Abstract
Bacteria engage in contact-dependent activities to coordinate cellular activities that aid their survival. Cells of Myxococcus xanthus move over surfaces by means of type IV pili and gliding motility. Upon direct contact, cells physically exchange outer membrane (OM) lipoproteins, and this transfer can rescue motility in mutants lacking lipoproteins required for motility. The mechanism of gliding motility and its stimulation by transferred OM lipoproteins remain poorly characterized. We investigated the function of CglC, GltB, GltA and GltC, all of which are required for gliding. We demonstrate that CglC is an OM lipoprotein, GltB and GltA are integral OM β-barrel proteins, and GltC is a soluble periplasmic protein. GltB and GltA are mutually stabilizing, and both are required to stabilize GltC, whereas CglC accumulate independently of GltB, GltA and GltC. Consistently, purified GltB, GltA and GltC proteins interact in all pair-wise combinations. Using active fluorescently-tagged fusion proteins, we demonstrate that GltB, GltA and GltC are integral components of the gliding motility complex. Incorporation of GltB and GltA into this complex depends on CglC and GltC as well as on the cytoplasmic AglZ protein and the inner membrane protein AglQ, both of which are components of the gliding motility complex. Conversely, incorporation of AglZ and AglQ into the gliding motility complex depends on CglC, GltB, GltA and GltC. Remarkably, physical transfer of the OM lipoprotein CglC to a ΔcglC recipient stimulates assembly of the gliding motility complex in the recipient likely by facilitating the OM integration of GltB and GltA. These data provide evidence that the gliding motility complex in M. xanthus includes OM proteins and suggest that this complex extends from the cytoplasm across the cell envelope to the OM. These data add assembly of gliding motility complexes in M. xanthus to the growing list of contact-dependent activities in bacteria.
Author Summary
Motility facilitates a wide variety of processes such as virulence, biofilm formation and development in bacteria. Bacteria have evolved at least three mechanisms for motility on surfaces: swarming motility, twitching motility and gliding motility. Mechanistically, gliding motility is poorly understood. Here, we focused on four proteins in Myxococcus xanthus that are essential for gliding. We show that CglC is an outer membrane (OM) lipoprotein, GltB and GltA are integral OM β-barrel proteins, and GltC is a soluble periplasmic protein. GltB, GltA and GltC are components of the gliding motility complex, and CglC likely stimulates the integration of GltB and GltA into the OM. Moreover, CglC, in a cell-cell contact-dependent manner, can be transferred from a cglC + donor to a ΔcglC mutant leading to stimulation of gliding motility in the recipient. We show that upon physical transfer of CglC, CglC stimulates the assembly of the gliding motility complex in the recipient. The data presented here adds to the growing list of cell-cell contact-dependent activities in bacteria by demonstrating that gliding motility can be stimulated in a contact-dependent manner by transfer of a protein that stimulates assembly of the gliding motility complexes.
Introduction
Bacteria interact extensively within and between species to coordinate cellular activities or efficiently compete. These interactions rely on diffusible factors or on direct cell-to-cell contacts [1,2]. Contact-dependent interactions include transfer of DNA or proteins by type IV secretion systems, killing involving the delivery of toxins by the type VI secretion systems, contact-dependent growth inhibition involving two-partner secretion systems, and stimulation of motility in Myxococcus xanthus [3–5]. Here, we focused on understanding the contact-dependent mechanism underlying stimulation of gliding motility in M. xanthus.
Bacterial motility facilitates a wide variety of processes including virulence, biofilm formation and development [6]. Bacteria have at least three mechanisms for motility on surfaces. Rotating flagella propel bacterial cells to bring about translocation [7]. Type IV pili undergo cycles of extension, adhesion to a substratum and retraction to bring about motility [8]. Bacterial cells can also move over surfaces without the aid of flagella and type IV pili by a mechanism referred to as gliding motility [9]. Bacterial flagella are homologous structures present in Gram-positive as well as–negative bacteria, mechanistically fairly well-understood and consists of four parts that together span from the cytoplasm, across the cell envelope to the cell surface [7]. Type IV pili are similarly widespread homologous structures and the machinery underlying type IV pili function also spans from the cytoplasm to the cell surface [8]. Gliding motility is present in Gram-positive as well as in -negative bacteria [9]. In contrast to the homologies observed for flagella and type IV pili systems, the machineries for gliding motility are non-homologous suggesting that gliding motility has evolved independently several times [6,9] and mechanistically, gliding motility is poorly understood.
M. xanthus is a rod-shaped, Gram-negative bacterium that has two genetically distinct motility systems that allow translocation on solid surfaces in the direction of the long axis of a cell [10]. One system depends on type IV and is often referred to as S-motility [10,11]. Gliding motility, often referred to as A-motility in M. xanthus, allows movement of single cells [10]. The force for gliding is generated along the cell length [12–14]. Consistently, several proteins that are required for gliding motility have been shown using fluorescently-tagged fusion proteins to localize to clusters that are distributed along the length of the ventral side of the cell [13,15–18]. Traction force is generated at the position of these clusters supporting the notion that these clusters represent gliding motility complexes [13]. The gliding motility complexes assemble at the leading cell pole, remain stationary with respect to the substratum as a cell is moving, and disassemble as they approach the lagging cell pole. During gliding M. xanthus cells deposit slime trails of unknown composition and the motility complex have been suggested to attach to the substratum via the slime [19].
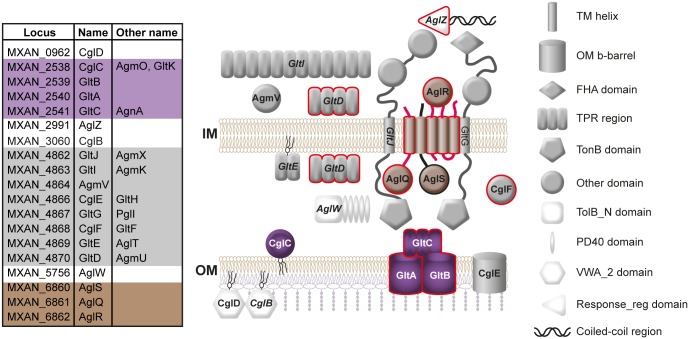
Numerous proteins involved in gliding motility have been described [13,17,18,20–22]. Genetic and cytological evidence suggests that gliding motility is driven by a protein complex that spans part or all of the cell envelope. This complex includes the AglR, AglQ and AglS proteins, which are homologs of MotA/TolQ/ExbB (AglR) and MotB/TolR/ExbD (AglQ and AglS) and form a proton channel in the inner membrane (IM) [13,18]. AglQ and AglR have been shown directly to localize to the clusters of motility complexes [13,23] (Fig 1). Additional proteins that localize to the cytoplasm, IM, periplasm or outer membrane (OM) have been suggested to be components of the gliding motility complex. These proteins include the 11 GltA-K proteins that are encoded by two gene clusters (Fig 1) and among which the eight GltA-H are paralogs of the NfsA-H proteins that are important for spore formation [17,24–27]. The NfsA-H proteins have been suggested to form a complex that spans the cell envelope and with protein localizing to the IM, periplasm and OM. Similarly, the GltA-K proteins have been suggested to form a complex that would span from the cytoplasm to the OM [17]. Several of the proteins required for gliding motility have been shown to interact based on pull down experiments [15,17] (Fig 1). However, only the cytoplasmic AglZ [16], CglF (GltF), which is reported to be a periplasmic as well as an IM protein, and GltD, which is reported to localize to the cytoplasm and periplasm, have been shown to localize to the clusters of motility complexes along the cell length [15,17] (Fig 1). GltD and AglR have also been reported to localize to a rotating helical structure [18,23]. Accordingly, two models for the gliding machinery in M. xanthus have been proposed. In both models, motility complexes are distributed along the length of the ventral side of the cell. In the focal adhesion model, the motility complex spans from the cytoplasm over the IM, periplasm, OM to the cell surface where it connects to the substratum via the slime and generate traction force [13,17,19]. In the helical rotor model, the motility complex spans the IM and periplasm and move on a rotating helix. These complexes are hypothesized to slow down close to the substratum in that way forming clusters and causing a deformation of the cell surface. These deformations, in turn, generate drag forces between the cell and the substratum [18,23]. An analysis of the biophysical properties of cell-substrate interactions during gliding, provided evidence in favor of a focal adhesion model [28]. A prediction from the focal adhesion complex model is that the gliding motility complex includes proteins that localize to the OM.
Fig 1. Subcellular localization of proteins required for gliding motility.
Synonyms for proteins required for gliding motility are indicated on the left. Proteins shown on a grey, brown or purple background in the left panel and in grey, brown or purple in the right panel are encoded together in the genome [17]; proteins in white are not encoded near other proteins shown here. Proteins outlined in red have been shown by fluorescence microscopy to localize in clusters along the cell body [here; [15–18]]; proteins that interact based on pull down experiments using M. xanthus cell extracts are indicated in italics [15]. CglB is an OM lipoprotein [29] and CglD is predicted to be an OM lipoprotein [20]. It is not known if they face towards the periplasm or are exposed on the cell surface. They are shown on the cell surface because they contain a von Willebrand domain (VWA_2), which is often involved in cell adhesion [30].
Even though several proteins that are essential for gliding motility have been predicted or directly shown to localize to the OM [17,29,31,32] (Fig 1), none of these proteins have been shown to be part of the gliding motility complex. Interestingly, several gliding motility mutants including cglC mutants can be transiently stimulated to move by gliding upon direct contact with cells that are wild-type (WT) for the corresponding genes [33]. This process, termed stimulation, depends on the contact-dependent physical transfer of OM lipoproteins between cells [29,34–36] in a process that may involve fusion of the OMs of donor and recipient [36,37]. Currently, it is not known how transfer of lipoproteins required for gliding results in stimulation of gliding in M. xanthus. Interestingly, in the type IV pili system of M. xanthus, lack of the Tgl lipoprotein results in a defect in type IV pili-dependent motility [33]. Type IV pili-dependent motility can be transiently restored in a tgl mutant by the contact-dependent physical transfer of Tgl from a tgl + donor [29,33]. In the recipient cells, transferred Tgl stimulates multimer formation by the PilQ secretin in the OM thereby allowing assembly of the type IV pili machinery [29,38,39].
Here, we identify the first OM components of the gliding machinery. We demonstrate that GltB and GltA are OM β-barrel proteins that form a subcomplex with the periplasmic GltC protein and that this subcomplex is an integral part of the gliding motility complex. Moreover, our data suggest that the OM lipoprotein CglC stimulates gliding by facilitating the OM integration of GltB and GltA. Also, our data demonstrate that assembly of the gliding motility complex depends on CglC, GltB, GltA and GltC as well as on AglZ in the cytoplasm and AglQ in the IM suggesting a non-hierarchical assembly pathway for the motility complex. Intriguingly, upon stimulation of a ΔcglC recipient by a cglC + donor, CglC instigates the assembly of the gliding motility complex in the recipient likely by facilitating the integration of GltB and GltA into the OM in the recipient. Therefore, transferred CglC directly stimulates assembly of the gliding motility complex.
