Abstract
Fungal pathogens have evolved antioxidant defense against reactive oxygen species produced as a part of host innate immunity. Recent studies proposed peroxidases as components of antioxidant defense system. However, the role of fungal peroxidases during interaction with host plants has not been explored at the genomic level. Here, we systematically identified peroxidase genes and analyzed their impact on fungal pathogenesis in a model plant pathogenic fungus, Magnaporthe oryzae. Phylogeny reconstruction placed 27 putative peroxidase genes into 15 clades. Expression profiles showed that majority of them are responsive to in planta condition and in vitro H2O2. Our analysis of individual deletion mutants for seven selected genes including MoPRX1 revealed that these genes contribute to fungal development and/or pathogenesis. We identified significant and positive correlations among sensitivity to H2O2, peroxidase activity and fungal pathogenicity. In-depth analysis of MoPRX1 demonstrated that it is a functional ortholog of thioredoxin peroxidase in Saccharomyces cerevisiae and is required for detoxification of the oxidative burst within host cells. Transcriptional profiling of other peroxidases in ΔMoprx1 suggested interwoven nature of the peroxidase-mediated antioxidant defense system. The results from this study provide insight into the infection strategy built on evolutionarily conserved peroxidases in the rice blast fungus.
In plant-pathogen interactions, one of the first defense responses in plants is rapid and transient production of reactive oxygen species (ROS)1,2,3. ROS generation is spatially and temporally regulated. ROS are typically produced in the apoplastic compartment within minutes following initiation of pathogenic infection4,5,6,7. ROS and intracellular redox changes can be used by pathogenic fungi for signalling purposes8 as well as development and pathogenicity9,10. Although O2– is the proximal product generated, the more stable H2O2 is the most abundant ROS. ROS can directly kill pathogens, produce cross-linked cell wall polymers to fortify physical barriers to pathogen entry, or act as a messenger in a cell signaling pathway, leading to pathogenesis-related (PR) gene expression and localized hypersensitive responses1,3,11,12,13,14.
Pathogens must effectively incapacitate production of host-driven ROS or detoxify ROS for successful infection of host cells15,16. Studies have demonstrated that H2O2 detoxification is an essential virulence determinant in fungal pathogens such as Candida albicans17, Ustilago maydis18, Alternaria alternata19,20, and Magnaporthe oryzae18,21,22,23. Furthermore, recent studies have identified a group of ROS-scavenging enzymes, peroxidases, as workhorses for the fungal antioxidant defense system24,25,26. Peroxidases (EC1.11.1.x) are enzymes that mediate electron transfer from H2O2 and organic peroxide to various electron acceptors. They are evolutionarily conserved and implicated in biological processes as diverse as immune responses and hormone regulation27,28,29,30. Examples of peroxidase enzymes include NAD(P)H oxidase, catalase, glutathione peroxidase, catalase peroxidase, ascorbate peroxidase, lignin peroxidase, and peroxiredoxin31.
Pathogenic fungi are becoming great threats to both plant and animal and are jeopardizing food security32. The hemibiotrophic fungus M. oryzae is a causal agent of the rice blast, one of the most devastating disease in cultivated rice33. This disease is estimated to destroy an amount of rice that would feed 60 millions of people annually33. The fungus is genetically tractable and can undergo infection-specific development in a laboratory setting. Since the full genome sequences of M. oryzae and rice are publicly available34, rice blast is a model system for studying plant-pathogen interactions.
The rice blast fungus experiences sequential developmental changes. The disseminated asexual spore, the conidium, attaches to the hydrophobic host surface upon hydration, and produces a cylindrical germ tube. The end of the germ tube forms a specialized dome-shaped infection structure called an appressorium. The mature heavily malanized appressorium mechanically penetrates the cuticular layer of host plants, using enormous turgor pressure (>8 Mpa)33,35. In a host plant, the fungus enters the host cytoplasm via a penetration peg, develops bulbous, infectious hyphae in the first-invaded plant cell, and, subsequently, grows into neighboring cells, presumably through the plasmodesmata36.
In M. oryzae, six peroxidase genes have been functionally characterized. Studies have provided evidence that both balancing the intracellular level of ROS and efficient removal of extracellular ROS are pivotal for early phase host infection. Genetic analysis of NADPH oxidase genes, NOX1 and NOX2, suggests that regulation of intracellular ROS is required for appressorium maturation and penetration peg formation37. The deletion mutant of a glutathione peroxidase gene, HYR1, is less tolerant to ROS and produces smaller lesions on rice plants24. The role of other putative peroxidase genes in antioxidant defense is unknown.
Mutants lacking the secreted large subunit catalase, CATB, are less pathogenic due to their involvement in strengthening the fungal cell wall rather than detoxifying host-derived H2O238. Deletion of the catalase peroxidase gene, CPXB, renders the mutant sensitive to exogenous H2O2, but it does not impair pathogenicity39. Finally, loss of the peroxiredoxin gene, TPX1, is required for pathogenicity, but not to neutralize plant-generated ROS40.
