Abstract
Recently, a series of acute swine erysipelas outbreaks occurred in Eastern China. Eight strains isolated from cases of septicemia were determined as serotype 1a, and 4 of the isolates were resistant to acriflavine. One isolate strain named HX130709 was attenuated on agar media containing acriflavine dye. The 432-bp hypervariable region in spaA gene of the field and attenuated strains were amplified and sequenced. It was further compared with the vaccine strain G4T10, and thus, the eight field strains can be divided into four spaA-types. The partial spaA gene analysis also showed that no point mutations occurred among different archived passages of HX130709 during the attenuation. Results of pulsed-field gel electrophoresis showed that eight distinct patterns with 22 to 30 DNA fragment bands were produced from field strains, and twelve distinct patterns with 23 to 27 DNA fragment bands were produced from different passages of the attenuated strains. Mouse pathogenicity test showed that the mortality of the mice infected with 104 CFU field strains was 100% and the attenuation of strain HX130709 occurred between 46 and 50 passages. All the field and attenuated strains were highly sensitive to β-lactam antibiotics, tetracyclines and macrolides. So, we can make conclusions that the acute swine erysipelas outbreaks in Eastern China were caused by serotype 1a E. rhusiopathiae strains with different biochemical characteristics, and the virulence of serotype 1a E. rhusiopathiae strains is unrelated with some point mutations in 432-bp hypervariable region of the spaA gene.
Keywords: acriflavine, attenuation, Erysipelothrix rhusiopathiae, PFGE, spa
Erysipelothrix rhusiopathiae (E. rhusiopathiae) is a small gram-positive, slender and straight or slightly rod bacterium that causes erysipelas in swine and a wide spectrum of diseases in other animals, like sheep, some fishes, reptiles and birds. E. rhusiopathiae is also an important pathogen with respect to erysipeloid, a skin disease in humans [22]. This bacterium has been isolated in most parts of the world, from sick and healthy animals (pork, seafood and chicken) and even from the environment they lived [1, 3]. Among the 15 serotypes of E. rhusiopathiae, serotypes 1a, 1b and 2 are the most important agents affecting swine industry [2, 5, 8, 14, 17, 22].
Due to the importance of swine erysipelas, vaccines with killed organisms and attenuated live vaccines were developed. In 1932, an acriflavine-resistant attenuated live vaccine was developed in Japan. This vaccine strain Koganei 65-0.15 (serotype 1a) was attenuated by 65 passages on agar media containing 0.15% of an acriflavine dye [15]. Though outbreaks of acute septicemia or subacute urticaria of erysipelas have decreased dramatically by using the live vaccine, a chronic form of erysipelas found during meat inspections in slaughterhouses has been increasing [11]. After 1985, about 2,000 pigs annually have been shown to have either acute or sub-acute infections [19]. In the United States, erysipelas cases were recorded with increasing frequency in both vaccinated and non-vaccinated pigs during the summer of 2001 [13]. Since swine erysipelas reappeared as a clinical problem in pig populations in Japan and the Midwestern United States, it has been considered as a reemerging disease that contributes substantially to economic losses in the swine industry [1, 2, 8].
It has been demonstrated that pulsed-field gel electrophoresis (PFGE) can be used to differentiate E. rhusiopathiae strains [12, 13]. Recent studies have focused on characterization of strains based on their spa type, whose gene encodes a surface protective antigen (Spa) [7, 9, 16]. The Spa proteins of E. rhusiopathiae can be classified into 3 molecular species, named SpaA (produced by serovars 1a, 1b, 2, 5, 8, 9, 12, 15, 16, 17 and N), SpaB (produced by serovars 4, 6, 11, 19 and 21) and SpaC (produced by serovar 18) [18]. More recently, a method for discrimination between a Japanese vaccine strain and the field strains was developed based on the nucleotide sequencing of a 432-bp hypervariable region in the spaA gene [11].
In 2009, sporadic outbreaks of acute swine erysipelas occurred, and one strain of E. rhusiopathiae was isolated from Siyang county of Jiangsu province. However, a wave of acute swine erysipelas outbreaks was seen in Eastern China, and seven other E. rhusiopathiae strains were isolated since the summer of 2013. In the present study, the spaA sequence analysis in combination with serotyping, PFGE, acriflavine resistance, mouse pathogenicity and antimicrobial susceptibility tests were used to identify and characterize recent field strains of E. rhusiopathiae from acute swine erysipelas. Then, one strain HX130709 was attenuated by 55 passages on agar media containing a gradually increasing concentration (0.0025% to 0.03%) of acriflavine dye and sequenced every five passages to see if any mutations occurred in the hypervariable region of spaA gene during the attenuation.
MATERIALS AND METHODS
Bacteria isolation: BHI-T 80 agar: BHI-T 80 agar and broth were used for isolation of E. rhusiopathiae strains from tissue samples. The strains were identified and confirmed as E. rhusiopathiae by polymerase chain reaction (PCR) [18] after Gram stain and microscopic examination. All strains were isolated from cases of acute swine erysipelas causing sudden death of fattening pigs and postpartum sows in 2009 (1 strain) and 2013 (7 strains) and were named after the isolation districts and time (Table 1).