Results
CglC, GltB, GltA and GltC are essential for gliding motility
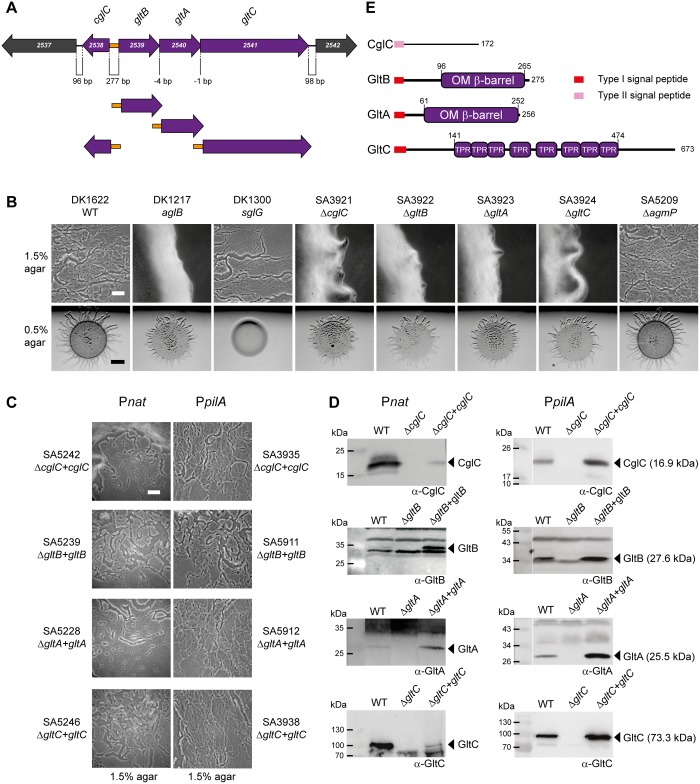
While studying the RomR response regulator that is required for full function of both motility systems in M. xanthus [40,41], we isolated miniHimar insertion mutations that caused defects in gliding motility. Two of these insertions mapped to MXAN_2540 and MXAN_2541 (Fig 2A). Previously, MXAN_2538, MXAN_2541 and MXAN_2542 were identified in transposon mutagenesis screens and suggested to be important for gliding motility [21,22]. Luciano et al. [17] showed that in-frame deletions of MXAN_2538, MXAN_2539, MXAN_2540 or MXAN_2541 caused gliding motility defects and named these genes gltK, gltB, gltA and gltC, respectively (Fig 1). Because MXAN_2538 was recently shown to be identical to cglC [20], we investigated these genes in more detail to gain insight into how CglC may stimulate gliding motility.
Fig 2. CglC, GltB, GltA and GltC are required for gliding motility.
(A) Genetic organization of cglC, gltB, gltA and gltC region. Numbers below diagram indicates distances between start and stop codons of flanking genes. Orange box indicates the intergenic region containing the divergent cglC and gltBAC promoters. The four complementation constructs with the native promoter (orange) are indicated below. (B, C) Motility phenotypes of the indicated mutants (B) and complementation strains (C). Cells were incubated at 32°C for 24 h on 0.5% and 1.5% agar to score for type IV pili-dependent and gliding motility, respectively. In-frame deletion mutants are shown in (B) and the corresponding complementation strains are shown in (C). In the rows labeled Pnat and PpilA, the complementing genes were expressed from the native promoter (marked orange in panel A) and the pilA promoter, respectively. Scale bar, 50μm on 1.5% agar and 2 mm on 0.5% agar. (D) Accumulation of CglC, GltB, GltA and GltC. Total cell lysates from exponentially growing cultures were separated by SDS-PAGE (proteins from 7×107 cells loaded per lane) and analyzed by immunoblotting using specific antibodies as indicated. In the left and right panels, the complementation strains express the relevant gene from the native and the pilA promoter, respectively. Arrowheads indicate the relevant protein with the calculated molecular mass without signal peptides in brackets. Molecular mass markers are indicated in the leftmost lane of the two panels. Note that GltC has a calculated molecular mass of 73.3 kDa but is consistently observed to run as a higher molecular mass protein by SDS-PAGE. (E) Domain structure of CglC, GltB, GltA and GltC.
To verify the results of the transposon mutagenesis screen and previously published data, in-frame deletions in each of the five genes from MXAN_2538-MXAN_2542 were generated in the fully motile WT strain DK1622. Motility assays using DK1622 and verified mutants for gliding (DK1217) and type IV pili-dependent motility (DK1300) as controls, demonstrated that WT as well as the five in-frame deletion mutants formed the rafts characteristic of type IV pili-dependent motility on 0.5% agar, which is favorable to type IV pili-dependent motility only [42], whereas DK1300 as expected did not form these rafts (Fig 2B). On 1.5% agar, which is favorable to gliding motility only, DK1622 displayed the single cells and slime trails characteristic of gliding motility at the edge of the colony. In contrast, neither the DK1217 control strain nor the mutants with in-frame deletions of MXAN_2538, MXAN_2539, MXAN_2540 or MXAN_2541 did. The in-frame deletion of MXAN_2542 did not have any effect on gliding motility. In agreement with these findings, it was recently reported using a Tn5 transposon insertion in MXAN_2542 that this gene is not essential for gliding [20]. Consistently, we found that MXAN_2539, MXAN_2540 or MXAN_2541 were transcribed as a single polycistronic mRNA that did not include MXAN_2542 using reverse transcription-PCR on total RNA isolated from exponentially growing WT cells (S1 Fig). Henceforth, the cglC, gltB, gltA and gltC nomenclature is used for MXAN_2538-_2541.
CglC, GltB, GltA and GltC bear the hallmarks indicative of cell envelope localization: CglC contains a type II lipoprotein signal peptide followed by a region that does not contain known domains (Fig 2E) in agreement with previous analyses [20]. GltB, GltA and GltC all contain a type I signal peptide. GltB and GltA are paralogs and have a domain homologous to the OM β-barrel domain of OmpA from Escherichia coli (S2 Fig) suggesting that GltB and GltA are integral OM proteins. GltC contains multiple TPR repeats that are frequently observed in proteins in multi-protein assemblies. Moreover, the GltB and GltA paralogs NfsB and NfsA are also predicted to be integral OM β-barrel proteins and localize to the OM when expressed in E. coli [25]. Also, the GltC paralog NfsC associates with the OM when expressed in E. coli [25].
To confirm that the gliding motility defects caused by the in-frame deletions of cglC, gltB, gltA and gltC were due to the lack of the corresponding protein and not a polar effect on downstream genes, we carried out complementation experiments in which cglC was cloned downstream of its native promoter and gltB, gltA and gltC separately cloned downstream of the promoter of the gltBAC operon (Fig 2A). The corresponding plasmids were integrated ectopically at the phage Mx8 attB site in the relevant mutants. The gliding defect in all four mutants was complemented by the ectopic copy of the corresponding WT gene (Fig 2B and 2C). Immunoblot analyses demonstrated that CglC and GltC accumulated at lower levels in the complementation strains than in WT (Fig 2D). In contrast, GltB and GltA accumulated at higher levels in the complementation strains than in WT despite using the same promoter for expression of gltB, gltA and gltC. In complementation strains in which cglC, gltB, gltA and gltC were expressed from the pilA promoter full complementation was also observed (Fig 2B and 2C). In these four strains, CglC and GltA accumulated at higher levels than in WT and GltB and GltC accumulated at levels similar to those in WT (Fig 2D). Thus, we conclude that CglC, GltB, GltA and GltC are required for gliding motility.
CglC, GltB, GltA and GltC localize to the cell envelope
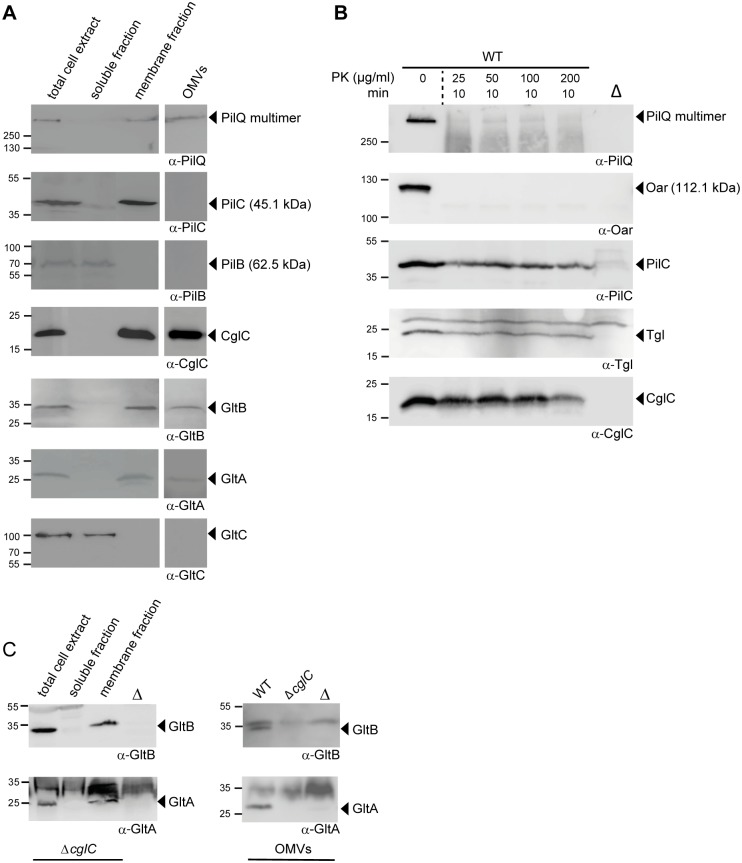
To determine the subcellular localization of CglC, GltB, GltA and GltC, fractionation experiments were performed in which total cell extract, the soluble fraction enriched for cytoplasmic and periplasmic proteins, the membrane fraction enriched for IM and OM proteins, and OM vesicles (OMVs) were isolated from exponentially growing M. xanthus cells. As shown in Fig 3A, the control proteins multimeric PilQ in the OM [39], PilC in the IM [43] and PilB in the cytoplasm [44] fractionated as expected, confirming the successful fractionation. CglC, GltB and GltA fractionated with the membrane fraction as well as with OMVs. GltC fractionated with the soluble fraction and was neither detected in the membrane fraction nor in OMVs. In combination with the sequence analyses, we conclude that CglC is an OM lipoprotein, GltB and GltA are integral OM β-barrel proteins, and GltC is a soluble periplasmic protein. These findings are in agreement with the observation that the GltB and GltA paralogs NfsB and NfsA localize to the OM when expressed in E. coli [25]. The GltC paralog NfsC also fractionates with the OM in E. coli and has been suggested to associate with the OM [25]. Importantly, GltC and NfsC only share 19%/35% identity/similarity supporting the notion that these two proteins may interact differently with the OM.
Fig 3. Subcellular localization of CglC, GltB, GltA and GltC.
(A) GltB, GltA and GltC localize to the OM and GltC is a soluble protein. Total WT cell extracts were separated into soluble and membrane fractions. Outer membrane vesicles (OMVs) were isolated from the cell free supernatant. Fractions were analyzed by immunoblotting using the indicated antibodies. Molecular mass markers are indicated to the left. The relevant proteins are indicated. (B) CglC is facing towards the periplasm. WT cells were treated with Proteinase K (PK) at the indicated concentrations for 10 min, followed by SDS-PAGE and immunoblotting. Lanes labeled Δ contain total cell extracts from the relevant in-frame deletion mutant. (C) CglC is important for OM incorporation of GltB and GltA. Left panel, total cell extracts from ΔcglC cells were separated into soluble and membrane fractions. Right panel, OMVs were isolated as in (A) from the indicated strains. All fraction were analyzed by immunoblotting using the indicated antibodies. Lanes labeled Δ contain total cell extracts from the relevant in-frame deletion mutants.
