Abstract
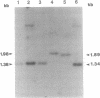
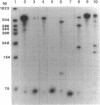
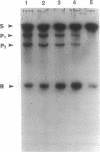
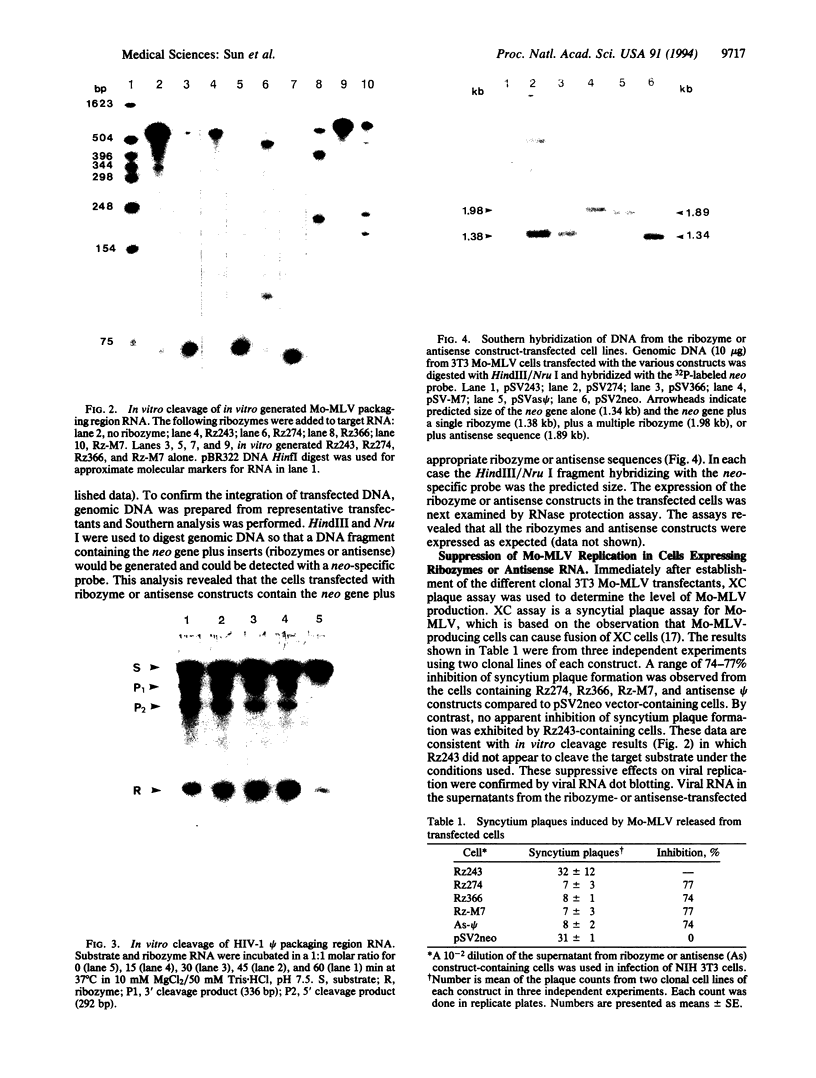
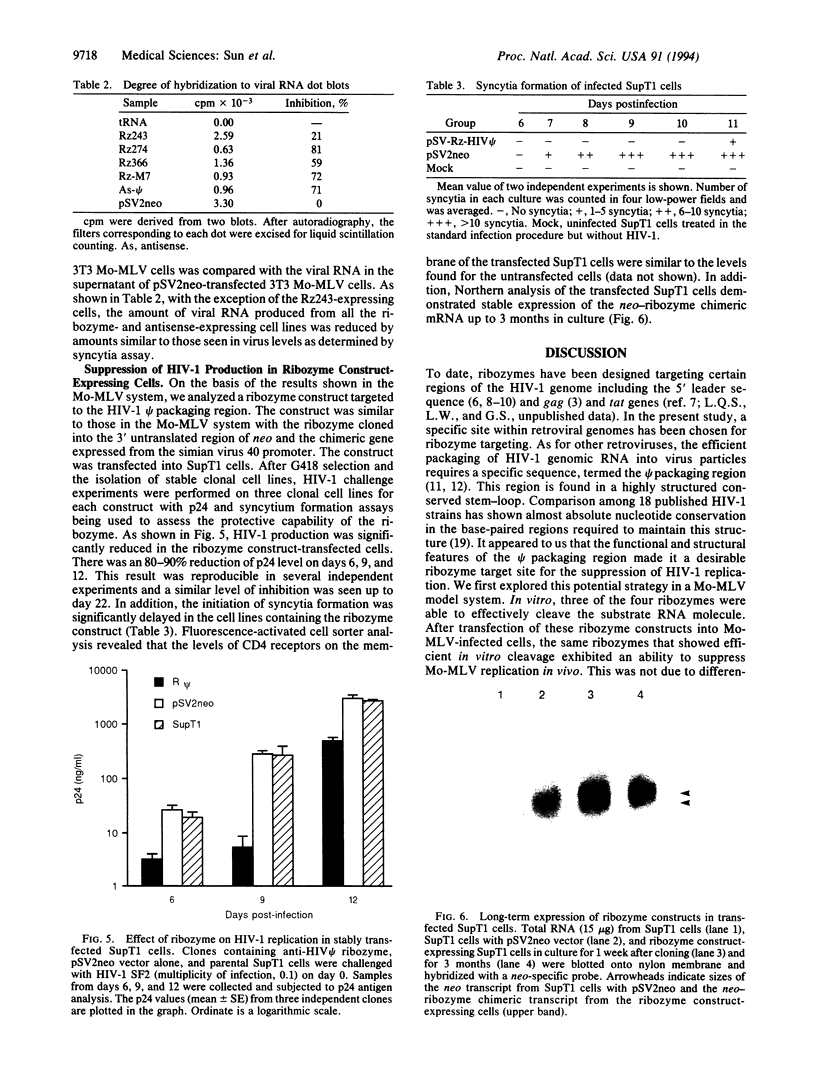
Several hammerhead ribozymes targeted to different sites within the retroviral packaging (psi) sequences of the Moloney murine leukemia virus (Mo-MLV) and the human immunodeficiency virus type 1 (HIV-1) were designed and shown to cleave target RNA in vitro at the chosen sites. The engineered ribozymes, as well as antisense sequence complementary to the Mo-MLV psi packaging region, were cloned into the 3' untranslated region of the neomycin-resistance gene (neo). This was coupled to the simian virus 40 early promoter within the pSV2neo vector. For the ribozymes against the Mo-MLV psi site, the constructs were transfected into Mo-MLV-infected and virus-producing mouse NIH 3T3 cells. With the exception of one of the single ribozymes (the one least effective in cutting target RNA in vitro), all of the constructs effectively (70-80%) suppressed retrovirus production. These results demonstrate a direct correlation between in vitro cleavage and in vivo ribozyme-mediated virus suppression. In addition, a ribozyme targeted to the HIV-1 psi packaging site was engineered into the same vector and transfected into the human T-cell line SupT1. The transfectants were cloned and then challenged with HIV-1. When compared to vector-transfected control cells, a significant reduction in HIV-1 production was observed as measured by p24 and syncytia formation assays. This study demonstrates a feasible approach to the suppression of retrovirus replication by targeting the psi packaging site with hammerhead ribozymes.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adam M. A., Miller A. D. Identification of a signal in a murine retrovirus that is sufficient for packaging of nonretroviral RNA into virions. J Virol. 1988 Oct;62(10):3802–3806. doi: 10.1128/jvi.62.10.3802-3806.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alford R. L., Honda S., Lawrence C. B., Belmont J. W. RNA secondary structure analysis of the packaging signal for Moloney murine leukemia virus. Virology. 1991 Aug;183(2):611–619. doi: 10.1016/0042-6822(91)90990-s. [DOI] [PubMed] [Google Scholar]