Table 1. Strains used in the study and eight field strains isolated from cases of septicemia causing acute death of fattening pigs and postpartum sows in Eastern China.
| Strains | Date Isolated | Area Isolated | GenBank Accession number |
Serotype | Country of Isolation |
|---|---|---|---|---|---|
| Serotype 1a (Fujisawa) | unknown | unknown | AB259652.1 | 1a | Japan |
| G4T10 | unknown | unknown | KJ645072 | 1a | unknown |
| SY091027 | 27/10/2009 | Siyang | KJ645080 | 1a | China |
| NZ130701 | 01/07/2013 | Nanzhang | KJ645073 | 1a | China |
| HX130709 | 09/07/2013 | Hexian | KJ645074 | 1a | China |
| XC130710 | 10/07/2013 | Xuancheng | KJ645075 | 1a | China |
| SH130723 | 23/07/2013 | Sihong | KJ645076 | 1a | China |
| YC130820a) | 20/08/2013 | Yancheng | KJ645077 | 1a | China |
| YC130828a) | 28/08/2013 | Yancheng | KJ645078 | 1a | China |
| YC131115a) | 15/11/2013 | Yancheng | KJ645079 | 1a | China |
a) Isolates YC130820, YC130828 and YC131115 were originated from three different farms in Yancheng.
G4T10 strain (serotype 1a), a widely used commercial vaccine in China, was used as a reference attenuated strain with acriflavine resistance.
Serotyping: Serotyping was done on all the 8 field E. rhusiopathiae strains as described previously with some modifications [21]. All the culture supernatants were prepared following the schedule provided by Dr. Tanja Opriessnig (Veterinary Diagnostic Laboratory, College of Veterinary Medicine, Iowa State University, Ames, Iowa, U.S.A.). In brief, a pure culture was grown at 37°C for 48 hr in 30 ml of BHI broth (Binhe Microorganism Reagent, Hangzhou, China) supplemented with 10% bovine serum (Tianhang Biological Technology, Deqing, China). The culture was treated with formalin solution (final concentration of 1%) (Xilong chemical corporation, Shantou, China), held at room temperature for 24 hr, harvested by centrifugation and washed three times in 1 × PBS buffer containing 5% formalin. Washed cells were suspended in 1 ml of distilled water and autoclaved at 121°C for 45 min. The supernatant was collected and delivered to Dr. Tanja Opriessnig’s lab for the agar gel precipitation test. Reactions were recorded after 24 hr.
Analysis of the 432-bp hypervariable region in the spaA gene of different strains: Primers Erko-1F (5′-GTGAAACACCGTATTTTAGTA-3′) and Erko-2R (5′- TTCAAGAAGTTCCTGTAGTTT-3′) were used to amplify the highly variable region of the spaA gene [11]. Briefly, genomic DNA of E. rhusiopathiae strains was prepared using bacterial DNA Kit (OMEGA, Norcross, GA, U.S.A.). Polymerase chain reaction (PCR) was performed as described elsewhere [18] under the following conditions with some modifications: PrimeSTAR HS DNA Polymerase (TaKaRa, Tokyo, Japan) was used to perform PCR; a denaturation step at 94°C for 3 min followed by 35 cycles of denaturation at 94°C for 45 sec; annealing at 55°C for 30 sec and extension at 72°C for 30 sec; and final extension of 5 min. For each strain, PCR was performed three times, and each time, the PCR products were ligated with the pMD19-T Simple Vector (TaKaRa), and then, three colonies of each ligation product were sent to Invitrogen (Life Technologies, Shanghai, China) for sequencing. The DNA sequences were analyzed as described previously [11].
Pulsed-field gel electrophoresis: Chromosomal DNAs from the strains were digested with SmaI, and PFGE was performed 3 times according to Opriessnig et al.[13]. Electrophoresis was carried out for 23 hr at 14°C and 6 V with pulse times from an initial 1.2 sec to a final 30 sec. PFGE patterns were detected by UV transillumination after staining by SYBR Gold nucleic acid gel stain (S11494, Life technologies). A lambda ladder PFG marker (N0340s, NEB, Ipswitch, MA, U.S.A.) was used as DNA size standard.
Acriflavine resistance test: The acriflavine resistance of all the eight strains was examined by streaking strains onto agar plates containing 0.04, 0.03, 0.02, 0.015, 0.01, 0.075, 0.005, 0.0025, 0.00125 or 0% acriflavine (Aladdin, Shanghai, China). The acriflavine agar plates were prepared by adding 1% acriflavine (dissolved in distilled water) to the sterilized basal medium (BHI-T 80 agar). The isolates were cultivated in 3 ml BHI-T 80 broth overnight at 37°C and were then streaked onto the agar plates. The vaccine strain (G4T10) was also streaked onto each plate as a positive control. The growth on the plates was observed after incubation at 37°C for 48 hr. Acriflavine resistance was determined by the highest concentration at which a strain showed almost the same growth as on the agar that did not contain acriflavine [8, 9, 11].
Attenuation of strain HX130709: Strain HX130709a (derived from strain HX130709) was attenuated by 55 passages on BHI-T 80 agar containing a gradually increasing concentration (0.0025% to 0.03%) of acriflavine dye. Bacteria were collected and stored under −70°C every five passages during the attenuation. Finally, the 432-bp partial spaA genes of all the archived strains were amplified for sequence analysis, and the virulence of each archived strain was confirmed in mice.