CglC faces the periplasmic side of the outer membrane
OM lipoproteins are commonly assumed to face towards the periplasm; however, several OM lipoproteins that are exposed on the cell surface have been identified [45–48]. To determine the orientation of CglC in the OM, intact WT M. xanthus cells were treated with limited amounts of Proteinase K. The OM β-barrel protein Oar [32,49,50] and the PilQ multimer in the OM were readily degraded (Fig 3B) while Tgl, which is an OM lipoprotein that faces the periplasm [51], and the IM protein PilC [43] showed little degradation in this assay. Importantly, CglC also only showed little degradation. Thus, we conclude that CglC is facing the periplasm.
GltB and GltA are mutually stabilizing and stabilize GltC
To analyze whether CglC, GltB, GltA or GltC may interact directly, we systematically determined the accumulation of each protein in the absence of each individual other protein using immunoblot analysis. CglC accumulated at WT levels in the absence of GltB, GltA or GltC and the absence of CglC did not affect the accumulation of GltB, GltA or GltC (Fig 4A and 4B). In the absence of GltB protein, GltA and GltC did not accumulate (Fig 4A and 4B). In the absence of GltA protein, GltB and GltC did not accumulate (Fig 4A and 4B). In the absence of GltC protein, GltB and GltA accumulated as in WT. These effects on protein accumulation were not caused by polar effects of the in-frame deletions because each of the in-frame deletion mutants was complemented by an ectopic copy of the relevant WT gene (Fig 2B and 2C and 2D). Additionally, the protein stability effects were not caused by the lack of gliding motility because GltB, GltA and GltC accumulated in the ΔcglC mutant. Similarly, GltB and GltA were stable in the absence of AglZ and AglQ (see below). We conclude that the two integral OM proteins GltB and GltA mutually stabilize each other as well as the periplasmic protein GltC suggesting that these three proteins interact directly. Supporting this conclusion, NfsA and NfsB are also mutually stabilizing and both are important for NfsC stability [24].
Fig 4. GltB and GltA are mutually stabilizing and stabilize GltC.
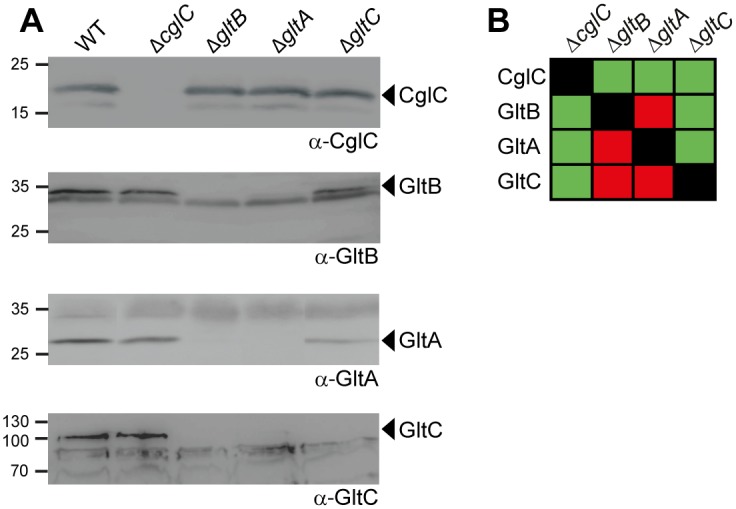
(A) GltB and GltA are mutually stabilizing and stabilize GltC. Total cell extracts from cells grown as described in Fig 2D were isolated from strains of the indicated genotypes and analyzed by immunoblotting using specific antibodies as indicated. Molecular mass markers are indicated to the left. The relevant proteins are indicated. (B) Summary of observed effects on protein stability. Green and red indicate no effect or negative effect, respectively.
In the M. xanthus type IV pili system, the OM lipoprotein Tgl functions as a pilotin to stimulate multimer formation by the PilQ secretin in the OM [29]. To assess whether the OM lipoprotein CglC is important for the OM integration of GltB and GltA, we isolated the soluble fraction, the membrane fraction and OMVs from WT and ΔcglC cells and analyzed for the subcellular localization of GltB and GltA. In the absence of CglC protein, GltB and GltA accumulated in the membrane fraction (Fig 3C, left panel); however, the accumulation of GltB and GltA in the OMVs was significantly reduced (Fig 3C, right panel). Because GltB and GltA accumulate at WT levels in the ΔcglC mutant (Fig 4A and 4B), these data strongly suggest that CglC is important for integration of GltB and GltA into the OM and that GltB and GltA accumulate in the IM in the absence of CglC.
GltB, GltA and GltC interact directly
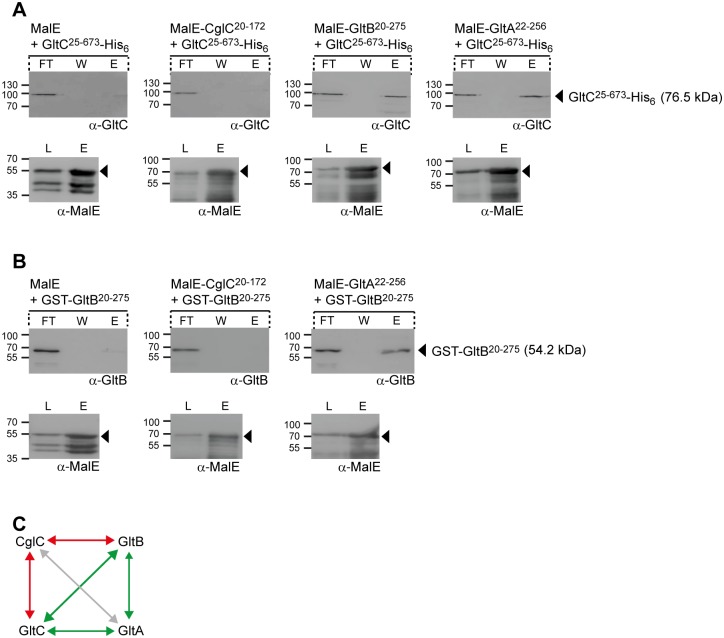
To test for direct interactions, soluble tagged variants of CglC, GltB, GltA and GltC without their signal peptides were expressed and purified under native conditions from E. coli, i.e. MalE-CglC20-172, MalE-GltB20-275, MalE-GltA22-256, GltC25-673-His6 and GST-GltB20-275. Surprisingly, the OM β-barrel proteins GltB20-275 and GltA22-256 fused to MalE and also to GST in the case of GltB20-275 were soluble whereas C-terminally His6–tagged GltB20-275 and N-terminally His6-tagged GltA20-256 were not soluble suggesting that the MalE and GST tags help to maintain GltB20-275 and GltA22-256 solubility.
To test for direct interactions, each MalE-tagged protein as well as MalE without an attached protein was incubated with an equal amount of GltC25-673-His6. Subsequently, the protein mixtures were loaded on an amylose-coupled matrix, bound proteins eluted by addition of maltose, separated by SDS-PAGE and analyzed by immunoblotting. In these experiments, GltC25-673-His6 specifically bound to the amylose matrix in the presence of MalE-GltB20-275 and MalE-GltA22-256 but not in the presence of MalE-CglC20-172 or MalE (Fig 5A and 5C).
Fig 5. GltB, GltA and GltC interact directly.
(A) GltC interacts with GltB and GltA. Purified MalE, MalE-CglC20-172, MalE-GltB20-275 or MalE-GltA22-256 were mixed with an equal amount of GltC25-673-His6, loaded on an amylose matrix, the matrix was washed and then bound proteins were eluted with 10 mM maltose. Samples before loading (L), from the flow through (FT), the washing step (W) and elution (E) were analyzed by immunoblotting using antibodies against GltC and MalE. In the upper row, arrowheads indicate GltC25-673-His6 with its calculated molecular mass and in the lower row, the individual MalE proteins. Molecular mass markers are indicated to the left. (B) GltB interacts with GltA. Purified MalE, MalE-CglC20-172 or MalE-GltA22-256 were mixed with an equal amount of GST-GltB20-275 and analyzed as described in A. Blots are marked as in A. (C) Summary of direct protein interactions. Green lines indicate interaction detected, red lines indicate interactions tested but not detected; grey line indicates interaction not tested.
In protein mixtures containing GST-GltB20-275 and MalE-CglC20-172, MalE-GltA22-256 or MalE, GST-GltB20-275 bound to the amylose matrix in the presence of MalE-GltA22-256 but not in the presence of MalE-CglC20-172 or MalE (Fig 5B). Therefore, GltB and GltA and GltC interact directly in all three pair-wise combinations whereas no interaction was observed between MalE-CglC20-172 and GltC25-673-His6 or GST-GltB20-275 (Fig 5C). These observations are in agreement with the in vivo protein stability experiments (Fig 4A and 4B). Because CglC20-172 and GltA22-256 could only be purified as soluble proteins when fused to MalE, we were unable to test for direct interactions between CglC and GltA. Our inability to detect an interaction between CglC and GltB despite the observation that CglC is important for OM integration of GltB and GltA indicate that such interactions may only be transient, that CglC interacts primarily with GltA, or this effect of CglC is indirect.
GltB, GltA and GltC are components of the gliding motility complexes
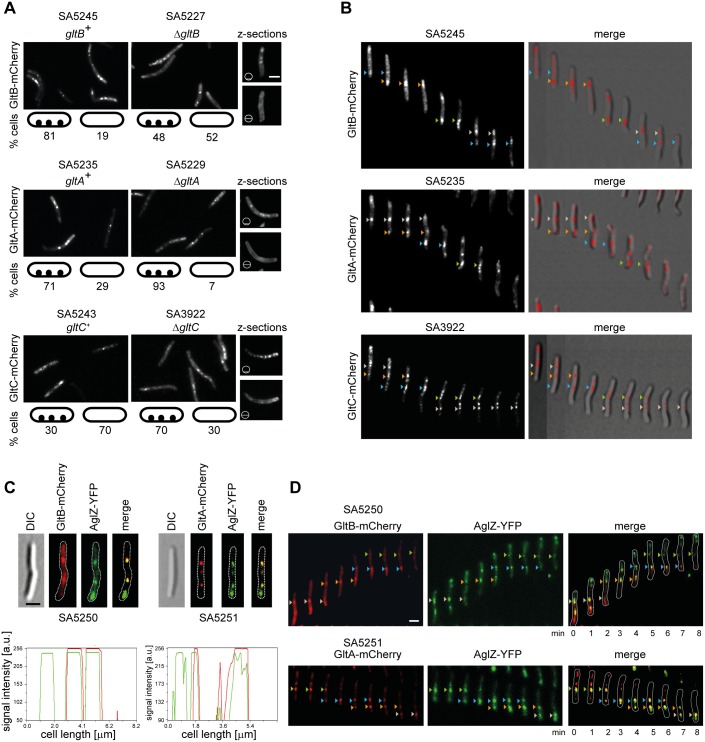
To determine if CglC, GltB, GltA and GltC are components of the gliding motility complexes, we generated fluorescent proteins in which mCherry was fused to the C-terminus of the four proteins and then expressed under the control of the native promoter from plasmids integrated at the Mx8 attB site. The ectopic expression of the relevant fusion proteins restored the motility defects in the ΔgltB, ΔgltA and ΔgltC mutants and were not negative dominant in a WT background (S3A Fig), demonstrating that the fusion proteins are active. In the case of CglC-mCherry, we observed that the full-length fusion protein was degraded and therefore this construct was not considered further. GltB-mCherry (S3B Fig) and GltA-mCherry (S3C Fig) accumulated at levels comparable to those of the native proteins whereas GltC-mCherry accumulated at reduced levels (S3D Fig). Expression of GltB-mCherry in the ΔgltB background restored accumulation of GltA and GltC (S3F and S3G Fig). Similarly, expression of GltA-mCherry in the ΔgltA background restored accumulation of GltB and GltC (S3E and S3G Fig). As opposed to the native proteins (Fig 4A and 4B), GltB-mCherry accumulated at WT levels in the ΔgltA background (S3B Fig) and GltA-mCherry accumulated at WT levels in the ΔgltB background (S3C Fig) suggesting that the mCherry tag protects GltB and GltA from proteolytic degradation in the absence of GltA and GltB, respectively.
