorescent ligand in nM and KD is the KD of fluorescent ligand in nM. The calculated KD values used were as calculated from the saturation binding experiments. The IC50 is calculated from the following equation:
where [A] is the concentration of competing drug and the IC50 is the molar concentration of ligand required to inhibit 50% of the specific binding of concentration [L] of the fluorescent ligand. We also used this equation to fit concentration-inhibition data where the affinity of the labelled ligand is unknown.
From association kinetic data, we obtained kon, koff and KD values from the following equation:
Where KD is the equilibrium dissociation constant, koff is the dissociation rate of the ligand in min−1 and kon is the association rate in M−1 min−1 and is calculated as follows:
Where [L] is the ligand concentration in M and kobs is calculated from global fitting of the data to the following monoexponential association function:
Where Ymax equals levels of binding at infinite time (t) and kobs is the rate of observed association.
We carried out statistical analysis using unpaired t-test or ANOVA as appropriate (P<0.05). The n values in the text refer to the number of separate repeat experiments. Based on our experience, a minimum of three repeat experiments, a power of 98% and a P value of 0.05, will give a standardized difference of interest (e.g. a difference in pKi values) of approximately 0.5 using the NanoBRET ligand binding assay.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank C. Corona and P. Meinsenheimer (Promega) for providing the alprenolol-TAMRA fluorescent ligand and J. Denman (The University of Nottingham) for assistance with generating the NanoLuc-adenosine receptor constructs. This project was funded by the Australian Research Council (ARC) Linkage Grant LP130100037. The University of Nottingham, Promega Corporation and BMG Labtech Pty Ltd provided funding as partner organizations of this grant. E.K.M.J. was funded by the Richard Walter Gibbon Medical Research Scholarship from The University of Western Australia. Work in S.J.H.’s laboratory was funded by the UK Medical Research Council (G0800006). K.D.G.P. was funded by an ARC Future Fellowship (FT100100271) and subsequently a National Health and Medical Research Council of Australia RD Wright Fellowship (1085842). S.J.H. thanks the Raine Foundation for a Visiting Research Professorship and currently holds an Adjunct Professorship of The University of Western Australia.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
M.B.R., T.M. and K.V.W. are employees of Promega Corporation, which has proprietary rights over the NanoBRET assay. S.J.H. is a former Director of the University of Nottingham spin out company CellAura Technologies that provided some of the fluorescent ligands.
References
- 1.Pfleger KDG, Eidne KA. Nat. Methods. 2006;3:165–174. doi: 10.1038/nmeth841. [DOI] [PubMed] [Google Scholar]
- 2.Jaeger WC, Armstrong SP, Hill SJ, Pfleger KD. Front. Endocrinol. (Lausanne) 2014;5:26. doi: 10.3389/fendo.2014.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfleger KD, Seeber RM, Eidne KA. Nat. Protoc. 2006;1:337–345. doi: 10.1038/nprot.2006.52. [DOI] [PubMed] [Google Scholar]
- 4.Kocan M, See HB, Seeber RM, Eidne KA, Pfleger KD. J. Biomol. Screen. 2008;13:888–898. doi: 10.1177/1087057108324032. [DOI] [PubMed] [Google Scholar]
- 5.Griss, et al. Nat. Chem. Biol. 2014;10:598–603. doi: 10.1038/nchembio.1554. [DOI] [PubMed] [Google Scholar]
- 6.Vernall AJ, Hill SJ, Kellam B. Br. J. Pharmcol. 2014;171:1073–1087. doi: 10.1111/bph.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall MP, et al. ACS Chem. Biol. 2012;7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker JG, et al. Br. J. Pharmacol. 2010;159:772–786. doi: 10.1111/j.1476-5381.2009.00488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.May LT, Bridge LJ, Stoddart LA, Briddon SJ, Hill SJ. FASEB J. 2011;25:3465–3476. doi: 10.1096/fj.11-186296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoddart LA, et al. Chem. Biol. 2012;19:1105–1115. doi: 10.1016/j.chembiol.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vernall AJ, et al. Org Biomol. Chem. 2013;11:5673–5682. doi: 10.1039/c3ob41221k. [DOI] [PubMed] [Google Scholar]
- 12.Vernall AJ, Stoddart LA, Briddon SJ, Hill SJ, Kellam B. J. Med. Chem. 2012;55:1771–1782. doi: 10.1021/jm201722y. [DOI] [PubMed] [Google Scholar]
- 13.Baker JG, Hill SJ. J. Pharmacol. Exp. Ther. 2007;320:218–228. doi: 10.1124/jpet.106.113589. [DOI] [PubMed] [Google Scholar]
- 14.Middleton RJ, et al. J. Med. Chem. 2007;50:782–793. doi: 10.1021/jm061279i. [DOI] [PubMed] [Google Scholar]
- 15.Calebiro D, et al. Proc. Natl. Acad. Sci. 2013;110:743–748. doi: 10.1073/pnas.1205798110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zwier JM, et al. J. Biomol. Screen. 2010;15:1248–1259. doi: 10.1177/1087057110384611. [DOI] [PubMed] [Google Scholar]
- 17.Cottet M, et al. Biochem. Soc. Trans. 2013;41:148–153. doi: 10.1042/BST20120237. [DOI] [PubMed] [Google Scholar]
- 18.Fernández-Dueñas V, et al. J. Neurochem. 2012;123:373–384. doi: 10.1111/j.1471-4159.2012.07956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dacres H, Michie M, Wang J, Pfleger KD, Trowell SC. Biochem. Biophys. Res. Commun. 2012;425:625–629. doi: 10.1016/j.bbrc.2012.07.133. [DOI] [PubMed] [Google Scholar]
- 20.Baker JG. Br. J. Pharmacol. 2005;144:317–322. doi: 10.1038/sj.bjp.0706048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.