Mouse pathogenicity tests: One hundred and sixty-five female ICR mice of six-week-age were purchased from the Laboratory Animal Centre of Nantong University (Nantong, China). The animals were cared for in accordance with the Guide for the Care and Use of Laboratory Animals (1996, published by National Academy Press, NW, Washington, DC, U.S.A.). The animal experiments were approved by the Institutional Animal Care and Ethics Committee of Nanjing Agricultural University (Approval No. IACECNAU-20100902).
A total of 90 female ICR mice of six-week-age were given subcutaneous injections in the right groin with 104 colony forming units (CFU)/ 0.1 ml of the isolates or the vaccine strain. A group of 10 mice served as an untreated control. Since a positive correlation between the isolate’s CFU and OD value had been built in advance (data not shown), the number of bacteria injected into each mouse was almost the same. Mice were observed every 12 hr to detect clinical signs of the disease for the subsequent 14 days [8].
Another 60 female ICR mice of six-week-age were given subcutaneous injections in the right groin with 104 CFU/ 0.1 ml of the isolate strain HX130709 (F0) and different passages of the attenuated strain HX130709a (F5, F10, F15, F20, F25, F30, F35, F40, F45, F50 and F55). A group of 5 mice served as an untreated control. Mice were observed as above.
Antimicrobial susceptibility test: Each isolate was cultured in 3 ml BHI-T 80 broth overnight at 37°C. Then, 0.1 ml of the bacterial culture of each isolate was dropped onto BHI-T 80 agar, after which, antibiotic susceptibility discs (Binhe Microorganism Reagent) were put onto the BHI-T 80 agar. The diameter of the bacteriostatic circle was measured after incubation at 37°C for 24 hr. The susceptibility was determined according to instructions of the Clinical and Laboratory Standards Institute (CLSI) [4].
RESULTS
Serotyping and acriflavine resistance: All the eight E. rhusiopathiae isolates were determined to be serotype 1a (Fig. 1), which was finished by the laboratory of Dr. Tanja Opriessnig (Veterinary Diagnostic Laboratory, College of Veterinary Medicine, Iowa State University). The results of the acriflavine resistant test are shown in Table 2. Out of the total 8 isolates, 3 (37.5%) showed culture growth on the BHI-T 80 agar containing 0.02% acriflavine, which was identical to the growth of G4T10 strain (positive control).
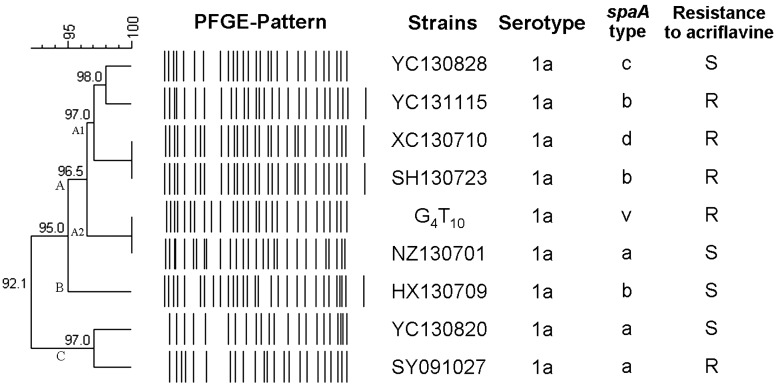
Fig. 1.
Genetic relationship between 8 E. rhusiopathiae field isolates and 1 vaccine strain and schematic representation of 8 different PFGE patterns obtained after restriction digestion with Sma I. The classification and divergence of isolates were calculated by the unweighted pair group method with averages from the PFGE results. At 5% divergence, 3 PFGE groups (A-C) were present; group A was subgrouped into A1/A2 at 3.5% divergence.
Table 2. Acriflavine resistance of E. rhusiopathiae field isolates and vaccine reference strains.
| Strain | MIC of Acriflavine (% w/v) |
Acriflavine resistancea) |
|---|---|---|
| SY091027 | 0.02 | R |
| NZ130701 | 0.005 | S |
| HX130709 | 0.0025 | S |
| XC130710 | 0.02 | R |
| SH130723 | 0.02 | R |
| YC130820 | 0.0025 | S |
| YC130828 | 0.005 | S |
| YC131115 | 0.0175 | R |
| G4T10 (vaccine) | 0.02 | R |
a) Resistant (R) or sensitive (S) to 0.01% acriflavine.
Sequence analysis of the 432-bp hypervariable region in the spaA gene: According to Nagai et al.[11], different nucleotide substitutions in the 432-bp hypervariable region on the spaA gene of the eight E. rhusiopathiae field strains and the Japanese official challenge strain Fujisawa comparing with the vaccine strain G4T10 are shown in Table 3. According to the different substitutions, spaA can be divided into four spaA-types. No nucleotide substitutions in the 432-bp hypervariable region on the spaA gene of all the archived attenuated strains were found comparing with the parental strain HX130709 (Table 4).