In the WT background, GltB-mCherry localized to multiple clusters along the cell length in the majority of cells and in the ΔgltB background 48% of cells contained these clusters (Fig 6A) suggesting that GltB-mCherry is more efficiently incorporated into these clusters in the presence of native GltB. GltA-mCherry localized to multiple clusters along the cell length in the majority of cells in the WT as well as in the ΔgltA background (Fig 6A). GltC-mCherry also localized to multiple clusters along the cell length and did so more efficiently in the ΔgltC background (Fig 6A). In microscopy z-stacks of these cells, the GltB-mCherry, GltA-mCherry and GltC-mCherry clusters were only visible when the focal plane was focused close to the substratum (Fig 6A). In time-lapse fluorescence microscopy, all three proteins behaved similarly and formed fixed clusters that remained stationary with respect to the substratum as cells moved, assembled towards the leading cell pole, and disassembled towards the lagging cell pole (Fig 6B) suggesting that all three proteins are incorporated into the gliding motility complex.
Fig 6. GltB and GltA are incorporated into gliding motility complexes.
(A) Localization of GltB-mCherry, GltA-mCherry GltC-mCherry in the indicated genetic backgrounds. Cells were transferred from exponentially growing cultures to a thin agar pad on a microscope slide and imaged by fluorescence microscopy. For the z-sections, the z positions are indicated by a barred circle. The localization patterns observed are indicated in the schematics and numbers represent % of cells with that pattern; n > 100. Scale bar, 2μm. (B) Time-lapse microscopy of cells containing GltB-mCherry, GltA-mCherry or GltC-mCherry. Cells of the indicated genotypes were treated as in (A) and imaged by time-lapse DIC and fluorescence microscopy at 60 s intervals. Same colored arrowheads indicate position of motility complex during cell movement. Left panel, fluorescence microscopy images; right panel, merged DIC and fluorescence microscopy images. (C) Fluorescence microscopy images and line scans of cells expressing GltB-mCherry or GltA-mCherry and AglZ-YFP. In the line scans, red lines refer to GltB/GltA-mCherry while green lines refer to AglZ-YFP. Cells were treated as in A. Scale bar, 2μm. (D) Time-lapse microscopy of cells containing GltB-mCherry or GltA-mCherry and AglZ-YFP. Cells containing the indicated fusions were treated as in (A) and imaged by time-lapse fluorescence microscopy at 60 s intervals. Same colored triangles indicate position of motility complex during cell movement. The corresponding linescans are shown in S4 Fig.
To further establish that GltB, GltA and GltC are integral components of the gliding motility complex, we generated strains to localize GltB or GltA in parallel with AglZ, which localizes to the leading cell pole and to the clusters of gliding motility complexes [16]. As shown in Fig 6C and 6D and S4 Fig, GltB-mCherry as well as GltA-mCherry colocalized with AglZ-YFP to clusters along the cell length in both snapshot analyses and time-lapse analyses but did not colocalize with AglZ-AFP at the leading cell pole. From these analyses and because GltB, GltA and GltC interact directly, we conclude that GltB, GltA and GltC are components of the gliding motility complex.
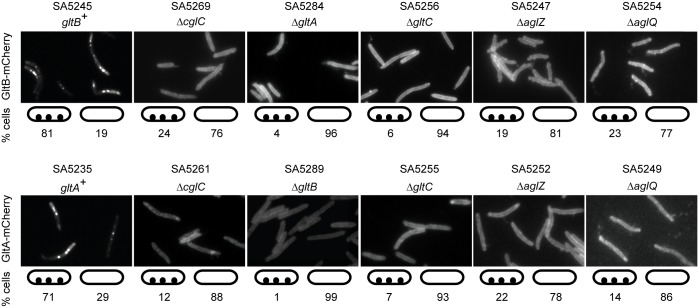
Incorporation of GltB and GltA into the gliding motility complex depends on proteins required for gliding motility
Next, we investigated the localization of GltB and GltA in the absence of other proteins required for gliding (Fig 7). Localization of GltB-mCherry to clusters along the cell length was strongly reduced in the absence of GltA, GltC, AglZ and AglQ and instead GltB-mCherry localized along the entire cell circumference suggesting that the fusion protein is homogeneously dispersed in the OM. GltA-mCherry localization to the clusters along the cell length was also dependent on GltB, GltC, AglZ and AglQ and in the absence of any one of these proteins GltA-mCherry also mostly localized along the entire cell circumference suggesting that the fusion protein is also homogeneously dispersed in the OM. GltB-mCherry and GltA-mCherry also mostly localized along the cell periphery in the absence of CglC, which our data suggest is important for OM integration of GltB and GltA, supporting the notion that in the absence of CglC, GltB and GltA are homogeneously dispersed in the IM. These observations confirm that GltB and GltA in the clusters along the cell length are incorporated into gliding motility complexes. Because native GltB and GltA (S5A Fig), as well as the GltB-mCherry and GltA-mCherry fusions (S5B Fig), accumulate as in WT in the absence of AglZ or AglQ, we conclude that incorporation of GltB and GltA, and by implication GltC, into the gliding motility complex depends on proteins that localize to the cytoplasm, IM, periplasm and OM.
Fig 7. GltB and GltA incorporation into motility complexes depends on other gliding motility proteins.
Cells were treated and analyzed as in Fig 6A. Note that all strains tested for GltB-mCherry and GltA-mCherry localization are gltB + and gltA +, respectively. Scale bar, 2μm.
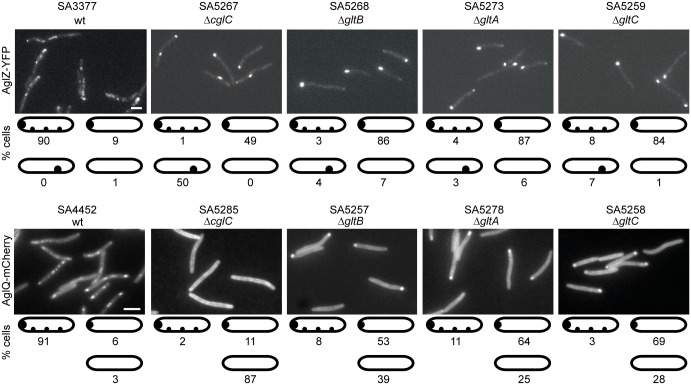
Incorporation of AglZ and AglQ into the gliding motility complex depends on CglC, GltB, GltA and GltC
To determine whether the incorporation of AglZ and AglQ into the gliding motility complex also depends on CglC, GltB, GltA and GltC, active AglZ-YFP and AglQ-mCherry fusions were expressed from their native sites in the four individual mutant backgrounds. As reported previously [16], AglZ-YFP localized to multiple clusters along the cell length in the majority of the cells as well as to a cell pole in the WT background (Fig 8). In contrast, in the four mutant backgrounds, AglZ-YFP predominantly formed a single large cluster that localized to a cell pole or somewhere along the cell length. These results were similar to those obtained with AglQ-mCherry, which also localize to multiple clusters along the cell length and a cell pole in the WT background [13] (Fig 8). In the absence of CglC, GltB, GltA or GltC, AglQ-mCherry mostly localized diffusely to the cell periphery as well as in a single cluster at a cell pole in the case of the ΔgltB, ΔgltA and ΔgltC mutants. In all four mutant backgrounds, the two fusion proteins accumulated as in WT (S6 Fig). We conclude that proteins that localize to the periplasm and OM are important for incorporation of AglZ as well as AglQ into the gliding motility complex.
Fig 8. AglZ-YFP and AglQ-mCherry incorporation into motility complexes depends on CglC, GltB, GltA and GltC.
Cells were treated and analyzed as in Fig 6A. Scale bar, 2μm.
CglC stimulates assembly of the gliding motility complex after transfer
Because CglC is important for the incorporation of GltB, GltA, AglZ, AglQ and likely also GltC into the gliding motility complexes, we hypothesized that transfer of CglC from a cglC + donor to a ΔcglC recipient instigates the assembly of intact motility complexes. To test this hypothesis, we performed stimulation assays in which the non-motile cglC + strain DK6204, which cannot be stimulated to move, served as a CglC donor as described [20] and five different ΔcglC strains served as recipients. DK6204 and the five ΔcglC strains had defects in gliding motility and had no single cells and slime trails at the colony edges (Fig 9A). Importantly, when the donor was mixed with a recipient, single cells and slime trails were readily observed at the colony edge, thus, verifying stimulation of the ΔcglC strains. As expected, GltB-mCherry, GltA-mCherry and AglQ-mCherry in the ΔcglC recipients mostly localized to the cell periphery in the absence of the donor (Fig 9B). Strikingly, all three proteins in the ΔcglC recipients in the presence of the donor strain localized in multiple clusters along the cell length in the majority of cells (Fig 9B). AglZ-YFP in the ΔcglC recipient in the absence of the donor mostly localized to a single cluster at a pole and also shifted towards formation of clusters along the cell length in the presence of the donor in the majority of cells (Fig 9B). Thus, during stimulation CglC from a donor brings about the assembly of intact and functional motility complexes in the recipient.
Fig 9. CglC stimulates formation of motility complexes after transfer.