There is still much to learn about peroxidases and fungal pathogenicity. First, are specific peroxidases particularly important for pathogenicity? Second, what are the relationships among peroxidases, sensitivity to exogenous H2O2, and pathogenicity? In an attempt to answer these questions, we performed comprehensive expression and functional analyses based on reconstruction of phylogeny for 27 putative peroxidase genes in M. oryzae.
Results
Phylogenetic analysis of peroxidase genes in M. oryzae
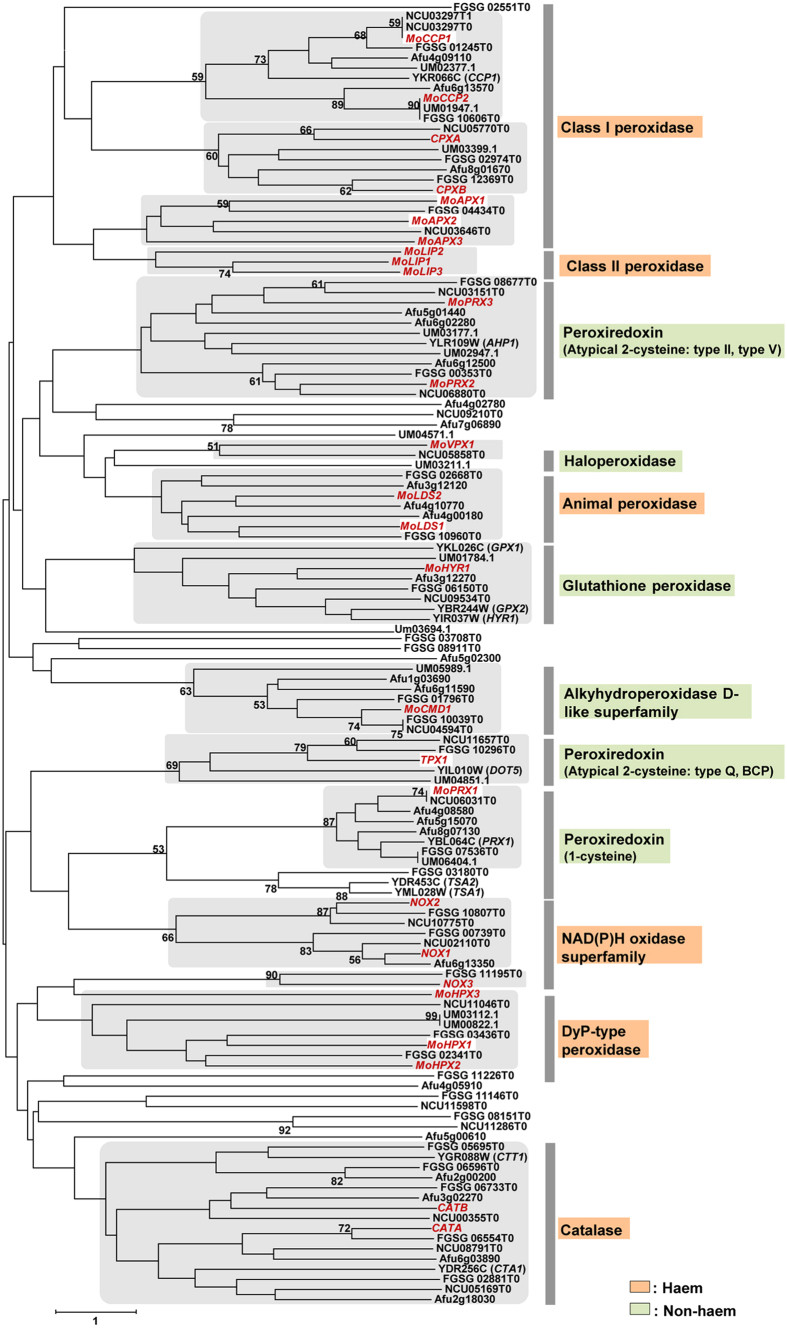
For systematic analysis of peroxidase genes in M. oryzae, we first investigated the evolutionary relationships among peroxidase genes in filamentous fungi by constructing a phylogenetic tree. Searching through the Fungal Peroxidase Database (fPoxDB; http://peroxidase.riceblast.snu.ac.kr/), a total of 896 peroxidase genes were identified from genomes of 35 species that included 29 fungi, 1 chromista, 3 metazoa, and 2 viridiplantae (Supplementary information [SI] Table S1). In M. oryzae, we found 27 putative peroxidase-encoding genes. Protein sizes ranged from 160 (MoPRX3) to 1171 (MoLDS2) amino acids (Supplementary Table S2). SignalP (http://www.cbs.dtu.dk/services/signalP/) predicted nine of them (MoAPX1, MoAPX2, CPXB, MoLIP1, MoLIP2, MoHPX1, MoHPX2, MoHPX3 and CATB) to contain signal peptides (Supplementary Table S2). Using amino acid sequences of putative peroxidases, including 27 M. oryzae peroxidases, the phylogeny of these peroxidases was reconstructed. The resulting phylogenetic tree revealed that the fungal peroxidases are not explicitly divided into distinct subgroups, with most nodes associated with subgroup branching and supported by low bootstrap values (Supplementary Fig. S1). Peroxidases sharing domain architecture were clustered into clades.
To focus our analysis on fungal peroxidases, we selected six representative fungi that included three plant pathogenic fungi (M. oryzae, Fusarium graminearum, Ustilago maydis), one human pathogenic fungus (Aspergillus fumigatus), and two saprophytic fungi (Neurospora crassa and Saccharomyces cerevisiae). Phylogenetic analysis with selected fungal species placed 27 M. oryzae peroxidase genes into 15 clades (shaded box in Fig. 1). Among the 27 peroxidase genes in M. oryzae, 16 genes (MoAPX3, CPXA, CPXB, MoCCP1, MoCCP2, NOX1, NOX2, CATB, CATA, MoLDS1, MoLDS2, MoPRX1, TPX1, MoPRX2, MoCMD1 and HYR1) were conserved in Ascomycota (Supplementary Table S3).
Figure 1. Phylogenetic analysis of putative peroxidase genes in six selected fungi.
A neighbor-joining tree was constructed based on the amino acid sequences of representative fungal peroxidase genes. Numbers at nodes represent bootstrap confidence values, or percentage of clade occurrence in 2,000 bootstrap replicates; only nodes supported by >50% bootstraps are shown. The scale bar represents the number of amino acid differences per site. Sub-clades containing Magnaporthe oryzae peroxidase genes are shaded, in which M. oryzae peroxidase genes and characterized genes from other fungi are depicted in bold red or black, respectively. Abbreviations for fungal species, followed by their GenBank accession numbers, are as follows: Mo, Magnaporthe oryzae; Sc, Saccharomyces cerevisiae; Fg, Fusarium graminearum; Nc, Neurospora crassa; Um, Ustilago maydis; Af, Alternaria fumigatus.
Expression profiling of 27 M. oryzae peroxidase genes during fungal development and under oxidative stress
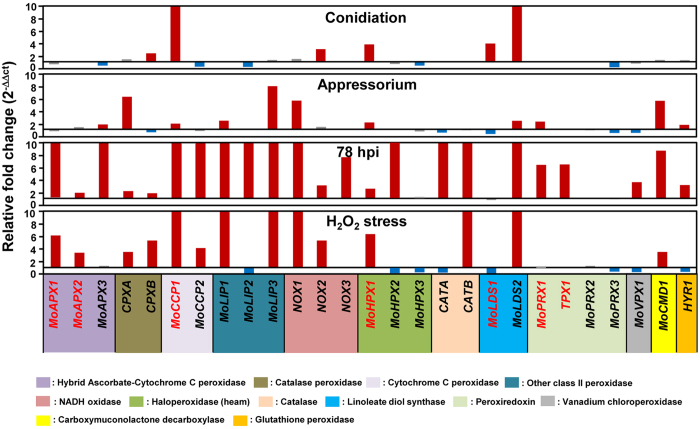
As a next step, we performed expression profiling of the 27 peroxidase-encoding genes using qRT-PCR during infection-related developmental stages that included conidiation, appressorium formation, and 78 h post incubation (hpi) on rice plants and under oxidative stress with 2.5 mM H2O2 (Fig. 2). Expression analysis revealed that most of the peroxidase genes were differentially expressed under the imposed conditions. Compared to expression in mycelia, only a few genes were upregulated in developmental samples, including the conidia and appressoria. However, we found that a majority of the genes (23 genes or 85.2%) were upregulated during the infection stage at 78 hpi. Fourteen genes (51.9%), the exceptions being MoAPX3, MoLIP2, NOX3, MoHPX2, CATA, MoPRX1, TPX1, MoVPX1 and HYR1, were also upregulated under H2O2 stress conditions (Fig. 2). Such an expression pattern suggested the possibility of peroxidase genes playing roles associated with ROS during plant infection.
Figure 2. Expression profiling of 27 M. oryzae peroxidase genes during infection-related developmental stages and infection (78 hpi) on rice, and under oxidative stress.
Upregulated genes (more than 1.5-fold) are indicated by red bars and downregulated genes (less than 0.5-fold) are denoted by blue bars. The genes not showing differential expression are marked in gray. Seven peroxidase genes were selected for functional analysis are highlighted as red.
Genetic analysis of peroxidase genes and fungal pathogenicity
Based on groupings from phylogenetic analysis, we prioritized seven peroxidase genes for functional analysis. We selected the genes from clades that did not contain previously characterized genes (Fig. 1). The seven genes included three genes (MoAPX1, MoAPX2 and MoCCP1) from class I peroxidase, one gene (MoHPX1) from Dye-type peroxidase, one gene (MoLDS1) from animal peroxidase, and two genes (MoPRX1 and TPX1) from peroxiredoxin peroxidase (Fig. 2 and Supplementary Table S1). Targeted gene disruption was conducted using the KJ201 strain as wild-type to genetically assess the impact of selected putative peroxidase genes on fungal development and pathogenicity. Knockout constructs were prepared using double-joint PCR and directly used for transformation of wild-type protoplasts. The resulting hygromycin-resistant colonies were screened by PCR, and the correct gene replacement event for each targeted gene was confirmed by Southern blot analysis (Supplementary Fig. S2).