Table 3. Substitutions in nucleotide and amino acid in a 432-bp hypervariable region on the spaA gene of 8 E. rhusiopathiae field strains compared with the corresponding sequence of the vaccine strain G4T10.
| Substitution Pattern/strain |
Substitutions (position [nucleotide], amino acid a) ) | No. of nucleotide substitutions |
spaA type | ||||
|---|---|---|---|---|---|---|---|
| 584 (A) | 590 (T) | 609 (T) | 769 (A) | 885 (A) | |||
| D>A | I>T | I>M | I>L | NS | |||
| Pattern 1 | |||||||
| NZ130701 | C | 1 | a | ||||
| YC130820 | C | 1 | a | ||||
| SY091027 | C | 1 | a | ||||
| Pattern 2 | |||||||
| HX130709 | G | 1 | b | ||||
| SH130723 | G | 1 | b | ||||
| YC131115 | G | 1 | b | ||||
| Pattern 3 | |||||||
| YC130828 | C | G | 2 | c | |||
| Pattern4 | |||||||
| XC130710 | C | G | 2 | d | |||
| Reference strain | |||||||
| Fujisawab) | C | 1 | |||||
a) Original amino acid > substituted amino acid; NS=no amino acid mutation; G=glycine; D=aspartic acid; N=asparagine; A=alanine; I=isoleucine; T=threonine; M=methionine; L= leucine; E=glutamic acid. b) Japanese official challenge strain.
Table 4. Nucleotide substitutions in a 432-bp hypervariable region on the spaA gene of 11 E. rhusiopathiae strains during the attenuation compared with the corresponding sequence of the original strain HX130709.
| Concentration of acriflavine (% w/v) |
Number of passage |
Bacteria collected for spaA sequencing |
No. of nucleotide substitutions |
spaA type |
|---|---|---|---|---|
| 0.0025 | F1-F4 | |||
| 0.00375 | F5-F7 | F5 | 0 | b |
| 0.005 | F8-F10 | F10 | 0 | b |
| 0.0125 | F11-F12 | |||
| 0.02 | F13-F27 | F15, F20, F25 | 0 | b |
| 0.0225 | F28-F38 | F30, F35 | 0 | b |
| 0.025 | F39-F45 | F40, F45 | 0 | b |
| 0.0275 | F46-F48 | |||
| 0.03 | F49-F55 | F50, F55 | 0 | b |
| Reference strain | ||||
| HX130709 | F0 | b |
PFGE and data analysis: Eight distinct PFGE patterns with 22 to 30 DNA fragment bands were produced from the genomic DNA of the 8 isolate strains and 1 vaccine strain G4T10 with SmaI digestion (Fig. 2). Strains XC130710 and SH130723 shared the same PFGE pattern. Genetic relationships among the isolates were compared, and the dendogram analysis was done (Fig. 1). Based on the dendogram analysis, 3 PFGE groups (A-C) were present at 5% divergence. At 3.5% divergence, group A was subgrouped into A1/A2. Most of the isolates (6/9) were within group A including the vaccine strain G4T10. Resistance to acriflavine and spaA-types showed no specific patterns and were randomly distributed. Data analysis of the homogeneity of the PFGE patterns showed that these 9 strains shared over 92.1% identity with each other.
Fig. 2.
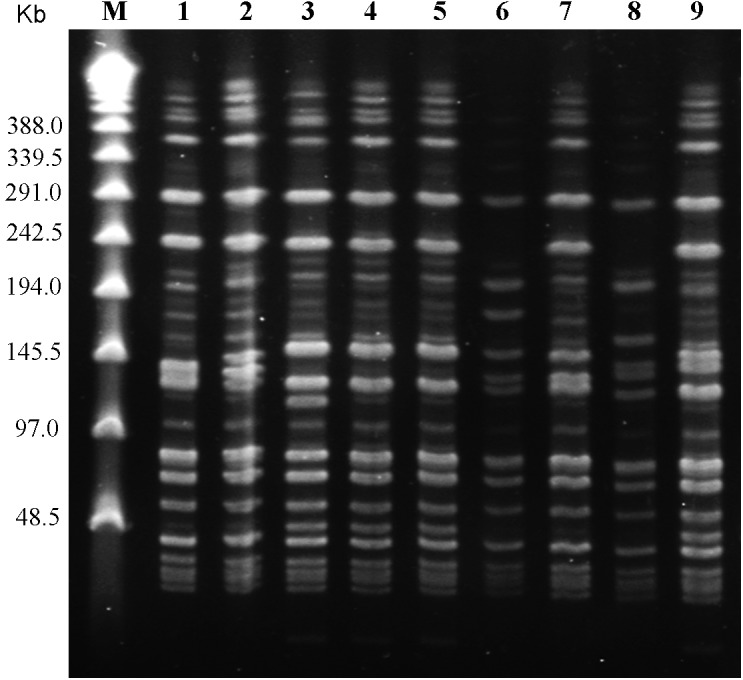
PFGE patterns produced from 8 erysipelas field isolates and 1 vaccine strain digested with Sma I. Lanes: 1, G4T10; 2, NZ130701; 3, HX130709; 4, XC130710; 5, SH130723; 6, YC130820; 7, YC130828; 8, SY091027; 9, YC131115; M, Lambda ladder PFG marker (N0340s, NEB).