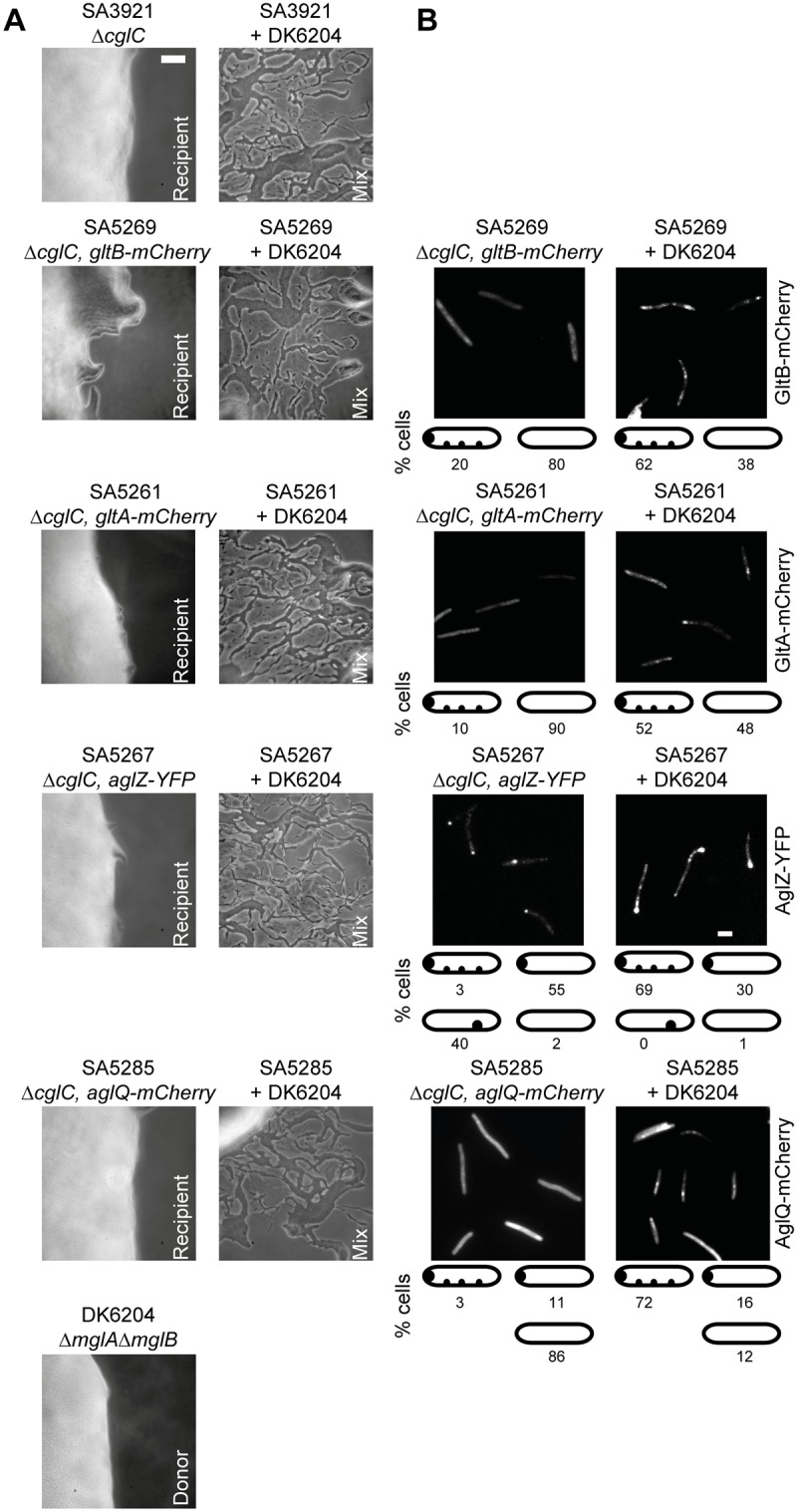
(A) Motility and stimulation phenotypes of the indicated strains and strain mixtures. Cells were incubated at 32°C for 48 h on 1.5% agar to score for gliding motility. Scale bar, 50μm. (B) Localization of GltB-mCherry, GltA-mCherry, AglZ-YFP and AglQ-mCherry in the ΔcglC mutant background incubated alone or in the presence of the non-motile CglC donor DK6204. Recipient and donor were mixed in a 1:1 ratio and incubated as described in (A), transferred to a thin agar pad on a microscope slide and imaged by fluorescence microscopy. Scale bar, 2μm.
Discussion
The machinery in M. xanthus that generates gliding motility has been hypothesized to span parts of the cell envelope or even the entire cell envelope. We report the functional characterization of the four proteins CglC, GltB, GltA and GltC all of which are required for gliding motility. We show that CglC is an OM lipoprotein that is facing towards the periplasm, GltB and GltA are integral OM β-barrel proteins, and GltC is a soluble periplasmic protein. The gliding motility machinery in M. xanthus is localized in clusters along the cell length. The localization of proteins required for gliding motility to such clusters is used as a readout for their incorporation into the motility machinery. We show that GltB, GltA and GltC localize to clusters along the cell length demonstrating that they are components of the gliding motility machinery. This conclusion is corroborated by several lines of evidence. First, GltB, GltA and GltC interact directly in all pair-wise combinations. Second, GltB and GltA colocalize with AglZ. Third, the localization of GltB and GltA to these clusters, in addition to being mutually dependent, was not only dependent on CglC and GltC but also on AglQ and AglZ that are bona fide components of the gliding motility machinery. Conversely, localization of AglZ and AglQ to these clusters, and therefore their incorporation into the gliding motility complex, was also dependent on CglC, GltB, GltA and GltC. In total, we conclude that GltB, GltA and GltC are integral components of the gliding motility machinery.
Previously identified components of the gliding motility machinery localize to the cytoplasm, IM and periplasm [13,15–18,23] (Fig 1). The identification of GltB and GltA as OM components of the gliding motility complex has implications for the understanding of the structure of this complex. The data presented are consistent with a focal adhesion model where the GltB/GltA/GltC subcomplex connects to proteins in the periplasm, IM and cytoplasm. Therefore, we assert that the gliding motility complex spans from the cytoplasm, over the IM and periplasm to the OM similarly to the machineries that drive flagella rotation and type IV pili extension/retraction. Notably, the motility complexes after assembly at the leading cell pole remain stationary with respect to the substratum as cells glide, therefore, this structural model of the gliding motility complex implies that these complexes “move through the peptidoglycan” [28]. None of the many proteins that are required for gliding motility contain peptidoglycan hydrolyzing or synthesizing domains. So, it remains an open question how the motility complexes “move through the peptidoglycan”. Similarly, it remains to be shown how the GltB/GltA/GltC subcomplex connect to the remaining proteins of the gliding machinery. However, several proteins including GltG and GltJ required for gliding motility have large periplasmic regions that could connect to the GltB/GltA/GltC subcomplex (Fig 1).
GltB and GltA mutually stabilize each other as well as GltC. Thus, if the GltB/GltA complex does not form, then these proteins and GltC become unstable. In the absence of AglZ or AglQ, GltB and GltA are not incorporated into the gliding motility complex, however, both proteins accumulate. The stability of GltB and GltA in the absence of AglZ or AglQ implies that GltB, GltA and GltC interact directly to form a subcomplex in the OM and periplasm under these conditions. In the absence of AglZ or AglQ, this subcomplex is dispersedly localized throughout the OM/periplasm and in the presence of AglQ and AglZ, this subcomplex is incorporated into the gliding motility complex. In the absence of GltB, GltA or GltC as well as in the absence of proteins predicted to localize to the cytoplasm, IM or periplasm [15,17] AglZ and AglQ mostly localize to a single cell pole suggesting that these two proteins engage in the formation of polarly localized subcomplexes in the absence of other components of the gliding machinery. Taken together these observations indicate that each component regardless of its cytoplasmic, IM, periplasmic or OM localization is crucial for the assembly of the gliding motility complex. This assembly may proceed via the connection of subcomplexes that are otherwise localized dispersedly to the OM/periplasm and IM/cytoplasm. This is in contrast to the assembly pathways of other motility complexes, where the assembly process essentially proceeds in a linear inside-out manner in the case of flagella systems [7] and in a linear outside-in manner in the case of type IV pili [38,52].
The role of CglC as an integral part of the gliding machinery remains unclear. In the case of the type IV pili machinery in M. xanthus, the OM lipoprotein Tgl functions as a pilotin and is important for assembly of the multimeric form of the PilQ secretin in the OM [29]. Because multimeric PilQ in the OM functions as an assembly platform for the remaining type IV pili machinery, lack of Tgl indirectly interferes with assembly of this entire machinery [38]. Similarly, the data presented here suggest that the sole function of CglC may be to facilitate the OM integration of GltB and GltA thereby allowing the assembly of the entire gliding motility machinery.
In 1977 Hodgkin and Kaiser observed that several gliding motility mutants including cglC mutants could be transiently stimulated to glide upon contact with a donor that was WT for the mutant gene [33] in a process that depends on the transfer of OM lipoproteins from the donor to the recipient [29]. A type II signal sequence that targets a protein to the OM is necessary and sufficient for transfer of that protein between cells [34,36]. The function of the transferred proteins and how this transfer results in stimulation of gliding has remained enigmatic. Here, we showed that in a ΔcglC mutant, the motility complexes do not assemble. However, upon stimulation, GltB, GltA, AglZ and AglQ localized to clusters along the cell length demonstrating that after stimulation, the gliding motility complexes assemble. Because our data suggest that CglC facilitates the OM incorporation of GltB and GltA, we propose that CglC provided from a donor to a recipient facilitates the OM incorporation of GltB and GltA in the recipient, thereby, allowing the remaining components of the machinery to assemble into functional gliding motility complexes. This is in striking analogy to the type IV pili system of M. xanthus in which the Tgl lipoprotein is transferred from a donor to a recipient and stimulates multimer formation of the PilQ secretin in the OM thereby allowing assembly of the type IV pili machinery [29,33,38].
The direct cell contacts required for OM lipoprotein transfer in M. xanthus depends on the cell surface receptor TraA [35,53]. TraA is highly polymorphic and transfer of OM lipoproteins depends on identical TraA proteins present in donor and recipient. Thus, the TraA polymorphism confers upon M. xanthus cells the ability to discriminate between self and non-self ensuring that OM lipoproteins are only shared with kin [53]. Recently, the molecular mechanisms underlying cell-cell contact-dependent activities in bacteria have started to be unraveled. The data presented here adds to this list by demonstrating that gliding motility can be stimulated in a contact-dependent manner by transfer of a protein that stimulates assembly of the gliding motility complexes.
Materials and Methods
Bacterial strains and cell growth
DK1622 was used as the WT M. xanthus strain and all M. xanthus strains used are derivatives of DK1622. M. xanthus strains used are listed in Table 1. Plasmids are listed in S1 Table. All plasmids were verified by sequencing. M. xanthus cells were grown at 32°C in 1% CTT broth [33] and on CTT agar plates supplemented with 1.5% agar. Kanamycin (50 μg/ml) or oxytetracycline (10 μg/ml) was added when appropriate. Plasmids were introduced into M. xanthus by electroporation. Site-specific integration of plasmids at the Mx8 attB site on the chromosome was confirmed by PCR. In frame deletions and gene replacements were generated as described [54,55].