- Chen C. J., Banerjea A. C., Harmison G. G., Haglund K., Schubert M. Multitarget-ribozyme directed to cleave at up to nine highly conserved HIV-1 env RNA regions inhibits HIV-1 replication--potential effectiveness against most presently sequenced HIV-1 isolates. Nucleic Acids Res. 1992 Sep 11;20(17):4581–4589. doi: 10.1093/nar/20.17.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dropulić B., Lin N. H., Martin M. A., Jeang K. T. Functional characterization of a U5 ribozyme: intracellular suppression of human immunodeficiency virus type 1 expression. J Virol. 1992 Mar;66(3):1432–1441. doi: 10.1128/jvi.66.3.1432-1441.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautsch J. W., Meier H. A short-term quantitative XC assay for murine leukemia virus. Virology. 1976 Jul 15;72(2):509–513. doi: 10.1016/0042-6822(76)90179-3. [DOI] [PubMed] [Google Scholar]
- Han L., Yun J. S., Wagner T. E. Inhibition of Moloney murine leukemia virus-induced leukemia in transgenic mice expressing antisense RNA complementary to the retroviral packaging sequences. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4313–4317. doi: 10.1073/pnas.88.10.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison G. P., Lever A. M. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J Virol. 1992 Jul;66(7):4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988 Aug 18;334(6183):585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- Homann M., Tzortzakaki S., Rittner K., Sczakiel G., Tabler M. Incorporation of the catalytic domain of a hammerhead ribozyme into antisense RNA enhances its inhibitory effect on the replication of human immunodeficiency virus type 1. Nucleic Acids Res. 1993 Jun 25;21(12):2809–2814. doi: 10.1093/nar/21.12.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever A., Gottlinger H., Haseltine W., Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989 Sep;63(9):4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo K. M., Biasolo M. A., Dehni G., Palú G., Haseltine W. A. Inhibition of replication of HIV-1 by retroviral vectors expressing tat-antisense and anti-tat ribozyme RNA. Virology. 1992 Sep;190(1):176–183. doi: 10.1016/0042-6822(92)91203-7. [DOI] [PubMed] [Google Scholar]