The PFGE patterns produced from the genomic DNA of the 12 archived strains during the attenuation with SmaI digestion are shown in Supplemental Fig. 1. The PFGE patterns of these 12 strains were very similar with each other. A few changes of patterns occurred between strains F10 and F15, but strains F15-F55 almost shared the same PFGE pattern. Based on the dendogram analysis, 2 PFGE groups (A-B) were present at 5.8% divergence (Fig. 3). Most of the strains (9/12) belonged to group A in which these 9 strains shared over 99.5% identity with each other.
Fig. 3.
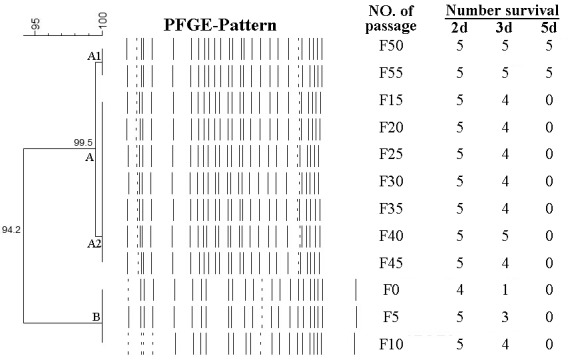
Genetic relationship among the 12 archived E. rhusiopathiae strains during the attenuation and their pathogenicity towards mice. The classification and divergence of the strains were calculated by the unweighted pair group method with averages from the PFGE results (Supplemental Fig. 1). At 5.8% divergence, 2 PFGE groups (A-B) were present. Most of the strains (9/12) were within group A in which these 9 strains shared over 99.5% identity with each other. Each mouse in different groups was injected with 0.1 ml PBS and 104 CFU strains F0-F55, respectively. Mice were observed daily to detect clinical signs of the disease for the subsequent 14 days. Only 3 groups survived for the end of the experiment after receiving subcutaneous injections of 104 CFU of strains F50 and F55 and PBS, while other groups injected with 104 CFU of strains F0-F45 died within 5 days.
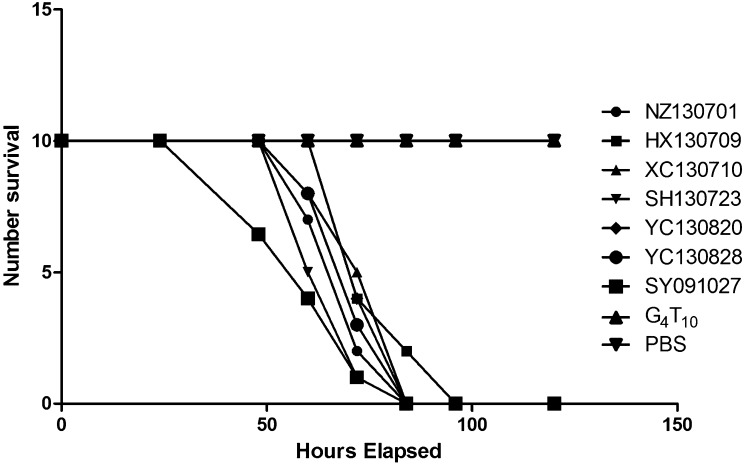
Pathogenicity towards mice: The results of the pathogenicity towards mice are shown in Figs. 3 and 4. Regardless of the acriflavine resistance, PFGE patterns or spaA types, all the 8 isolates and the early passages of the attenuated strains (F5, F10, F15, F20, F25, F30, F35, F40 and F45) showed high levels of virulence in mice. All mice died within 5 days after receiving subcutaneous injections of 104 CFU of the strains, while all those injected with the attenuated strains (G4T10, F50 and F55) and PBS survived for the end of the experiment.
Fig. 4.
Pathogenicity of the 8 E. rhusiopathiae field strains towards mice. Each mouse of the untreated control group received a subcutaneous injection of 0.1 ml PBS in the right groin and was observed every 12 hr as others. Almost every mouse injected with 104 CFU E. rhusiopathiae field strains died within 72 hr, while all mice injected with 104 vaccine strain G4T10 and PBS survived for the end of the experiment.
Antimicrobial susceptibility: Diameters of the bacteriostatic circles of the 8 isolates, vaccine strain G4T10 and the attenuated strain HX130709a (F55) against the 15 antimicrobial agents are shown in Table 5. All the strains were highly sensitive to PC-G, AMP, CEZ, CTX, EM, ROXM and AMO. Half of the isolates (HX130709, XC130710, SH 130723 and YC131115) were resistant to SM, TC, DOXY and LCM, and the other isolates (NZ130701, YC130820, YC130828 and SY091027) were moderately resistant to these antibiotics. KM and SXZ showed no activity against any of the strains.