Table 1. M. xanthus strains used in this work 1 .
| Strain | Relevant characteristics | Reference |
|---|---|---|
| DK1622 | Wild type | [70] |
| DK1217 | aglB | [71] |
| DK1300 | sglG1 | [71] |
| DK8615 | ΔpilQ | [72] |
| DK10417 | ΔpilC | [73] |
| DK10405 | Δtgl::Tc | [39] |
| SA2273 | Pnat-romR-GFP (pSH1208) | [62] |
| MxH2265 | ΔaglZ | [74] |
| SA3377 | aglZ-yfp (pSL65*) | [75] |
| SA5293 | ΔaglQ | This work |
| SA5309 | Pnat-romR-GFP (pSH1208), MXAN_2541::miniHimarΩ3908 | This work |
| SA5347 | Pnat-romR-GFP (pSH1208), MXAN_2540::miniHimarΩ3927 | This work |
| SA3921 | ΔcglC | This work |
| SA3922 | ΔgltB | This work |
| SA3923 | ΔgltA | This work |
| SA3924 | ΔgltC | This work |
| SA5544 | Δoar | This work |
| SA3935 | ΔcglC, PpilA-cglC (pDK110) | This work |
| SA5911 | ΔgltB, PpilA-gltB (pDK111) | This work |
| SA5912 | ΔgltA, PpilA-gltA (pDK112) | This work |
| SA3938 | ΔgltC, PpilA-gltC (pDK113) | This work |
| SA4452 | aglQ-mCherry | This work |
| SA5209 | ΔMXAN_2542 | This work |
| SA5242 | ΔcglC, Pnat-cglC (pBJA31) | This work |
| SA5239 | ΔgltB, Pnat-gltB (pBJA32) | This work |
| SA5228 | ΔgltA, Pnat-gltA (pBJA33) | This work |
| SA5246 | ΔgltC, Pnat-gltC (pBJA34) | This work |
| SA5245 | Pnat-gltB-mCherry (pBJA35) | This work |
| SA5269 | ΔcglC, Pnat-gltB-mCherry (pBJA35) | This work |
| SA5227 | ΔgltB, Pnat-gltB-mCherry (pBJA35) | This work |
| SA5284 | ΔgltA, Pnat-gltB-mCherry (pBJA35) | This work |
| SA5256 | ΔgltC, Pnat-gltB-mCherry (pBJA35) | This work |
| SA5235 | Pnat-gltA-mCherry (pBJA36) | This work |
| SA5261 | ΔcglC, Pnat-gltA-mCherry (pBJA36) | This work |
| SA5289 | ΔgltB, Pnat-gltA-mCherry (pBJA36) | This work |
| SA5229 | ΔgltA, Pnat-gltA-mCherry (pBJA36) | This work |
| SA5255 | ΔgltC, Pnat-gltA-mCherry (pBJA36) | This work |
| SA5247 | ΔaglZ, Pnat-gltB-mCherry (pBJA35) | This work |
| SA5254 | ΔaglQ, Pnat-gltB-mCherry (pBJA35) | This work |
| SA5252 | ΔaglZ, Pnat-gltA-mCherry (pBJA36) | This work |
| SA5249 | ΔaglQ, Pnat-gltA-mCherry (pBJA36) | This work |
| SA5243 | Pnat-gltC-mCherry (pBJA37) | This work |
| SA5244 | ΔgltC, Pnat-gltC-mCherry (pBJA37) | This work |
| SA5267 | ΔcglC, aglZ-yfp (pSL65*) | This work |
| SA5268 | ΔgltB, aglZ-yfp (pSL65*) | This work |
| SA5273 | ΔgltA, aglZ-yfp (pSL65*) | This work |
| SA5259 | ΔgltC, aglZ-yfp (pSL65*) | This work |
| SA5285 | ΔcglC, aglQ-mCherry | This work |
| SA5257 | ΔgltB, aglQ-mCherry | This work |
| SA5278 | ΔgltA, aglQ-mCherry | This work |
| SA5258 | ΔgltC,aglQ-mCherry | This work |
| SA5250 | Pnat-gltB-mCherry, aglZ-yfp (pBJA35, pSL65*) | This work |
| SA5251 | Pnat-gltA-mCherry, aglZ-yfp (pBJA36, pSL65*) | This work |
1 Plasmids mentioned in parentheses were integrated at the chromosomal Mx8 attB site with the exception of those marked with an asterisk (*), which were integrated at the native site by a single homologous recombination event. Constructs were transcribed from the native promoter (Pnat) or the pilA promoter (PpilA) as indicated.
Transposon mutagenesis
The transposon miniHimar(Kan) on the plasmid pMiniHimar, which is a non-replicating plasmid in M. xanthus (X. Duan and H.B. Kaplan, personal communication), was introduced into the strain SA2273 by electroporation. Transformants were selected on the basis of their resistance to kanamycin and individually transferred to plates with 0.5% CTT supplemented with 1.5% agar and 50 μg kanamycin/ml and incubated at 32°C to test for gliding motility defects. After 24 hrs, mutant strains were scored for motility defects. A total of 15,000 transformants were isolated and screened for gliding motility defects. Among these, 36 were deficient in gliding motility. The transposon insertion sites were identified by arbitrary PCR with subsequent sequencing as described [56,57]. Sequences were examined against the M. xanthus genome using BLASTn [58] to identify insertion sites. The entire collection of mutants will be described elsewhere.
Motility assays
Cells from exponentially growing cultures were concentrated to density of 5 × 109 cells/ml in TPM buffer (10 mM Tris-HC1 pH 7.5, 1 mM KH2PO4, 8 mM MgSO4). 5 μl of the cell suspensions were spotted on 0.5% CTT plates containing 0.5% agar or 1.5% agar [42] and incubated at 32°C for 24 h. Colony edges were visualized with a Leica MZ75 stereomicroscope equipped with a Leica DFC280 camera or a Leica DM6000B microscope equipped with a Cascade II camera.
Operon mapping
Total RNA was isolated from exponentially growing M. xanthus DK1622 cells in 1.0% CTT using a hot phenol extraction method [59]. Subsequently, RNA was treated with DNase I (Ambion) and purified with the RNeasy kit (Qiagen). cDNA was synthesized from 1.0 μg total RNA using the High capacity cDNA Archive kit (Applied Biosystems) and random hexamer primers. Total RNA and genomic DNA was used as negative and positive controls, respectively. SYBR Green PCR Master Mix (Applied Biosystems) was added to cDNA from the reverse transcription of 30 ng RNA together with 100 nM each of the two primers. The RT-PCR reaction was performed on a Mastercycler personal (Eppendorf).
Stimulation assay
Cells were treated as described for motility assay. Donor and recipient cell suspensions were spotted separately as well as in a 1:1 mixture on 0.5% CTT plates containing 1.5% agar and incubated at 32°C for 48 h. Colony edges were imaged as described for motility assays after 48 hrs of incubation. To visualize AglZ-YFP and AglQ-mCherry, cells from the edges of the colonies were harvested and resuspended in 30 μl CTT, spotted on 1.5% agar pads supplemented with TPM, covered with a cover slip and imaged as described for fluorescence microscopy.
Fractionation experiments
Cells were fractionated as described [60]. Briefly, cells were grown exponentially to a density of 5 × 108 cells/ml. Cells were harvested by centrifugation at 8000×g at RT for 10 min and resuspended in 50 mM Tris-HCl pH 7.6 containing “complete Protease inhibitor cocktail” (Roche). Cells were disrupted by sonication and samples were centrifuged at 3000×g at 4°C for 10 min to remove cell debris. Subsequently, the supernatant was centrifuged at 45.000×g at 4°C for 30 min to separate soluble and insoluble (membrane-associated) components. The resulting supernatant is enriched in soluble cytoplasmic and periplasmic proteins. The pellet containing a crude envelope fraction was washed with 50 mM Tris-HCl, resuspended in 50 mM Tris-HCl pH 7.2, 2% Triton X-100, 10 mM MgCl2 and incubated overnight with gentle shaking at 4°C and then subjected to centrifugation at 45.000×g for 30 min at 4°C. The resulting supernatant is enriched in membrane proteins. OMVs were isolated as described [32]. Briefly, culture supernatants were passed through a 0.2μm vacuum filter (Millipore). The resulting filtrate was centrifuged at 150000×g for 2 h at 4°C to recover membrane vesicles. The supernatant was carefully removed and the vesicle pellet was resuspended in 50 mM Tris-HCl pH 8.0 and centrifuged at 150000×g for 2 h at 4°C to concentrate vesicles.
Protease accessibility experiments
1 ml of exponentially growing M. xanthus cells in CTT were directly incubated with Proteinase K (Sigma) with gentle shaking at 32°C for 10 min at the indicated concentrations. 100 μl of solution 1 (1 tablet “complete mini-protease inhibitor cocktail” (Roche) dissolved in 1 ml CTT) was added to stop the reaction. The cell suspension was centrifuged at 16.000×g at 4°C for 5 min. The cell pellet was washed with 1 ml of solution 2 (1 tablet “complete mini-protease inhibitor cocktail” dissolved in 10ml CTT) and resuspended in solution 1 to a calculated density of 1×1010 cells/ml. Samples were incubated at 100°C for 15 min and mixed with SDS loading buffer containing protease inhibitors (1 tablet “Complete mini-protease inhibitor cocktail” dissolved in 1ml SDS lysis buffer) to a density of 5×109 cells/ml.
Immunoblotting
Cells from exponentially growing cultures were concentrated and resuspended in SDS lysis buffer to a density of 2.5 × 109 cells/ml. Proteins from the same number of cells were loaded per lane. Immunoblots were performed using standard procedures [61] with polyclonal rabbit α-CglC, α-GltB, α-GltA, α-GltC, α-PilB [44], α-PilC [43], α-PilQ [43], α-Oar, α-MalE (New England Biolabs), α-mCherry (Roche) and secondary anti-rabbit immunoglobulin G peroxidase conjugate (Sigma). For YFP-tagged protein detection, monoclonal anti-GFP mouse antibodies (Roche) and peroxidase-conjugated rabbit anti-mouse immunoglobulin G secondary antibodies (DakoCytomation) were used. Immunoblots were developed using Luminata Western HRP Substrate (Merck Millipore). The α-CglC, α-GltB, α-GltA, α-GltC antibodies were generated against purified His6-tagged proteins (see below). α-Oar antibodies were generated against the Oar peptides AB1 (237GTLEGTRKGIREEGT) and AB2 (1009SVDGDVNKNFKNPLS). Oar has a length of 1037 amino acid residues without the signal peptide.
Protein purification
To purify GltC25-673-His6, MalE-CglC20-172, MalE-GltB20-275, MalE-GltA22-256 and GST-GltB20-275 the plasmids pBJA9, pBJA26, pBJA27, pBJA28, pBJA29 were introduced into E. coli Rosetta 2 [F-ompT hsdSB(rB-mB-) gal dcm pRARE2] (Novagen). Cultures were grown in LB medium at 37°C to OD600 = 0.7. Protein expression was induced by addition of IPTG to 0.1mM final concentration. Proteins were expressed overnight at 18°C. Cells were harvested by centrifugation at 9.000×g for 10 min at RT and resuspended in lysis buffer containing “EDTA-free complete protease inhibitor cocktail” (Roche). For MalE- and GST-tagged proteins, CB1 lysis buffer (20 mM Tris-HCl pH7.4, 200 mM NaCl, 1 mM DTT, 1 mM EDTA) was used while for His6-tagged proteins the lysis buffer was (50 mM NaH2PO4, 300 mM NaCl pH 7.5, 1 mM DTT). Harvested cells were disrupted by sonication and centrifuged at 20.500×g for 30 min at 4°C to remove debris.
MalE-tagged proteins were purified on amylose matrix column (New England Biolabs) and eluted with CB1 buffer supplemented with 10 mM maltose. GltC25-673-His6 was purified on Ni2+-NTA-agarose column as recommended by the manufacturer (Qiagen) and eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0). GST-GltB20-275 was purified on GST-bind column (Novagen) and eluted with CB1 buffer supplemented with 10 mM reduced glutathione.
His6-CglC20-172, GltB20-275-His6 and His6-GltA22-256 were purified from inclusion bodies. The plasmids pBJA1, pBJA10, pBJA3 were introduced into E. coli Rosetta 2. Cultures were grown as described. Cells were resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl pH 7.5, 1 mM DTT) containing “EDTA-free complete Protease inhibitor cocktail” (Roche), sonicated and harvested as described. The cell pellet was resuspended in buffer B (100 mM NaH2PO4, 10 mM TrisHCl, 8 M urea, pH 8.0) and incubated overnight at 4°C. The mixture was centrifuged at 20.500×g for 30 min at 4°C to remove cell debris. Proteins were purified on Ni2+-NTA-agarose column as described by the manufacturer (Qiagen). The column was washed with buffer C (buffer B; pH 6.3) and proteins were eluted with buffer D (buffer B, pH 5.9) and buffer E (buffer B, pH 4.5). The purified proteins were dialyzed against buffer B containing 10% glycerol, frozen in liquid nitrogen, stored at -80°C and used for polyclonal rabbit antibody generation.