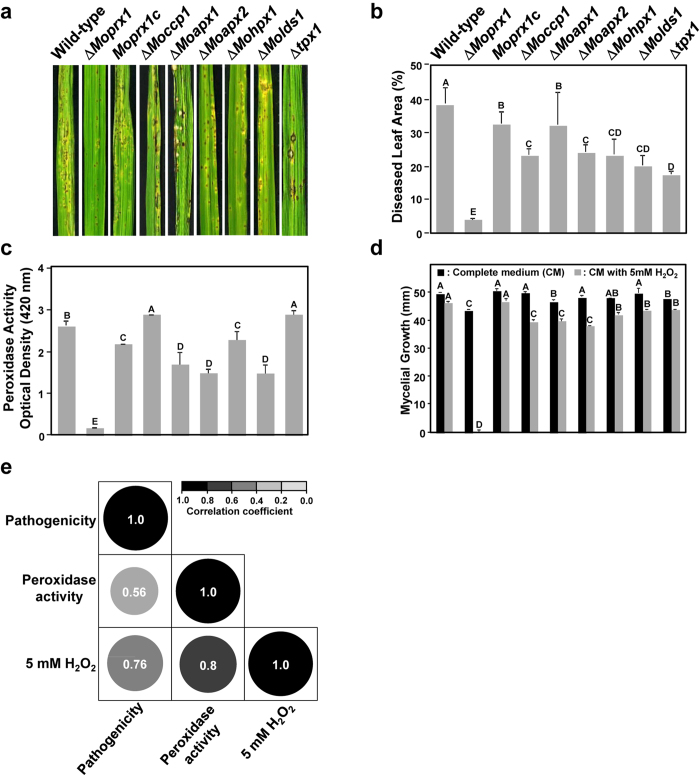
Deletion of seven genes, one gene at a time, had minor effects on vegetative growth (Fig. 3d), conidiation, conidial germination, and appressorium formation. The exception was the deletion of TPX1, which caused a substantial decrease in conidia production (Supplementary Table S4). When we spray-inoculated 3-week-old susceptible rice seedlings with spores from wild-type and deletion mutant strains (~5 × 104 spores/ml), all seven deletion mutants (ΔMoprx1, ΔMoccp1, ΔMoapx1, ΔMoapx2, ΔMohpx1, ΔMolds1, and Δtpx1) showed reduced pathogenicity compared to the wild-type, in terms of diseased leaf area (DLA; Duncan’s multiple range test, P < 0.05; Fig. 3a,b). In particular, we found that deletion of MoPRX1 resulted in the most dramatic reduction in both number and size of lesions on rice leaves.
Figure 3. Pathogenicity, peroxidase activity and mycelial growth of the strains.
(a) Disease symptoms on rice leaves, wild-type and seven peroxidase gene deletion mutants, including ΔMoprx1, ΔMoccp1, ΔMoapx1, ΔMoapx2, ΔMohpx1, ΔMolds1, and Δtpx1, and one complement strain Moprx1c. The diseased leaves were collected at 7 dpi. (b) Disease leaf area (DLA) percentage measured using the Image J software. (c) Extracellular peroxidase activity by ABTS oxidizing assay. (d) Mycelial growth of seven deletion mutants under oxidative stress. (e) Pearson correlation coefficients among DLA from pathogenicity tests, peroxidase activity, and mycelial growth on 5 mM H2O2; data generated from the wild-type and seven peroxidase gene deletion mutants. Error bars represent SD of the mean of three independent experiments. Bars with the same letters are not significantly different (Duncan’s multiple comparisons test, P < 0.05).
Peroxidase activity, sensitivity to H2O2, and pathogenicity
The above observation that all the mutants were less pathogenic than the wild-type prompted us to explore the relationships among peroxidase activity, sensitivity to exogenous H2O2, and pathogenicity. Peroxidase activity was quantified by measuring absorbance of 2, 2´-azino-bis (3-ethylbenzthiazoline-6-sulphonate; ABTS) at 420 nm in culture filtrates of seven deletion mutants and the wild-type. In our assay for peroxidase activity, the deletion mutant strains ΔMoprx1, ΔMoapx1, ΔMoapx2, ΔMohpx1, and ΔMolds1 showed reduced peroxidase activity compared to the wild-type (Fig. 3c). In particular, the mutant ΔMoprx1 lost almost all peroxidase activity. We then went on to assess the sensitivity of the deletion mutants to exogenous H2O2 (Fig. 3d). All seven mutants were sensitive to H2O2, suggesting that their ability to cope with oxidative stress was compromised to varying degrees. Again, ΔMoprx1 showed hypersensitivity to the treatment of H2O2. Such dramatic phenotypic defects of the ΔMoprx1 mutant could be complemented by reintroducing a functional copy of the MoPRX1 gene (Fig. 3 and Supplementary Fig. S2).
When Pearson’s correlation coefficients were computed among pathogenicity (DLA), peroxidase activity, and sensitivity to H2O2; some of the coefficients were significant. The highest correlation was between peroxidase activity and sensitivity to H2O2 (r = 0.8). The next highest correlation was between pathogenicity and sensitivity to H2O2 (r = 0.76). The correlation between pathogenicity and peroxidase activity (r = 0.56) was relatively low (Fig. 3e).
MoPRX1 as a conserved peroxidase
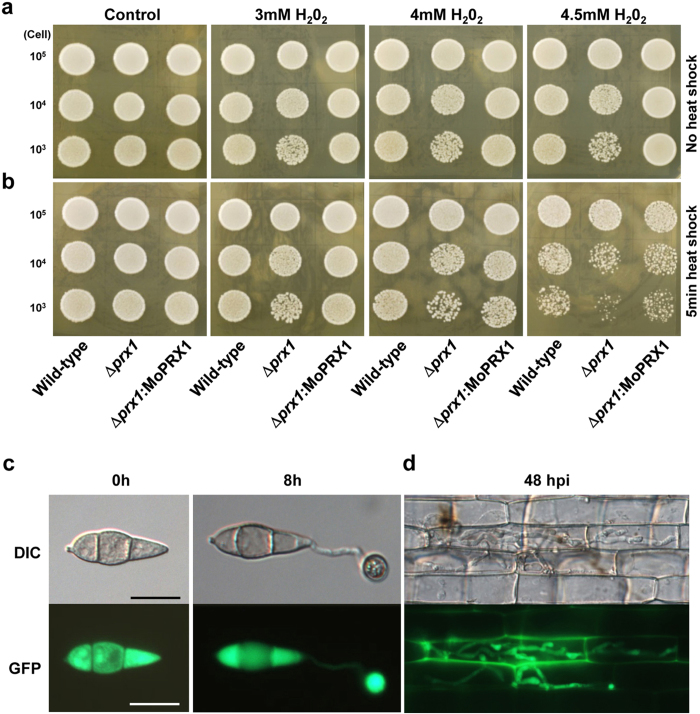
In response to the general trend observed in our comprehensive approach, we investigated peroxidase-mediated fungal pathogenesis through an in-depth analysis of MoPRX1. MoPRX1 (224 aa) is predicted to contain two conserved domains, the thioredoxin fold domain (IPR012335) and the C-terminal peroxiredoxin domain (IPR09479). MoPRX1 is highly homologous to PRX1 in S. cerevisiae, which is a member of the peroxiredoxin family with 1-cysteine (proteins that contain 1 conserved cysteine directly involved in catalysis). We used a deletion mutant in the yeast gene PRX1 to test the peroxidase activity of MoPRX1. PRX1 in S. cerevisiae is localized in mitochondria and is capable of removing organic hydroperoxides and H2O2, providing protection against oxidative stresses. Thus, the yeast mutant of PRX1 is considerably less tolerant to H2O2 treatment and heat-shock is known to exacerbate this phenotype41. When a full-length cDNA of MoPRX1 was introduced into a yeast strain lacking PRX1 (YBL064c), the MoPRX1 gene could restore the tolerance of the mutant to exogenous H2O2 to the wild-type level with or without heat-shock, indicating that MoPRX1 is a functional ortholog of yeast PRX1 (Fig. 4a,b).