Table 5. Antimicrobial susceptibility testing results of 8 E. rhusiopathiae field strains, the vaccine strain G4T10 and the attenuated strain HX130709a.
| Antibiotical susceptibility discs a) |
Diameter of bacteriostatic circle (mm) b) | No. of resistant strains (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| G4T10 | NZ | XC | SH | YC | YC | YC | SY | HX | HX | ||
| 130701 | 130710 | 130723 | 130820 | 130828 | 131115 | 091027 | 130709 | 130709a | |||
| PC-G | 40/S | 42/S | 42/S | 40/S | 38/S | 39/S | 42/S | 38/S | 40/S | 54/S | |
| AMO | 40/S | 39/S | 42/S | 40/S | 39/S | 40/S | 42/S | 40/S | 43/S | 55/S | |
| AMP | 36/S | 38/S | 36/S | 34/S | 38/S | 36/S | 40/S | 34/S | 36/S | 46/S | |
| CEZ | 32/S | 35/S | 35/S | 34/S | 34/S | 32/S | 36/S | 33/S | 34/S | 41/S | |
| CTX | 37/S | 36/S | 36/S | 42/S | 34/S | 34/S | 38/S | 33/S | 37/S | 47/S | |
| SM | 17/S | 12/I | 6.5/R | 6.5/R | 14/I | 14/I | 6.5/R | 12/I | 6.5/R | 23/S | 4 (40) |
| KM | 6.5/R | 6.5/R | 6.5/R | 6.5/R | 6.5/R | 6.5/R | 6.5/R | 6.5/R | 6.5/R | 6.5/R | 10 (100) |
| TC | 26/S | 30/S | 9/R | 10/R | 27/S | 27/S | 12/R | 26/S | 14/R | 39/S | 4 (40) |
| DOXY | 25/I | 30/S | 11/R | 14/R | 25/I | 26/I | 18/R | 27/I | 15/R | 36/S | 4 (40) |
| EM | 32/S | 35/S | 33/S | 34/S | 24/S | 32/S | 30/S | 35/S | 39/S | 44/S | |
| ROXM | 30/S | 31/S | 31/S | 30/S | 22/I | 30/S | 29/S | 27/S | 30/S | 42/S | |
| LCM | 36/S | 33S | 11.5/R | 14/R | 6.5/R | 32/S | 9/R | 30/S | 13/R | 42/S | 5 (50) |
| SXZ | 6.5/R | 6.5/R | 10/R | 8/R | 6.5/R | 6.5/R | 6.5/R | 6.5/R | 6.5/R | 10/R | 10 (100) |
| ERFX | 36/S | 6.5/R | 16/I | 11/R | 17/I | 19/I | 12/R | 34/S | 15/I | 10/R | 4 (40) |
| CIP | 30/S | 6.5/R | 20/I | 18/I | 18.5/I | 18.5/I | 17/I | 36/S | 18/I | 16/I | 1 (10) |
a) penicillin (PC-G), amoxicillin (AMO), ampicillin (AMP), cefazolin (CEZ), cefotaxime (CTX), streptomycin (SM), kanamycin (KM), tetracycline (TC), doxycycline (DOXY), erythromycin (EM), roxithromycin (ROXM), lincomycin (LCM), sulfisoxazole (SXZ), enrofloxacin (ERFX) and ciprofloxacin (CIP). b) The diameter of antibiotic susceptibility discs is 6.35 ± 0.50 mm. S=susceptible; I=intermediate; R=resistance.
Compared with strain HX130709, the attenuated strain HX130709a (F55) was much more sensitive to these antibiotics, except for KM, SXZ, ERFX and CIP.
DISCUSSION
Swine erysipelas used to be widely spread twenty years ago in China, which was later well controlled due to the wide use of antibiotics. In the present study, all the eight E. rhusiopathiae isolates were from cases of septicemia and were determined to be serotype 1a.
The acriflavine resistance has been used as one of the tools for discriminating the vaccine strain from field isolates because the live vaccine strain could grow in media containing at least 0.02% acriflavine, while the field strains are usually sensitive to acriflavine. Some acriflavine-resistant E. rhusiopathiae strains have been isolated from slaughter pigs affected by chronic arthritis [8, 9, 14]. It’s worth mentioning that among the eight E. rhusiopathiae strains isolated from septicemia cases in this study, 3 (37.5%) virulent isolates (SY091027, XC130710 and SH130723) showed culture growth on the BHI-T 80 agar containing 0.02% acriflavine as the vaccine strain G4T10 did.
Borrathybay et al.[6] proved that SpaA plays an important role in enhancing the virulence of E. rhusiopathiae strains. A method for discrimination between a Japanese vaccine strain and the field strains was developed based on the nucleotide sequencing of a 432-bp hypervariable region in the spaA gene [11]. In this study, spaA gene analysis showed that no mutations occurred among different passages of strain HX130709a comparing with the parental strain HX130709, which further confirmed that the virulence of serotype 1a E. rhusiopathiae strains isolated from acute swine erysipelas outbreaks in Eastern China is unrelated with some point mutations in the hypervariable region of the spaA gene.
Prevalence of Met-203 type spaA variant in E. rhusiopathiae isolates is increasing in Japan [20]. Interestingly, four of the eight isolates in our study belonged to Met-203 type, and the other four field isolates belonged to Ala-195 type (Supplemental Table 1). A further study should be carried out to investigate the prevalence of Met-203 type spaA variant in E. rhusiopathiae isolates in China.
Based on the dendogram analysis, the nine strains shared over 92.1% identity with each other, showing no correlations with spaA-type and resistance to acriflavine (Fig. 1).