Pull-down experiments
Purified proteins were dialyzed against CB1 buffer. Protein concentration was measured by using a Bradford assay (BioRad). In pull-down experiments, 300 μg of each protein were mixed in a total volume of 1 ml of buffer CB1 and incubated at 4°C for 1 h. Subsequently, these mixtures were incubated with an amylose matrix (New England Biolabs) for 2 h at 4°C, followed by a washing step with 10-times the matrix volume of buffer CB1. Proteins were eluted with buffer CB1 supplemented with 10 mM maltose.
Fluorescence microscopy
DIC and fluorescence microscopy was performed as described [62]. Briefly, cells from exponentially growing cultures were spotted on 1.5% agar pads supplemented with TPM or A50 (10 mM MOPS pH 7.2, 1 mM CaCl2, 1 mM MgCl2, 50 mM NaCl, 1.5% agar) and covered with a cover slip. Cells were incubated at RT for 15 min to attach to the agar surface. Microscopy was performed using a Leica DMI6000B microscope with an adaptive focus control, a motorized stage, a temperature-controlled stage and a Hamamatsu Flash 4.0 camera. Images were recorded with Leica MM AF software package and processed with Metamorph (Molecular Devices).
Sequence analysis of gliding machinery components
We performed sequence analysis of all of the gliding machinery components included in Fig 1. Type I and type II signal peptides were identified using the SignalP [63] and LipoP [64] webservers, respectively. Trans-membrane α-helices were identified using the DAS webserver [65]. OM β-barrel domains were identified using HHomp [66]. The HMMER3 software package [67] was used in conjunction with the Pfam26 domain library [68] for domain architecture analysis with default gathering thresholds. In the event of domain overlaps, the highest scoring domain model was chosen for the final domain architecture.
Supporting Information
DNA fragments labeled 1, 3, 5 or 7 cover intergenic regions while DNA fragments labeled 2, 4, 6 or 8 are fragments internal to genes. Numbers indicated above each lane correspond to that particular DNA fragment. Genomic DNA, total RNA and cDNA were used as templates in the indicated reactions.
(TIF)
The primary amino acid sequences of GltB and GltA without their signal peptide was analyzed using the software BOCTOPUS [69] to predict the topology of the β-strands in the β-barrel. Trans-membrane β-strands are indicated by black arrows overlapping the OM (grey). Note that the N-terminal 80 residues of GltB and 45 residues of GltA are not included.
(TIF)
(A) Gliding motility assay on 1.5% agar. Images of the colony edges were taken after 24 h incubation at 32°C. Scale bar = 50μm. (B-G) Immunoblot analysis of M. xanthus strains expressing GltB-mCherry, GltA-mCherry or GltC-mCherry under the control of native promoter at the Mx8 attB site. Bands corresponding to the fusion proteins are marked with grey triangles while native proteins are marked with black triangles.
(TIF)
Cells containing the indicated fusions were treated as in Fig 6 and imaged by time-lapse fluorescence microscopy at 60 s intervals. Same colored triangles indicate position of focal adhesion during cell movement. In the line scans, red lines refer to GltB/GltA-mCherry while green lines refer to AglZ-YFP. The upper set of line scans represent the GltB/AglZ strain and the lower set of line scans represent the GltA/AglZ strain. Note that the fluorescence images are identical to those shown in Fig 6D.
(TIF)
(A, B) Immunoblot analysis of the accumulation of GltB and GltA in ΔaglZ and ΔaglQ strains (A) and of GltB-mCherry or GltA-mCherry under the control of native promoter at the Mx8 attB site in ΔaglZ and ΔaglQ strains (B). Bands corresponding to the native proteins and fusion proteins are marked with black and grey triangles, respectively together with their calculated molecular masses.
(TIF)
Immunoblot with GFP—or mCherry primary antibodies. Bands corresponding to the fusion proteins are marked with the arrow heads together with the calculated molecular mass of the fusion proteins.
(TIF)
(DOC)
Acknowledgments
We thank Dr. Chris van der Does and Dr. Josh Shrout for many helpful discussions and Dr. Nuria Gomez Santos for constructing SA5544.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the Max Planck Society (http://www.mpg.de/) and the German Research Foundation (http://www.dfg.de/en/) to LSA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bassler BL, Losick R (2006) Bacterially speaking. Cell 125: 237–246. [DOI] [PubMed] [Google Scholar]
- 2. Konovalova A, Søgaard-Andersen L (2011) Close encounters: contact-dependent interactions in bacteria. Mol Microbiol 81: 297–301. 10.1111/j.1365-2958.2011.07711.x [DOI] [PubMed] [Google Scholar]
- 3. Hayes CS, Aoki SK, Low DA (2010) Bacterial contact-dependent delivery dystems. Annu Rev Genet 44: 71–90. 10.1146/annurev.genet.42.110807.091449 [DOI] [PubMed] [Google Scholar]
- 4. Ruhe ZC, Low DA, Hayes CS (2013) Bacterial contact-dependent growth inhibition. Trends Microbiol 21: 230–237. 10.1016/j.tim.2013.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pathak DT, Wei X, Wall D (2012) Myxobacterial tools for social interactions. Res Microbiol 163: 579–591. 10.1016/j.resmic.2012.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jarrell KF, McBride MJ (2008) The surprisingly diverse ways that prokaryotes move. Nat Rev Micro 6: 466–476. [DOI] [PubMed] [Google Scholar]
- 7. Macnab RM (2003) How bacteria assemble flagella. Ann Rev Microbiol 57: 77–100. [DOI] [PubMed] [Google Scholar]
- 8. Pelicic V (2008) Type IV pili: pluribus unum? Mol Microbiol 68: 827–837. 10.1111/j.1365-2958.2008.06197.x [DOI] [PubMed] [Google Scholar]
- 9. McBride MJ (2001) Bacterial gliding motility: Multiple mechanisms for cell movement over surfaces. Ann Rev Microbiol 55: 49–75. [DOI] [PubMed] [Google Scholar]
- 10. Hodgkin J, Kaiser D (1979) Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): Two gene systems control movement. Mol Gen Genet 171: 177–191. [Google Scholar]
- 11. Wu SS, Kaiser D (1995) Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus . Mol Microbiol 18: 547–558. [DOI] [PubMed] [Google Scholar]
- 12. Sliusarenko O, Zusman DR, Oster G (2007) The motors powering A-motility in Myxococcus xanthus are distributed along the cell body. J Bacteriol 189: 7920–7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun M, Wartel M, Cascales E, Shaevitz JW, Mignot T (2011) Motor-driven intracellular transport powers bacterial gliding motility. Proc Natl Acad Sci U S A 108: 7559–7564. 10.1073/pnas.1101101108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun H, Yang Z, Shi W (1999) Effect of cellular filamentation on adventurous and social gliding motility of Myxococcus xanthus . Proc Natl Acad Sci USA 96: 15178–15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nan BY, Mauriello EMF, Sun IH, Wong A, Zusman DR (2010) A multi-protein complex from Myxococcus xanthus required for bacterial gliding motility. Mol Microbiol 76: 1539–1554. 10.1111/j.1365-2958.2010.07184.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mignot T, Shaevitz JW, Hartzell PL, Zusman DR (2007) Evidence that focal adhesion complexes power bacterial gliding motility. Science 315: 853–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luciano J, Agrebi R, Le Gall AV, Wartel M, Fiegna F, Ducret A, et al. (2011) Emergence and modular evolution of a novel motility machinery in bacteria. PLoS Genet 7: e1002268 10.1371/journal.pgen.1002268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nan B, Chen J, Neu JC, Berry RM, Oster G, Zusman DR (2011) Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc Natl Acad Sci USA 108: 2498–2503. 10.1073/pnas.1018556108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ducret A, Valignat M-P, Mouhamar F, Mignot T, Theodoly O (2012) Wet-surface–enhanced ellipsometric contrast microscopy identifies slime as a major adhesion factor during bacterial surface motility. Proc Natl Acad Sci USA 109: 10036–10041. 10.1073/pnas.1120979109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pathak DT, Wall D (2012) Identification of the cglC, cglD, cglE, and cglF genes and their role in cell contact-dependent gliding motility in Myxococcus xanthus . J Bacteriol 194: 1940–1949. 10.1128/JB.00055-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Youderian P, Burke N, White D, Hartzell PL (2003) Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner . Mol Microbiol 49: 555–570. [DOI] [PubMed] [Google Scholar]
- 22. Yu R, Kaiser D (2007) Gliding motility and polarized slime secretion. Mol Microbiol 63: 454–467. [DOI] [PubMed] [Google Scholar]
- 23. Nan BY, Bandaria JN, Moghtaderi A, Sun IH, Yildiz A, Zusman DR (2013) Flagella stator homologs function as motors for myxobacterial gliding motility by moving in helical trajectories. Proc Natl Acad Sci USA 110: E1508–E1513. 10.1073/pnas.1219982110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Müller FD, Schink CW, Hoiczyk E, Cserti E, Higgs PI (2012) Spore formation in Myxococcus xanthus is tied to cytoskeleton functions and polysaccharide spore coat deposition. Mol Microbiol 83: 486–505. 10.1111/j.1365-2958.2011.07944.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holkenbrink C, Hoiczyk E, Kahnt J, Higgs PI (2014) Synthesis and assembly of a novel glycan layer in Myxococcus xanthus spores. J Biol Chem 289: 32364–32378. 10.1074/jbc.M114.595504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Agrebi R, Wartel M, Brochier-Armanet C, Mignot T (2015) An evolutionary link between capsular biogenesis and surface motility in bacteria. Nat Rev Micro 13: 318–326. [DOI] [PubMed] [Google Scholar]
- 27. Wartel M, Ducret A, Thutupalli S, Czerwinski F, Le Gall AV, Mauriello EM, et al. (2013) A versatile class of cell surface directional motors gives rise to gliding motility and sporulation in Myxococcus xanthus. PLoS Biol 11: e1001728 10.1371/journal.pbio.1001728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Balagam R, Litwin DB, Czerwinski F, Sun M, Kaplan HB, Shaevitz JW, et al. (2014) Myxococcus xanthus gliding motors are elastically coupled to the substrate as predicted by the focal adhesion model of gliding motility. PLoS Comput Biol 10: e1003619 10.1371/journal.pcbi.1003619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nudleman E, Wall D, Kaiser D (2005) Cell-to-cell transfer of bacterial outer membrane lipoproteins. Science 309: 125–127. [DOI] [PubMed] [Google Scholar]
- 30. Whittaker CA, Hynes RO (2002) Distribution and evolution of von Willebrand/Integrin A domains: Widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell 13: 3369–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rodriguez AM, Spormann AM (1999) Genetic and molecular analysis of cglB, a gene essential for single-cell gliding in Myxococcus xanthus . J Bacteriol 181: 4381–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kahnt J, Aguiluz K, Koch J, Treuner-Lange A, Konovalova A, Huntley S, et al. (2010) Profiling the outer membrane proteome during growth and development of the social bacterium Myxococcus xanthus by selective biotinylation and analyses of outer membrane vesicles. J Proteome Res 9: 5197–5208. 10.1021/pr1004983 [DOI] [PubMed] [Google Scholar]
- 33. Hodgkin J, Kaiser D (1977) Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus . Proc Natl Acad Sci USA 74: 2938–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wei X, Pathak DT, Wall D (2011) Heterologous protein transfer within structured myxobacteria biofilms. Mol Microbiol 81: 315–326. 10.1111/j.1365-2958.2011.07710.x [DOI] [PubMed] [Google Scholar]