Figure 4. MoPRX1 complements S. cerevisiae PRX1 and localized in cytoplasm.
For yeast complementation test, cells were cultured overnight, diluted in phosphate-buffered saline at a final concentration adjusted to 2 × 103 cells/μl, and aliquoted in 100-μl samples for each strain. Diluted 10 μl aliquots (10 μl containing 103, 104, and 105 cells/μl) of wild-type (BY4742), Δprx1 (YBL064C), and Δprx1:MoPRX1 were exposed to 3, 4 and 4.5 mM H2O2 on YPD agar plates after (a) with or (b) without 5-min heat shock at 50 °C. (c) Cellular localization of MoPRX1::GFP fusion protein in a conidium and appressoria of M. oryzae. (d) Infectious hypha expressing MoPRX1::GFP on rice sheaths at 48 hpi. DIC images were captured using a 20-ms exposure to transmitted light with a DIC filter. Fluorescence images were captured using a 400-ms exposure to absorbed light using a GFP filter. Bar = 10 μm.
Despite functional conservation, however, MoPRX1 was not localized to mitochondria in M. oryzae. When a fusion construct of the MoPRX1 promoter-MoPRX1-GFP was prepared and introduced into ∆Moprx1, a strong fluorescent signal was observed in the cytoplasm of conidia and appressoria (Fig. 4c). The PRX1 of a human pathogenic fungus, Candida albicans, was reported to translocate from the cytoplasm to the nucleus during hyphal transition. However, cytoplasmic localization of the protein was consistently observed, even in infectious hyphae of the rice blast fungus invading rice plant cells (Fig. 4c).
Roles of MoPRX1 during early phase of host infection
To elucidate the contribution of MoPRX1, as a peroxidase, to fungal pathogenicity, we monitored wild-type and ΔMoprx1 during the early phase of host infection using the rice sheath assay. When rice sheath cells were inoculated with fungal strains, the infection process was severely delayed in ΔMoprx1. In contrast to the wild-type and complementation strain (Moprx1c), which penetrated and colonized the first-invaded cell at 24 hpi, the mutant barely elaborated a penetration peg. At 48 hpi, infectious growth of ΔMoprx1 was mostly confined to one to two host cells, while that of the wild-type involved a larger number of cells (Fig. 5a). A similar pattern of delay in the early infection phase was observed in experiments using onion (Allium cepa L.) cells as a surrogate (Supplementary Fig. S3). Our observations identified a reduced number of small lesions from rice leaves spray-inoculated with ΔMoprx1 (Fig. 3a).
Figure 5. MoPRX1 detoxifies host ROS and is involved in rice root infection.
(a) Sheath assays with compatible Oryza sativa L. cv. Nakdongbyeo showed delayed and restricted growth of ΔMoPrx1 in rice cells. Sheaths were observed at 24 and 48 hpi inoculated with 2 × 104 conidia/ml. (b) ROS detection by DAB (3,3´-diaminobenzidine) staining, and (c) H2DCFDA (5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate acetyl ester) staining in inoculated rice sheaths at 48 hpi. DIC images were captured using a 20-ms exposure to transmitted light with a DIC filter. Fluorescence images were captured using a 200-ms exposure to absorbed light using a GFP filter. Bar = 50 μm. (d) Root infection from rice seedlings (Oryza sativa L. cv. Nakdongbyeo) by wild-type, ΔMoprx1 and MoPRX1c strains. White arrow indicates the infection site on the root surface.
One possible explanation for such delayed infection is that the mutant was inherently defective in the appressorium-mediated penetration process, including penetration peg formation. To probe this possibility, conidial suspensions from ΔMoprx1, Moprx1c, and the wild-type were infiltrated into rice leaves. This allowed the fungus to gain access to plant tissues without appressorium-mediated penetration. In an infiltration experiment, ΔMoprx1 produced smaller disease lesions compared to the wild-type and Moprx1c, suggesting that MoPRX1 plays roles in the post-penetration phase (Supplementary Fig. S3d).
Based on our results on the peroxidase activity and H2O2 sensitivity of ΔMoprx1, we hypothesized that MoPRX1 is involved in coping with host-derived ROS during early infection. To test this, accumulated H2O2 at the infection sites was examined by 3, 3´-diaminobenzidine (DAB) staining at 48 hpi, using the sheath assay. The results showed that rice sheath cells infected by wild-type and Moprx1c were not stained by DAB, whereas the rice cells infected with ΔMoprx1 were strongly stained, indicating a high concentration of H2O2 (Fig. 5b). We confirmed the accumulation of ROS using the dye 2´, 7´-dichlorodihydrofluorescein diacetate (H2DCFDA, Life technology D-399). H2DCFDA staining also showed fluorescence in ΔMoprx1 infection sites, signifying ROS accumulation, but not in the wild-type or Moprx1c (Fig. 5c). This association of ROS accumulation with the fungus lacking MoPRX1 suggests that MoPRX1 is implicated in direct and/or indirect detoxification of ROS.
A previous study has shown that M. oryzae can infect root tissue, and that this infection requires the fungus to undergo various types of programmed development, which is typical of a root-infecting pathogen42. The authors of that study also identified a tissue-specific virulence factor, MgFOW1. Therefore, our goal was to determine if MoPRX1 is required for root infection. Our pathogenicity assay using roots of rice seedlings showed that ΔMoprx1 cannot cause disease symptoms on root tissues, in contrast to the wild-type causing root browning (Fig. 5d). This suggests that MoPRX1 is a virulence factor required for infection of multiple tissues.
Transcription of other peroxidase genes can be perturbed by deletion of MoPRX1
The observation that ROS accumulation substantially increases in the absence of MoPRX1 led us to examine the expression of a further 26 peroxidase genes using qRT-PCR in the ΔMoprx1 background, compared to the wild-type. We included in this experiment MoPRX1 to confirm absence of its transcripts in the mutant strain. Expression of MoAPX1, MoLIP3, NOX1, NOX2, MoHPX2, MoHPX3, MoLDS1, MoPRX2, and MoVPX1 was upregulated (>1.5-fold), whereas expression of MoLIP1, MoLIP2, CATA, CATB and MoPRX3 was downregulated (<0.5-fold) as shown in supplementary Fig. S4. Expression of the remaining genes remained unchanged. No MoPRX1 transcript was detected, as expected. CII peroxidases (MoLIP1, MoLIP2) and catalase (CATB), which are predicted to be secreted peroxidases having signal peptides (Supplementary Table S2), were downregulated in ΔMoprx1 (Supplementary Fig. S4). Our results showed that deletion of MoPRX1 can alter transcript levels of other peroxidase genes and thus extracellular peroxidase activity.
Discussion
Successful infection of hosts requires pathogens to be equipped with means to breach their defense strategies15,16,22. The most prominent response of host plants to pathogen attack is a burst of ROS production. In response, pathogens have evolved antioxidant defense systems43. Recently, several studies have suggested that at least some of the peroxidases are integral components of such an antioxidant defense system in M. oryzae24,37,38,39,40. Searching of the Fungal Peroxidase Database (fPoxDB)44 revealed 27 putative peroxidase genes encoded in the genome of M. oryzae, leaving the general role of peroxidases in fungal pathogenesis open to question. We classified those 27 putative peroxidase genes based on our phylogenetic analysis, and conducted functional analysis of a subset of the 27 genes.
Our expression profiling of 27 peroxidase genes showed that the majority (23 genes or 85%) of peroxidase genes were induced in infected plant samples (78 hpi), and 14 genes were upregulated under H2O2 stress (Fig. 2). Seven peroxidase genes were upregulated during conidiation and 11 were upregulated during appressoria formation. Considering that the host environment involved high levels of ROS, these data suggest that transcription of many peroxidases in M. oryzae is responsive to ROS, including H2O2.