- Mann R., Baltimore D. Varying the position of a retrovirus packaging sequence results in the encapsidation of both unspliced and spliced RNAs. J Virol. 1985 May;54(2):401–407. doi: 10.1128/jvi.54.2.401-407.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojwang J. O., Hampel A., Looney D. J., Wong-Staal F., Rappaport J. Inhibition of human immunodeficiency virus type 1 expression by a hairpin ribozyme. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10802–10806. doi: 10.1073/pnas.89.22.10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi J. J., Cantin E. M., Sarver N., Chang P. F. The potential use of catalytic RNAs in therapy of HIV infection and other diseases. Pharmacol Ther. 1991;50(2):245–254. doi: 10.1016/0163-7258(91)90016-f. [DOI] [PubMed] [Google Scholar]
- Sarver N., Cantin E. M., Chang P. S., Zaia J. A., Ladne P. A., Stephens D. A., Rossi J. J. Ribozymes as potential anti-HIV-1 therapeutic agents. Science. 1990 Mar 9;247(4947):1222–1225. doi: 10.1126/science.2107573. [DOI] [PubMed] [Google Scholar]
- Sioud M., Drlica K. Prevention of human immunodeficiency virus type 1 integrase expression in Escherichia coli by a ribozyme. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7303–7307. doi: 10.1073/pnas.88.16.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrilow D., Takayama Y., Symonds G. A rapid protocol for the introduction of large, multiple oligonucleotide-mediated insertions. Biotechniques. 1992 Jul;13(1):42–45. [PubMed] [Google Scholar]
- Weerasinghe M., Liem S. E., Asad S., Read S. E., Joshi S. Resistance to human immunodeficiency virus type 1 (HIV-1) infection in human CD4+ lymphocyte-derived cell lines conferred by using retroviral vectors expressing an HIV-1 RNA-specific ribozyme. J Virol. 1991 Oct;65(10):5531–5534. doi: 10.1128/jvi.65.10.5531-5534.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Ojwang J., Yamada O., Hampel A., Rapapport J., Looney D., Wong-Staal F. A hairpin ribozyme inhibits expression of diverse strains of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6340–6344. doi: 10.1073/pnas.90.13.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]