- 35. Pathak DT, Wei X, Bucuvalas A, Haft DH, Gerloff DL, Wall D (2012) Cell contact-dependent outer membrane exchange in myxobacteria: genetic determinants and mechanism. PLOS Genet 8:: e1002626 10.1371/journal.pgen.1002626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ducret A, Fleuchot B, Bergam P, Mignot T (2013) Direct live imaging of cell–cell protein transfer by transient outer membrane fusion in Myxococcus xanthus . eLife 2: e00868 10.7554/eLife.00868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wei X, Vassallo CN, Pathak DT, Wall D (2014) Myxobacteria produce outer membrane-enclosed tubes in unstructured environments. J Bacteriol 196: 1807–1814. 10.1128/JB.00850-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Friedrich C, Bulyha I, Søgaard-Andersen L (2014) Outside-in assembly pathway of the type IV pilus system in Myxococcus xanthus . J Bacteriol 196: 378–390. 10.1128/JB.01094-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nudleman E, Wall D, Kaiser D (2006) Polar assembly of the type IV pilus secretin in Myxococcus xanthus . Mol Microbiol 60: 16–29. [DOI] [PubMed] [Google Scholar]
- 40. Keilberg D, Wuichet K, Drescher F, Søgaard-Andersen L (2012) A response regulator interfaces between the Frz chemosensory system and the MglA/MglB GTPase/GAP module to regulate polarity in Myxococcus xanthus . PLoS Genet 8: e1002951 10.1371/journal.pgen.1002951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang Y, Guzzo M, Ducret A, Li Y-Z, Mignot T (2012) A dynamic response regulator protein modulates G-protein–dependent polarity in the bacterium Myxococcus xanthus . PLoS Genet 8: e1002872 10.1371/journal.pgen.1002872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shi W, Zusman DR (1993) The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc Natl Acad Sci USA 90: 3378–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bulyha I, Schmidt C, Lenz P, Jakovljevic V, Höne A, Maier B, et al. (2009) Regulation of the type IV pili molecular machine by dynamic localization of two motor proteins. Mol Microbiol 74: 691–706. 10.1111/j.1365-2958.2009.06891.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jakovljevic V, Leonardy S, Hoppert M, Søgaard-Andersen L (2008) PilB and PilT are ATPases acting antagonistically in type IV pilus function in Myxococcus xanthus . J Bacteriol 190: 2411–2421. 10.1128/JB.01793-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dashper SG, Hendtlass A, Slakeski N, Jackson C, Cross KJ, Brownfield L, et al. (2000) Characterization of a novel outer membrane hemin-binding protein of Porphyromonas gingivalis . J Bacteriol 182: 6456–6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jin S, Joe A, Lynett J, Hani EK, Sherman P, Chan VL (2001) JlpA, a novel surface-exposed lipoprotein specific to Campylobacter jejuni, mediates adherence to host epithelial cells. Mol Microbiol 39: 1225–1236. [DOI] [PubMed] [Google Scholar]
- 47. Leuzzi R, Serino L, Scarselli M, Savino S, Fontana MR, Monaci E, et al. (2005) Ng-MIP, a surface-exposed lipoprotein of Neisseria gonorrhoeae, has a peptidyl-prolyl cis/trans isomerase (PPIase) activity and is involved in persistence in macrophages. Mol Microbiol 58: 669–681. [DOI] [PubMed] [Google Scholar]
- 48. Drummelsmith J, Whitfield C (2000) Translocation of group 1 capsular polysaccharide to the surface of Escherichia coli requires a multimeric complex in the outer membrane. EMBO J 19: 57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bhat S, Zhu X, Patel RP, Orlando R, Shimkets LJ (2011) Identification and localization of Myxococcus xanthus porins and lipoproteins. PLoS ONE 6: e27475 10.1371/journal.pone.0027475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Martinez-Canamero M, Munoz-Dorado J, Farez-Vidal E, Inouye M, Inouye S (1993) Oar, a 115-Kilodalton membrane protein required for development of Myxococcus xanthus . J Bacteriol 175: 4756–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rodriguez-Soto JP, Kaiser D (1997) Identification and localization of the Tgl protein, which is required for Myxococcus xanthus social motility. J Bacteriol 179: 4372–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Siewering K, Jain S, Friedrich C, Webber-Birungi MT, Semchonok DA, Binzen I, et al. (2014) Peptidoglycan-binding protein TsaP functions in surface assembly of type IV pili. Proc Natl Acad Sci USA 111: E953–961. 10.1073/pnas.1322889111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pathak DT, Wei X, Dey A, Wall D (2013) Molecular recognition by a polymorphic cell surface receptor governs cooperative behaviors in bacteria. PLOS Genetics 9: e1003891 10.1371/journal.pgen.1003891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shi XQ, Wegener-Feldbrugge S, Huntley S, Hamann N, Hedderich R, Søgaard-Andersen L (2008) Bioinformatics and experimental analysis of proteins of two-component systems in Myxococcus xanthus . J Bacteriol 190: 613–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Konovalova A, Löbach S, Søgaard-Andersen L (2012) A RelA-dependent two-tiered regulated proteolysis cascade controls synthesis of a contact-dependent intercellular signal in Myxococcus xanthus . Mol Microbiol 84: 260–275. 10.1111/j.1365-2958.2012.08020.x [DOI] [PubMed] [Google Scholar]
- 56. Das S, Noe JC, Paik S, Kitten T (2005) An improved arbitrary primed PCR method for rapid characterization of transposon insertion sites. J Microbiol Methods 63: 89–94. [DOI] [PubMed] [Google Scholar]
- 57. Pilhofer M, Bauer AP, Schrallhammer M, Richter L, Ludwig W, Schleifer KH, et al. (2007) Characterization of bacterial operons consisting of two tubulins and a kinesin-like gene by the novel two-step gene walking method. Nucleic Acids Res 35: e135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ (1990) Basic Local Alignment Search Tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 59. Overgaard M, Wegener-Feldbrugge S, Søgaard-Andersen L (2006) The orphan response regulator DigR is required for synthesis of extracellular matrix fibrils in Myxcoccus xanthus . J Bacteriol 188: 4384–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Thomasson B, Link J, Stassinopoulos AG, Burke N, Plamann L, Hartzell PL (2002) MglA, a small GTPase, interacts with a tyrosine kinase to control type IV pili-mediated motility and development of Myxococcus xanthus . Mol Microbiol 46: 1399–1413. [DOI] [PubMed] [Google Scholar]
- 61. Sambrook J, Russel DW (2001) Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 62. Leonardy S, Freymark G, Hebener S, Ellehauge E, Sogaard-Andersen L (2007) Coupling of protein localization and cell movements by a dynamically localized response regulator in Myxococcus xanthus . EMBO J 26: 4433–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 64. Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A (2003) Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci 12: 1652–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cserzo M, Wallin E, Simon I, vonHeijne G, Elofsson A (1997) Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Engineering 10: 673–676. [DOI] [PubMed] [Google Scholar]
- 66. Remmert M, Linke D, Lupas AN, Soding J (2009) HHomp--prediction and classification of outer membrane proteins. Nucleic Acids Res 37: W446–451. 10.1093/nar/gkp325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. (2010) The Pfam protein families database. Nucleic Acids Research 38: D211–222. 10.1093/nar/gkp985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. (2012) The Pfam protein families database. Nucleic Acids Research 40: D290–301. 10.1093/nar/gkr1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hayat S, Elofsson A (2012) BOCTOPUS: improved topology prediction of transmembrane β barrel proteins. Bioinformatics 28: 516–522. 10.1093/bioinformatics/btr710 [DOI] [PubMed] [Google Scholar]
- 70. Kaiser D (1979) Social gliding is correlated with the presence of pili in Myxococcus xanthus . Proc Natl Acad Sci U S A 76: 5952–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hodgkin J, Kaiser D (1979) Genetics of gliding motility in Myxococcus xanthus (Myxobacterales)—Genes controlling movement of single cells. Molecular & General Genetics 171: 167–176. [Google Scholar]
- 72. Wall D, Kolenbrander PE, Kaiser D (1999) The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for type IV pilus biogenesis, social motility, and development. J Bacteriol 181: 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wu SS, Kaiser D (1997) Regulation of expression of the pilA gene in Myxococcus xanthus . J Bacteriol 179: 7748–7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang RF, Bartle S, Otto R, Stassinopoulos A, Rogers M, Plamann L, et al. (2004) AglZ is a filament-forming coiled-coil protein required for adventurous gliding motility of Myxococcus xanthus . J Bacteriol 186: 6168–6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Leonardy S, Miertzschke M, Bulyha I, Sperling E, Wittinghofer A, Søgaard-Andersen L (2010) Regulation of dynamic polarity switching in bacteria by a Ras-like G-protein and its cognate GAP. EMBO J 29: 2276–2289. 10.1038/emboj.2010.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA fragments labeled 1, 3, 5 or 7 cover intergenic regions while DNA fragments labeled 2, 4, 6 or 8 are fragments internal to genes. Numbers indicated above each lane correspond to that particular DNA fragment. Genomic DNA, total RNA and cDNA were used as templates in the indicated reactions.
(TIF)
The primary amino acid sequences of GltB and GltA without their signal peptide was analyzed using the software BOCTOPUS [69] to predict the topology of the β-strands in the β-barrel. Trans-membrane β-strands are indicated by black arrows overlapping the OM (grey). Note that the N-terminal 80 residues of GltB and 45 residues of GltA are not included.
(TIF)
(A) Gliding motility assay on 1.5% agar. Images of the colony edges were taken after 24 h incubation at 32°C. Scale bar = 50μm. (B-G) Immunoblot analysis of M. xanthus strains expressing GltB-mCherry, GltA-mCherry or GltC-mCherry under the control of native promoter at the Mx8 attB site. Bands corresponding to the fusion proteins are marked with grey triangles while native proteins are marked with black triangles.
(TIF)
Cells containing the indicated fusions were treated as in Fig 6 and imaged by time-lapse fluorescence microscopy at 60 s intervals. Same colored triangles indicate position of focal adhesion during cell movement. In the line scans, red lines refer to GltB/GltA-mCherry while green lines refer to AglZ-YFP. The upper set of line scans represent the GltB/AglZ strain and the lower set of line scans represent the GltA/AglZ strain. Note that the fluorescence images are identical to those shown in Fig 6D.
(TIF)
(A, B) Immunoblot analysis of the accumulation of GltB and GltA in ΔaglZ and ΔaglQ strains (A) and of GltB-mCherry or GltA-mCherry under the control of native promoter at the Mx8 attB site in ΔaglZ and ΔaglQ strains (B). Bands corresponding to the native proteins and fusion proteins are marked with black and grey triangles, respectively together with their calculated molecular masses.
(TIF)
Immunoblot with GFP—or mCherry primary antibodies. Bands corresponding to the fusion proteins are marked with the arrow heads together with the calculated molecular mass of the fusion proteins.
(TIF)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.