Results of the drug susceptibility test showed that all the strains were highly sensitive to a variety of antibiotics, such as PC-G, AMP, CEZ, CTX, EM, ROXM and AMO, and also resistant to KM and SXZ, which well explained why swine erysipelas was easy to control with the use of antibiotics. However, nearly half of the isolates showed resistance to LCM, SM, TC, DOXY, SXZ and ERFX, because these antibiotics were widely added in the pig feeds in China [23].
With the emergence of new types of E. rhusiopathiae strains which are resistant to the widely used antibiotics, vaccines seem to be a better choice in controlling swine erysipelas. However, attenuated vaccine strains carry the risk of regaining their virulence thus, posing a hazard to susceptible swine, and the inactivated vaccines are rarely used because of the higher vaccination cost [22]. So, it is extremely urgent to develop more effective and safer subunit or gene engineering vaccines with virulence factor. However, the attenuation mechanism of E. rhusiopathiae strains remains unknown. Though spaA gene analysis showed that no mutations occurred during the attenuation, the mutations probably occurred in different important points of genome. So, all the archived strains (F5, F10, F15, F20, F25, F30, F35, F40, F45, F50 and F55) and the original strain HX130709 (F0) were analyzed using PFGE to detect whether any genome changes occurred during the attenuation. The PFGE patterns of the above 12 strains were very similar with each other (Supplemental Fig. 1). A few changes of the PFGE patterns occurred between strains F10 and F15, but strains F15-F55 shared over 99.5% identity with each other (Fig. 3). Surprisingly, though strains F15-F55 shared almost the same PFGE patterns, only strains F50 and F55 were avirulent (Fig. 3). This indicates the attenuation occurred between F46 and F50 and the mutations of SmaI Restriction Enzyme cutting site in the genome of the stains did not lead to any change of the virulence. It’s possible that the mutations in the genome which directly lead to virulence loss cannot be detected by PFGE with SmaI digestion. In this case, comparing the entire genome of wild strain and the attenuated strain may be a good way to understand the attenuation mechanism. But, there is also another possibility that the strain virulence was decreased during the attenuation mainly by phenotype changes instead of genotype changes [10].
In this study, we conclude that the acute swine erysipelas outbreaks in Eastern China were caused by serotype 1a E. rhusiopathiae strains with different biochemical characteristics, and the virulence of serotype 1a E. rhusiopathiae strains is unrelated with some point mutations in hypervariable region of the spaA gene. Meanwhile, the attenuated strain HX130709a was obtained from a highly virulent clinical strain HX130709 after 55 passages on agar plates containing acriflavine. Also, its virulence had been confirmed in mice, which lays the foundation for a further study of virulence factor of E. rhusiopathiae with proteomics and transcriptomic technologies.
Supplementary Material
Acknowledgments
This study was supported by public welfare Grants from the Ministry of Agriculture, the People’s Republic of China (201203039) and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The authors thank Dr. Tanja Opriessnig (Veterinary Diagnostic Laboratory, College of Veterinary Medicine, Iowa State University, Ames, Iowa, U.S.A.) for offering to test serovars of the E. rhusiopathiae isolates.
REFERENCES
- 1.Bender J. S., Shen H. G., Irwin C. K., Schwartz K. J., Opriessnig T.2010. Characterization of Erysipelothrix species isolates from clinically affected pigs, environmental samples, and vaccine strains from six recent swine erysipelas outbreaks in the United States. Clin. Vaccine Immunol. 17: 1605–1611. doi: 10.1128/CVI.00206-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender J. S., Irwin C. K., Shen H. G., Schwartz K. J., Opriessnig T.2011. Erysipelothrix spp. genotypes, serotypes, and surface protective antigen types associated with abattoir condemnations. J. Vet. Diagn. Invest. 23: 139–142. doi: 10.1177/104063871102300126 [DOI] [PubMed] [Google Scholar]
- 3.Brooke C. J., Riley T. V.1999. Erysipelothrix rhusiopathiae: bacteriology, epidemiology and clinical manifestations of an occupational pathogen. J. Med. Microbiol. 48: 789–799. doi: 10.1099/00222615-48-9-789 [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute (CLSI)2009. Performance standards for antimicrobial disk susceptibility tests, approved standard − 10th ed., M02-A10, CLSI, Wayne. [Google Scholar]
- 5.Eamens G. J., Forbes W. A., Djordjevic S. P.2006. Characterisation of Erysipelothrix rhusiopathiae isolates from pigs associated with vaccine breakdowns. Vet. Microbiol. 115: 329–338. doi: 10.1016/j.vetmic.2006.02.015 [DOI] [PubMed] [Google Scholar]