For the biotrophic pathogens, Claviceps purpurea and Ustilago maydis, the transcription factor genes CPTF1 and YAP1, respectively, were demonstrated to be required to cope with oxidative stresses18,45. In M. oryzae, an ortholog of YAP1, MoAP1, was also shown to mediate oxidative stress responses21. Loss of these genes universally resulted in attenuated virulence. It is tempting to speculate that these transcription factors, in combination with others, would form a regulatory network shared by peroxidase genes in M. oryzae, explaining the common in planta transcriptional response among peroxidase genes. In support of this notion, microarray experiments on a YAP1 deletion mutant showed that at least two peroxidase genes were regulated by YAP1 in U. maydis.
We conducted functional analysis using targeted gene deletion for a set genes selected among the 27 genes. We basically selected genes from clades that did not contain previously characterized genes. Such an approach, in combination with knowledge of previously characterized genes, allowed us to evaluate the roles of peroxidase genes in fungal pathogenesis. Interestingly, all deletion mutants of the seven genes displayed attenuated virulence compared to the wild-type.
We also observed that loss of the selected M. oryzae peroxidase genes resulted in decreased peroxidase activity and reduced tolerance to exogenous H2O2. Our correlation analysis of these attributes (pathogenicity, peroxidase activity, and sensitivity to exogenous H2O2) suggested that sensitivity to exogenous H2O2 could be a good proxy for pathogenicity defects (r = 0.76). It is noteworthy that deletion of 13 peroxidase genes to date (6 from previous studies and 7 from this study) led to attenuated virulence, without an exception. Such unanimous results suggest that all peroxidases are required for fungal pathogenesis, although their contributions vary widely.
Among the previously characterized genes, NOX1 and NOX2 were involved in regulation at the intracellular ROS level. For the remaining four genes, deletion of individual genes rendered the mutants sensitive to exogenous H2O2. This sensitivity to H2O2 correlated with the degree of fungal virulence, with the exception of CPXB, deletion of which had no effect on virulence. The reason that these mutants were impaired in virulence was not the loss of ability to detoxify or suppress host-driven ROS, except for HYR1. Nevertheless, fortifying the cell wall, as shown in the study of CATB, can be, though indirect, a means of coping with the fatal ROS-rampant environment. In most of the mutants in our work developmental changes, including conidiation, conidial germination, and appressorium formation, were not significantly impaired. It is likely that all of the mutants were defective in the penetration or post-penetration phase. We confirmed that, at least in ΔMoprx1, the penetration process was not compromised. Considering the reduction in peroxidase activity and H2O2 tolerance, this suggests that the mutants were less pathogenic due to defects in coping directly or indirectly with host-driven ROS.
During our study, one of the mutants, ΔMoprx1, drew our attention because it was almost non-pathogenic to rice plants. MoPRX1 was, based on sequence similarity, predicted as a gene encoding peroxiredoxin. In mammals, peroxiredoxins are known to be versatile. They possess antioxidant and chaperone-like activities, and, therefore, protect cells from oxidative insults46. Moreover, they might directly interact with transcriptional factors such as c-Myc and NF-κB in the nucleus, and be secreted by some cells30,47. Such versatility of mammalian counterparts and the dramatic defect in ΔMoprx1 pathogenicity encouraged us to examine this mutant in more detail.
MoPRX1 was homologous to PRX1 (YBL064C) in S. cerevisiae (Supplementary Table S1and S3). PRX1 exerts protective antioxidant roles in a cell through its peroxidase activity in detoxifying ROS48,49,50. In S. cerevisiae, PRX1 expression is induced in response to oxidative stress and activation of respiratory pathways51. Similarly, the PRX1 gene in the human pathogenic fungi, A. fumigatus and C. albicans, was also induced after H2O2 exposure52,53. Unlike S. cerevisiae and human pathogenic fungi, however, we found that MoPRX1 expression was not induced in the presence of exogenous H2O2, but during appressorial development and infection at 78 hpi (Fig. 2). Furthermore, MoPRX1 seemed to be present in cytoplasm regardless of developmental changes. In silico analysis of S. cerevisae PRX1 and MoPRX1 using PSortII54, TargetP55, and Mitoprot56 predicted unanimously that PRX1 would be localized to mitochondria while MoPRX1 to cytoplasm and mitochondrial-targeting signal (MTS)57 was not found in MoPRX1 , suggesting that cytoplasmic localization of MoPRX1 is less likely to be artifact.
This was in contrast to S. cerevisiae PRX1, which was shown to localize to mitochondria and C. albicans PRX1 that translocated from cytoplasm to the nucleus during hyphal transition. Despite such differences, MoPRX1 could complement hypersensitivity of the PRX1 deletion mutant of S. cerevisiae, indicating functional conservation as a peroxiredoxin. Although expression of MoPRX1 was not induced in response to exogenous H2O2, the deletion mutant was extremely sensitive to this exogenous H2O2, suggesting that MoPRX1 might be constitutively expressed to function at the front line against oxidative stresses. Another interesting speculation regarding MoPRX1 is that it mainly functions as a general protective barrier against ROS in cytoplasm, while its orthologs detoxify ROS in tight association with metabolic or developmental cell changes. Our data suggest that the functions of PRX1 as a peroxiredoxin have been co-opted depending on the lifestyle of the fungal species.
Using DAB and H2DCFDA staining’s of rice sheaths during infection; we showed that MoPRX1 is required to incapacitate host-derived ROS in planta. Our localization analysis of MoPRX1 raised the question of how MoPRX1 removes host-derived ROS while they are present in the fungal cytoplasm. This is reminiscent of recent reports from studies focusing on DES1 and HYR122,24. DES1 and HYR1 were not secreted into the extracellular milieu, yet they could detoxify/suppress ROS generated by the host plant as a defense response. One possible explanation is that MoPRX1 regulated other ROS-related genes, such as peroxidases and laccases, as suggested in the case of DES1 and HYR1. In support of this, we found that deletion of MoPRX1 resulted in loss of extracellular peroxidase and laccase activity (Supplementary Fig. S5), as well as causes perturbations in the transcription of other peroxidase genes.
In humans, a peroxiredoxin was shown to function as a molecular chaperone58,59,60. Alternatively, it is, therefore, possible that MoPRX1 also acts as a molecular chaperone that is involved in processing secreted proteins. The second possibility is not mutually exclusive to the first and should be an interesting topic for future studies.
In conclusion, we systematically identified and characterized peroxidase genes in the rice blast fungus. Combining a phylogenetic approach with expression profiling and gene deletion, our work provided not only a comprehensive view of the contributions made by peroxidase genes to fungal pathogenesis but also insights into infection strategy built on evolutionarily conserved peroxidases in the fungus. Furthermore, our study posed important questions regarding how the peroxidase-mediated system is regulated at the transcriptional level and the relationships among peroxidases and novel factors, such as DES1. These issues should be addressed in future studies to elucidate ROS-scavenging pathways in fungi pathogenic to plants.
Materials and Methods
Identification of peroxidase genes
Putative peroxidase-encoding genes were retrieved from the Fungal Peroxidase Database (fPoxDB; http://peroxidase.riceblast.snu.ac.kr/), which is a fungi-oriented peroxidase genomics platform44. The collected protein sequences were used to conduct phylogenetic analysis. The protein sequences were aligned with ClustalW in the MEGA6.0 software with default parameters61. Phylogenetic trees were constructed using the neighbor-joining method in the MEGA6.0 software. The peroxidase gene protein structure was obtained from the InterPro database (http://www.ebi.ac.kr/interpro).
Fungal strains and culture conditions
Magnaporthe oryzae KJ201 was obtained from the Center for Fungal Genetic Resources (CFGR) at Seoul National University, Seoul, Korea, and used as the wild-type in this study. This strain and all generated transformants were cultured on oatmeal agar medium (50-g oatmeal per liter with 2% agar (w/v)) or V8-Juice agar medium (4% V8 Juice, pH 6.8) at 25 °C under continuous fluorescent light, cultured in complete liquid medium (0.6% yeast extract, 0.6% casamino acid, and 1% glucose) at 25 °C for 3–4 days with agitation (120 rpm) for genomic DNA extraction. For RNA extraction, all materials were prepared as described previously62 (Supplementary Table S5). Hygromycin B-resistant and geneticin-resistant transformants generated by fungal transformation were selected on solid TB3 agar medium (0.3% yeast extract, 0.3% casamino acid, 1% glucose, 20% sucrose (w/v), and 0.8% agar) supplemented with 200 ppm hygromycin B and 800 ppm geneticin. The wild-type and transformants were cultured on complete agar medium (CM), minimal agar medium (MM), C starvation, and N starvation medium to observe growth and colony characteristics63. Cell wall biogenesis was examined by imposing stress conditions under 200 ppm Congo Red (CR, Aldrich, 860956) supplementation in CM agar medium. Oxidative stress conditions were elicited in CM agar medium, and amended to final concentrations of 2.5, 5, and 10 mM H2O2.
Analysis of transcript levels
Quantitative real-time RT-PCR (qRT-PCR) was employed to measure transcript levels. Total RNA samples and first-strand cDNA were prepared as described previously62. qRT-PCR followed Park et al.62 using each primer pair (Supplementary Table S6). All reactions were performed with more than two biological and three experimental replicates. The β-tubulin gene was used as the internal control for normalization. All amplification curves were analyzed with a normalized reporter threshold of 0.1 to obtain the threshold cycle (Ct) values. The comparative ΔΔCt method was applied to evaluate relative quantities of each amplified sample product. Fold changes were calculated as 2−ΔΔCt 62,64. We applied a fold-change cutoff of ≥1.5 for upregulation, and ≤0.5 for downregulation.
Targeted deletion of seven peroxidase genes and ΔMoprx1 complementation in M. oryzae
Based on seven peroxidase gene sequences in the M. oryzae genome, the 5´ (1.2–1.5-kb) and 3´ (1.2–1.5-kb) flanking regions were amplified using primer pairs from each gene: _UF and _UR for the 5´ flanking, and _DF and _DR for the 3´ flanking regions (Supplementary Table S6) from KJ201 genomic DNA. The 1.4-kb HPH marker cassette was amplified from pBCATPH65 using primers HPH_F and HPH_R. These three amplicons were fused by double-joint PCR66, and the resulting mutant constructs were amplified using the nested primer pair (_UNF and _DNR; as shown in Supplementary Table S6). Fungal protoplasts from wild-type KJ201 were directly transformed using the double-joint PCR product following purification using the standard polyethylene glycol method. Putative gene deletion mutants were screened, and the candidate gene deletion mutants were subsequently purified by single conidia isolation. Southern blot analysis was performed to confirm deletion mutants.
ΔMoprx1 complementation was achieved by amplifying a 3.6-kb fragment containing the MoPRX1 open reading frame (ORF), and a 1.3-kb of the 5´ and 1.2 kb of the 3´ flanking regions from wild-type genomic DNA using the MoPRX1_UF and MoPRX1_DR primers. Geneticin resistance fragment was amplified from PII9922 vector plasmid with primers HPH_F (2.1 kb) and HPH_R (2.1 kb) (Supplementary Table S6). The purified 3.6-kb complementation construct was co-transformed with the geneticin fragment into ΔMoprx1 protoplasts. Putative complemented transformants were selected on TB3 plates amended with 800 ppm geneticin. After genetic purification by single conidium isolation, complements were confirmed by MoPRX1 gene expression through RT-PCR (Supplementary Fig. S2).
Nucleic acids manipulation and Southern blotting
Most molecular biology-related techniques, including clone preparation, plasmid DNA, restriction enzyme digestion, and Southern blot analysis, were performed as described previously67. Genomic DNA and total RNA were extracted following Park et al.67 and Park et al.62, respectively. For PCR screening of generated transformants, genomic DNA was extracted using a rapid and safe DNA extraction method68.
In vitro growth assays, monitoring of infectious growth, and pathogenicity assays
Vegetative growth was measured on CM and MM agar plates at 7 and 12 days post incubation (dpi) with three replicates. Melanization, conidiation, conidial size, conidial germination, and infection assays on rice sheath cells, onion epidermis, and rice seedlings were conducted as described previously69,70. For pathogenicity test,10 ml conidial suspension (5 × 104 spores/ml) containing Tween 20 (250 ppm) was used to spray onto susceptible 3-week-old rice seedlings (Oryza sativa cv.Nakdongbyeo). Disease severity was measured at 7 dpi and disease leaf area (DLA) was measured for more accurate evaluation. The disease area and healthy leaf area were measured using the Image J software (http://imagej.nih.gov/ij/) from equal areas of infected leaves of each strain. All experiments were replicated a minimum of three times. For infiltration, a wound inoculation experiment was performed by cutting leaves from plants, which were subsequently wounded with a needle tip prior to inoculation with 40-μl conidial suspension; the same concentration used in spray inoculation. Leaves were placed in a moist box and incubated in a growth chamber. Photographs were taken at 5 dpi. For rice root inoculation, mycelial blocks were plugged on the root surface of seedlings placed on water agar and incubated in a sealed growth chamber. Lesions were observed and photographs were taken at 5 dpi.
Staining of H2O2 accumulation in host cells
DAB (3,3´ diaminobenzidine, Sigma, D-8001) staining was conducted as described previously22. Excised sheath samples were incubated in 1 mg/ml DAB solution at room temperature for 8 h, and destained with clearing solution (ethanol: acetic acid = 94:4, v/v) for 1 h. H2DCFDA staining was also performed with excised rice sheaths following Huang et al.24. Inoculated excised rice sheaths were incubated for 1 h in 5–20 mM H2DCFDA dissolved in DMSO at room temperature, washed with 0.1 mM KCl, 0.1 mM CaCl2 (pH 6.0), and maintained at room temperature for 1 h before observation. Fluorescence and DIC micrographs were generated using a Zesis Axio Imager AI fluorescence microscope (Carl Zeiss, Oberkochen, Germany). UV light and eGFP filter were used to detect phenolic compounds and ROS signals after staining with H2DCFDA.
Yeast strain and complementation assays
The S. cerevisiae strains YBL064c (ΔPrx1) and BY4742 (wild-type) were obtained from EUROSCARF (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/), and maintained on YPD medium. The MoPRX1 ORF was amplified from first-strand cDNAs of the wild-type with primers MoPRX1_ORF_F_HindIII and Moprx1_ORF_R_XbaI (Supplementary Table S6), and cloned into the HindIII and XbaI sites of pYES2 (Invitrogen, Carlsbad, CA, USA). The isolated plasmid pSY259 was transformed into the yeast strain YBL064c (ΔPrx1), using the lithium acetate method71. Ura3+ transformants were isolated, and the presence of the MoPRX1 ORF in the transformants was double confirmed by PCR. For complementation assays, cells were cultured overnight, diluted in phosphate-buffered saline and cells were counted using a hemocytometer to adjust the required concentration. The final concentration was adjusted to 2 × 103 cells/μl, and aliquoted in 100-μl samples for each strain. Samples were heated to 50 °C for different time intervals until lethal heat shock was reached, cooled on ice, and 10-μl samples were plated on YPD agar medium. The plates were incubated at 30 °C for 2 days. Cells were counted and survival rate was measured. Oxidant chemical sensitivity was also tested using 10-μl aliquot patch assays containing 103, 104, and 105 cells/μl from overnight cultures, spotted on YPD agar medium supplemented with 3, 4, and 4.5 mM H2O2, respectively.