- 6.Borrathybay E., Gong F. J., Zhang L., Nazierbieke W.2014. Role of surface protective antigen A in the pathogenesis of Erysipelothrix rhusiopathiae strain C43065. J. Microbiol. Biotechnol. DOI: [DOI] [PubMed] [Google Scholar]
- 7.Imada Y., Goji N., Ishikawa H., Kishima M., Sekizaki T.1999. Truncated surface protective antigen (SpaA) of Erysipelothrix rhusiopathiae serotype 1a elicits protection against challenge with serotypes 1a and 2b in pigs. Infect. Immun. 67: 4376–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imada Y., Takase A., Kikuma R., Iwamaru Y., Akachi S., Hayakawa Y.2004. Serotyping of 800 strains of Erysipelothrix isolated from pigs affected with erysipelas and discrimination of attenuated live vaccine strain by genotyping. J. Clin. Microbiol. 42: 2121–2126. doi: 10.1128/JCM.42.5.2121-2126.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makino S., Yamamoto K., Murakami S., Shirahata T., Uemura K., Sawada T., Wakamoto H., Morita H.1998. Properties of repeat domain found in a novel protective antigen, SpaA, of Erysipelothrix rhusiopathiae. Microb. Pathog. 25: 101–109. doi: 10.1006/mpat.1998.0216 [DOI] [PubMed] [Google Scholar]
- 10.Muramatsu M., Sawada T., Seto K.1975. Recovery of virulence of attenuated strain Koganei by passage in broth media containing Tween 80. Annu. Rep. Natl. Vet. Assay Lab. 12: 3–8. [Google Scholar]
- 11.Nagai S., To H., Kanda A.2008. Differentiation of Erysipelothrix rhusiopathiae strains by nucleotide sequence analysis of a hypervariable region in the spaA gene: discrimination of a live vaccine strain from field isolates. J. Vet. Diagn. Invest. 20: 336–342. doi: 10.1177/104063870802000313 [DOI] [PubMed] [Google Scholar]
- 12.Okatani A. T., Uto T., Taniguchi T., Horisaka T., Horikita T., Kaneko K., Hayashidani H.2001. Pulsed-field gel electrophoresis in differentiation of erysipelothrix species strains. J. Clin. Microbiol. 39: 4032–4036. doi: 10.1128/JCM.39.11.4032-4036.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Opriessnig T., Hoffman L. J., Harris D. L., Gaul S. B., Halbur P. G.2004. Erysipelothrix rhusiopathiae: genetic characterization of midwest US isolates and live commercial vaccines using pulsed-field gel electrophoresis. J. Vet. Diagn. Invest. 16: 101–107. doi: 10.1177/104063870401600202 [DOI] [PubMed] [Google Scholar]
- 14.Ozawa M., Yamamoto K., Kojima A., Takagi M., Takahashi T.2009. Etiological and biological characteristics of Erysipelothrix rhusiopathiae isolated between 1994 and 2001 from pigs with swine erysipelas in Japan. J. Vet. Med. Sci. 71: 697–702. doi: 10.1292/jvms.71.697 [DOI] [PubMed] [Google Scholar]
- 15.Seto K., Nishimura Y., Fujiki M., Azechi H., Suzuki K.1971. Studies on acriflavine-fast attenuated Erysipelothrix insidiosa. Comparison on pathogenicity and immunogenicity between mice and pigs. Jpn. J. Vet. Sci 33: 161–171. doi: 10.1292/jvms1939.33.161 [DOI] [PubMed] [Google Scholar]
- 16.Shimoji Y., Mori Y., Fischetti V. A.1999. Immunological characterization of a protective antigen of Erysipelothrix rhusiopathiae: identification of the region responsible for protective immunity. Infect. Immun. 67: 1646–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi T., Nagamine N., Kijima M., Suzuki S., Takagi M., Tamura Y., Nakamura M., Muramatsu M., Sawada T.1996. Serovars of Erysipelothrix strains isolated from pigs affected with erysipelas in Japan. J. Vet. Med. Sci. 58: 587–589. doi: 10.1292/jvms.58.587 [DOI] [PubMed] [Google Scholar]
- 18.To H., Nagai S.2007. Genetic and antigenic diversity of the surface protective antigen proteins of Erysipelothrix rhusiopathiae. Clin. Vaccine Immunol. 14: 813–820. doi: 10.1128/CVI.00099-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.To H., Sato H., Tazumi A., Tsutsumi N., Nagai S., Iwata A., Nagano T.2012. Characterization of Erysipelothrix rhusiopathiae strains isolated from recent swine erysipelas outbreaks in Japan. J. Vet. Med. Sci. 74: 949–953. doi: 10.1292/jvms.11-0533 [DOI] [PubMed] [Google Scholar]
- 20.Uchiyama M., Yamamoto K., Ochiai M., Yamamoto T., Hirano F., Imamura S., Nagai H., Ohishi K., Horiuchi N., Kijima M.2014. Prevalence of Met-203 type spaA variant in Erysipelothrix rhusiopathiae isolates and the efficacy of swine erysipelas vaccines in Japan. Biologicals 42: 109–113. doi: 10.1016/j.biologicals.2013.12.002 [DOI] [PubMed] [Google Scholar]
- 21.Wood R. L., Harrington R., Jr1978. Serotypes of Erysipelothrix rhusiopathiae isolated from swine and from soil and manure of swine pens in the United States. Am. J. Vet. Res. 39: 1833–1840. [PubMed] [Google Scholar]
- 22.Wood R. L.1999. Erysipelas. pp. 419–430. In: Diseases of Swine, 8th ed. (Straw, B. E, D’Allaire, S., Mengeling, M. L. and Taylor, D. J. eds.), Iowa State University Press, Ames. [Google Scholar]
- 23.Yunpeng W., Yue M.2008. Potential public hazard of using antibiotics in livestock industry. Chin. J. Antibiot. 33: 519–523. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.