Measurement of extracellular peroxidase and laccase activities
Extracellular enzyme activity was measured using 3-day-old CM liquid culture filtrate. The measurement was performed by following Chi et al.22. Peroxidase and laccase activities were measured by combining 1 ml of reaction mixture (50 mM sodium acetate buffer, pH 5.0 and 20 mM ABTS [Sigma, A1888]) with 200-μl culture filtrate, followed by incubation for 5 min at 25 °C. Absorbance was evaluated at a 420-nm wavelength using a spectrophotometer. Three independent biological experiments, with three replicates per experiment, were performed for each test. A mycelial block was placed on CM medium containing 200 ppm Congo Red agar plate for 9 dpi to measure peroxidase secretion. Laccase activity was also monitored on 0.2 mM ABTS agar plate assays with or without 0.5 mM copper sulfate at 4 dpi.
Cellular localization of MoPRX1::GFP
The MoPRX1::eGFP fusion construct was generated by double-joint PCR. A 2.3-kb genomic fragment, including the putative promoter and full MoPRX1 ORF region, was amplified with primers MoPRX1_UF and MoPRX1_ORF_R_eGFP (Supplementary Table S6). The eGFP ORF (0.7 kb) with terminator (0.3 kb) was amplified with primers eGFP_F and NC_Term_R from the SK2707 plasmid as a template. The resulting PCR products were fused by double-joint PCR66 using primers MoPRX1_5NF and NC_Term_NR. The eGFP fusion construct was introduced into ΔMoprx1 by co-transformation with pII9922 plasmid, which carried the geneticin-resistance gene. MoPRX1::eGFP cellular localization was observed in conidia, appressoria and infectious hypha within rice cells at 48 hpi using a fluorescence microscope (Carl Zeiss Microscope Division, Oberkochen, Germany) with a GFP filter.
Additional Information
How to cite this article: Mir, A. A. et al. Systematic characterization of the peroxidase gene family provides new insights into fungal pathogenicity in Magnaporthe oryzae. Sci. Rep. 5, 11831; doi: 10.1038/srep11831 (2015).
Supplementary Material
Acknowledgments
This work was supported by the National Research Foundation of Korea grant funded by the Ministry of Science, ICT & Future Planning (NRF-2014R1A2A1A10051434), the Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ011154), Rural Development Administration, Republic of Korea. Albely is grateful for a graduate fellowship through the Brain Korea 21 PLUS Program.
Footnotes
Author Contributions Conceived and designed the experiments: A.A.M., S.Y.P. and Y.H.L. Performed the experiments: A.A.M., S.Y.P., M.A.S. and S.K. Analyzed the data: A.A.M., S.Y.P., J.C., J.J., S.K. and Y.H.L. Contributed reagents/materials/analysis tools: A.A.M., S.Y.P., M.A.S. and S.K. wrote the paper: A.A.M., S.Y.P., J.J. and Y.H.L.
References
- Wojtaszek P. Oxidative burst: an early plant response to pathogen infection. Biochem. J. 322 (Pt 3), 681–92 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolwell G. P. & Wojtaszek P. Mechanisms for the generation of reactive oxygen species in plant defence-a broad perspective. Physiol. Mol. Plant Pathol. 51, 347–366 (1997). [Google Scholar]
- Lamb C. & Dixon R. A. The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 251–275 (1997). [DOI] [PubMed] [Google Scholar]
- Levine A., Tenhaken R., Dixon R. & Lamb C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79, 583–93 (1994). [DOI] [PubMed] [Google Scholar]
- Grant M. et al. The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant. J. 23, 441–50 (2000). [DOI] [PubMed] [Google Scholar]
- Doke N. Involvement of superoxide anion generation in the hypersensitive response of potato tuber tissues to infection with an incompatible race of Phytophthora infestans and to the hyphal wall components. Physiol. Plant Pathol. 23, 345–357 (1983). [Google Scholar]
- Auh C.-K. & Murphy T. M. Plasma membrane redox enzyme is involved in the synthesis of O2- and H2O2 by Phytophthora elicitor-stimulated rose cells. Plant Physiol. 107, 1241–1247 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller J., Meyer A. J. & Tudzynski P. Redox-sensitive GFP2: use of the genetically encoded biosensor of the redox status in the filamentous fungus Botrytis cinerea. Mol. Plant Pathol. 13, 935–947 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller J. & Tudzynski P. Reactive Oxygen Species in Phytopathogenic Fungi: Signaling, Development, and Disease. Ann. Rev. Phytopathol. 49, 369–390 (2011). [DOI] [PubMed] [Google Scholar]
- Tudzynski P., Heller J. & Siegmund U. Reactive oxygen species generation in fungal development and pathogenesis. Curr. Opin. Microbiol. 15, 653–659 (2012). [DOI] [PubMed] [Google Scholar]
- Huckelhoven R. Cell wall-associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 45, 101–27 (2007). [DOI] [PubMed] [Google Scholar]
- Jones J. D. & Dangl J. L. The plant immune system. Nature 444, 323–9 (2006). [DOI] [PubMed] [Google Scholar]
- Torres M. A., Jones J. D. & Dangl J. L. Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 37, 1130–4 (2005). [DOI] [PubMed] [Google Scholar]
- Bradley D. J., Kjellbom P. & Lamb C. J. Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel, rapid defense response. Cell 70, 21–30 (1992). [DOI] [PubMed] [Google Scholar]
- Agrios G. N. Frontiers and challenges in plant pathology communications. Phytopathology 82, 32–34 (1992). [Google Scholar]
- Chai L. Y. A. et al. Fungal strategies for overcoming host innate immune response. Med. Mycol. 47, 227–236 (2009). [DOI] [PubMed] [Google Scholar]
- Enjalbert B., MacCallum D. M., Odds F. C. & Brown A. J. Niche-specific activation of the oxidative stress response by the pathogenic fungus Candida albicans. Infect. Immun. 75, 2143–51 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina L. & Kahmann R. An Ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell 19, 2293–309 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. H., Yang S. L. & Chung K. R. The YAP1 homolog-mediated oxidative stress tolerance is crucial for pathogenicity of the necrotrophic fungus Alternaria alternata in citrus. Mol. Plant-Microbe Interact 22, 942–52 (2009). [DOI] [PubMed] [Google Scholar]
- Kim K. H. et al. TmpL, a transmembrane protein required for intracellular redox homeostasis and virulence in a plant and an animal fungal pathogen. PLoS Pathog. 5, e1000653 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M. et al. The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 7, e1001302 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi M. H., Park S. Y., Kim S. & Lee Y. H. A novel pathogenicity gene is required in the rice blast fungus to suppress the basal defenses of the host. PLoS Pathog. 5, e1000401 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samalova M., Meyer A. J., Gurr S. J. & Fricker M. D. Robust anti-oxidant defences in the rice blast fungus Magnaporthe oryzae confer tolerance to the host oxidative burst. New Phytol. 201, 556–573 (2014). [DOI] [PubMed] [Google Scholar]
- Huang K., Czymmek K. J., Caplan J. L., Sweigard J. A. & Donofrio N. M. HYR1-mediated detoxification of reactive oxygen species is required for full virulence in the rice blast fungus. PLoS Pathog. 7, e1001335 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz K. J. et al. The function of peroxiredoxins in plant organelle redox metabolism. J. Exp. Bot. 57, 1697–709 (2006). [DOI] [PubMed] [Google Scholar]
- Missall T. A., Pusateri M. E. & Lodge J. K. Thiol peroxidase is critical for virulence and resistance to nitric oxide and peroxide in the fungal pathogen, Cryptococcus neoformans. Mol. Microbiol. 51, 1447–58 (2004). [DOI] [PubMed] [Google Scholar]
- Edgar R. S. et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature 485, 459–64 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S. G., Woo H. A., Kil I. S. & Bae S. H. Peroxiredoxin functions as a peroxidase and a regulator and sensor of local peroxides. J. Biol. Chem. 287, 4403–4410 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourquet S., Huang M. E., D’Autreaux B. & Toledano M. B. The dual functions of thiol-based peroxidases in H2O2 scavenging and signaling. Antioxid. Redox Signal. 10, 1565–76 (2008). [DOI] [PubMed] [Google Scholar]
- Ishii T., Warabi E. & Yanagawa T. Novel roles of peroxiredoxins in inflammation, cancer and innate immunity. J. Clin. Biochem. Nutr. 50, 91–105 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passardi F. et al. PeroxiBase: the peroxidase database. Phytochemistry 68, 1605–11 (2007). [DOI] [PubMed] [Google Scholar]
- Fisher M. C. et al. Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–94 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot N. J. On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 57, 177–202 (2003). [DOI] [PubMed] [Google Scholar]
- Dean R. A. et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434, 980–6 (2005). [DOI] [PubMed] [Google Scholar]
- Howard R. J., Ferrari M. A., Roach D. H. & Money N. P. Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc. Natl. Acad. Sci. U S A 88, 11281–4 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankanala P., Czymmek K. & Valent B. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19, 706–24 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan M. J., Wang Z. Y., Jones M. A., Smirnoff N. & Talbot N. J. Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc. Natl. Acad. Sci. U S A 104, 11772–7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skamnioti P., Henderson C., Zhang Z., Robinson Z. & Gurr S. J. A novel role for catalase B in the maintenance of fungal cell-wall integrity during host invasion in the rice blast fungus Magnaporthe grisea. Mol. Plant-Microbe Interact 20, 568–80 (2007). [DOI] [PubMed] [Google Scholar]
- Tanabe S. et al. The role of catalase-peroxidase secreted by Magnaporthe oryzae during early infection of rice cells. Mol. Plant-Microbe Interact 24, 163–71 (2011). [DOI] [PubMed] [Google Scholar]
- Fernandez J. & Wilson R. A. Characterizing roles for the glutathione reductase, thioredoxin reductase and thioredoxin peroxidase-encoding genes of Magnaporthe oryzae during rice blast disease. PLoS ONE 9, e87300 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson J. F., Whyte B., Bissinger P. H. & Schiestl R. H. Oxidative stress is involved in heat-induced cell death in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U S A 93, 5116–21 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesma A. & Osbourn A. E. The rice leaf blast pathogen undergoes developmental processes typical of root-infecting fungi. Nature 431, 582–6 (2004). [DOI] [PubMed] [Google Scholar]
- Aguirre J., Hansberg W. & Navarro R. Fungal responses to reactive oxygen species. Med. Mycol. 44, S101–S107 (2006). [DOI] [PubMed] [Google Scholar]
- Choi J. et al. fPoxDB: fungal peroxidase database for comparative genomics. BMC Microbiol. 14, 117 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathues E. et al. CPTF1, a CREB-like transcription factor, is involved in the oxidative stress response in the phytopathogen Claviceps purpurea and modulates ROS level in its host Secale cereale. Mol. Plant-Microbe Interact 17, 383–393 (2004). [DOI] [PubMed] [Google Scholar]
- Rhee S. G., Chae H. Z. & Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic. Biol. Med. 38, 1543–52 (2005). [DOI] [PubMed] [Google Scholar]
- Riddell J. R., Wang X. Y., Minderman H. & Gollnick S. O. Peroxiredoxin 1 stimulates secretion of proinflammatory cytokines by binding to TLR4. J. Immunol. 184, 1022–1030 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk R., Griffin P. & Nathan C. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407, 211–5 (2000). [DOI] [PubMed] [Google Scholar]
- Hofmann B., Hecht H. J. & Flohe L. Peroxiredoxins. Biol. Chem. 383, 347–64 (2002). [DOI] [PubMed] [Google Scholar]
- Peshenko I. V. & Shichi H. Oxidation of active center cysteine of bovine 1-Cys peroxiredoxin to the cysteine sulfenic acid form by peroxide and peroxynitrite. Free Radic. Biol. Med. 31, 292–303 (2001). [DOI] [PubMed] [Google Scholar]
- Pedrajas J. R., Miranda-Vizuete A., Javanmardy N., Gustafsson J. A. & Spyrou G. Mitochondria of Saccharomyces cerevisiae contain one-conserved cysteine type peroxiredoxin with thioredoxin peroxidase activity. J. Biol. Chem. 275, 16296–301 (2000). [DOI] [PubMed] [Google Scholar]
- Lessing F. et al. The Aspergillus fumigatus transcriptional regulator AfYap1 represents the major regulator for defense against reactive oxygen intermediates but is dispensable for pathogenicity in an intranasal mouse infection model. Eukaryot. Cell 6, 2290–302 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasa K. et al. Characterization of a putative thioredoxin peroxidase Prx1 of Candida albicans. Mol. Cells 33, 301–307 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K. & Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24, 34–35 (1999). [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S. & von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016 (2000). [DOI] [PubMed] [Google Scholar]
- Claros M. G. & Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241, 779–786 (1996). [DOI] [PubMed] [Google Scholar]
- Rapaport D. Finding the right organelle - Targeting signals in mitochondrial outer-membrane proteins. Embo Rep. 4, 948–952 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H. H. et al. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell 117, 625–35 (2004). [DOI] [PubMed] [Google Scholar]
- Lee W. S. et al. Human peroxiredoxin 1 and 2 are not duplicate proteins - The unique presence of Cys(83) in Prx1 underscores the structural and functional differences between Prx1 and Prx2. J. Biol. Chem. 282, 22011–22022 (2007). [DOI] [PubMed] [Google Scholar]
- Welch W. J. & Brown C. R. Influence of molecular and chemical chaperones on protein folding. Cell Stress Chaperon 1, 109–115 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. Y. et al. Global expression profiling of transcription factor genes provides new insights into pathogenicity and stress responses in the rice blast fungus. PLoS Pathog. 9, e1003350 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot N. J., McCafferty H. R. K., Ma M., Koore K. & Hamer J. E. Nitrogen starvation of the rice blast fungus Magnaporthe grisea may act as an environmental cue for disease symptom expression. Physiol. Mol. Plant P 50, 179–195 (1997). [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–8 (2001). [DOI] [PubMed] [Google Scholar]
- Yun S. H. Molecular genetics and manipulation of pathogenicity and mating determinants in Mycosphaerella zeae-maydis and Cochliobolus heterostrophus Ithaca, Ph.D. thesis, Cornell University, NY, U.S.A (1998). [Google Scholar]
- Yu J.H. et al. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41, 973–81 (2004). [DOI] [PubMed] [Google Scholar]
- Park S. Y., Milgroom M. G., Han S. S., Kang S. & Lee Y. H. Diversity of pathotypes and DNA fingerprint haplotypes in populations of Magnaporthe grisea in Korea over two decades. Phytopathology 93, 1378–1385 (2003). [DOI] [PubMed] [Google Scholar]
- Chi M. H., Park S. Y., Kim S. & Lee Y. H. A quick and safe method for fungal DNA extraction. Plant Pathol. J. 25, 108–111 (2009). [Google Scholar]
- Kim S. et al. Homeobox transcription factors are required for conidiation and appressorium development in the rice blast fungus Magnaporthe oryzae. PLoS Genet. 5, e1000757 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga H., Dohi K., Nakayachi O. & Mori M. A novel inoculation methods of Magnaporthe grisea for cytological observation of the infection process using intact leaf sheaths of rice plants. Physiol. Mol. Plant. Pathol. 64, 67–72 (2004). [Google Scholar]
- Shiestl R. H. & Gietz R. D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16, 339–346 (1989). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.