Abstract
The planarian Dugesia japonica has amazing ability to regenerate a head from the anterior ends of the amputated stump with maintenance of the original anterior-posterior polarity. Although planarians present an attractive system for molecular investigation of regeneration and research has focused on clarifying the molecular mechanism of regeneration initiation in planarians at transcriptional level, but the initiation mechanism of planarian head regeneration (PHR) remains unclear at the protein level. Here, a global analysis of proteome dynamics during the early stage of PHR was performed using isobaric tags for relative and absolute quantitation (iTRAQ)-based quantitative proteomics strategy, and our data are available via ProteomeXchange with identifier PXD002100. The results showed that 162 proteins were differentially expressed at 2 h and 6 h following amputation. Furthermore, the analysis of expression patterns and functional enrichment of the differentially expressed proteins showed that proteins involved in muscle contraction, oxidation reduction and protein synthesis were up-regulated in the initiation of PHR. Moreover, ingenuity pathway analysis showed that predominant signaling pathways such as ILK, calcium, EIF2 and mTOR signaling which were associated with cell migration, cell proliferation and protein synthesis were likely to be involved in the initiation of PHR. The results for the first time demonstrated that muscle contraction and ILK signaling might played important roles in the initiation of PHR at the global protein level. The findings of this research provide a molecular basis for further unraveling the mechanism of head regeneration initiation in planarians.
Introduction
Planarians have an extraordinary regenerative ability through the activation of pluripotent stem cells (also called neoblasts) [1–3]. Strikingly, planarian can regenerate a head at the anterior-facing wounds within a week after amputation. This process includes several distinct events: closure of the wound, cell apoptosis, blastema formation, and blastema cell differentiation [4–7]. More specifically, wound healing is activated to cover the exposed tissues in the first 30 min–3 h after amputation. Subsequently, neoblasts at the wound site undergo an initial mitotic peak within the first 6 h, and proceed the migration towards the wound at about 18 h, and then a spatially restricted mitotic response at 48–72 h to promote blastema formation [5, 8, 9]. Conversely, cell apoptosis at the wound site is tightly coordinated with the proliferative reaction, so as to ensure the appropriate proportions in the regenerating planarian [7]. A study reported that wound closure in planarians after amputation was facilitated by muscle contraction [10], but the molecular mechanism that triggers wound closure is still unknown. In this study, we firstly focus on the mechanism of wound healing at the early stage of planarian head regeneration (PHR) after amputation at the proteome level.
Planarian regeneration is a complex process, regulated by various signaling pathways. For instance, extracellular signal-related kinase (ERK) signaling and c-Jun N-terminal kinase (JNK) signaling were highly activated at the wound site following amputation and were necessary for planarian regeneration by regulating the formation and differentiation of blastema [11, 12]. Furthermore, the regenerating tissues of the planarian re-establish proper anterior-posterior (AP) polarity involving canonical Wnt/β-catenin and Hedgehog (Hh) signaling [13–15], and proper dorsal-ventral (DV) polarity involving BMP signaling [16]. Recent studies revealed that Wnt/β-catenin and Hh signaling in planarians were necessary for posterior polarity (tail regeneration), and its inhibition resulted in head regeneration at the wound site [14, 15, 17–19]. A recent study also confirmed the notion that posterior β-catenin signaling interfered with head tissue differentiation in planarians through negatively modulating ERK signaling [20]. Although signaling pathways involving blastema formation and proper polarity have been extensively studied during the regeneration in planarians, those important pathways for priming head regeneration in planarians need to be further excavated. Studies have reported that integrin-linked kinase (ILK) signaling played important roles in wound healing, cell adhesion and migration, and tissue regeneration [21–23], but no research is performed on the role of ILK signaling in PHR as well as the expression changes of proteins associated with ILK signaling in PHR.
Previous researches have focused on the characterization of the regeneration transcriptome of the planarian Schmidtea mediterrane and Dugesia japonica [24–26]. However, it fails to reflect the influence of post-transcriptional modifications and protein interactions. Delightedly, isobaric tags for relative and absolute quantitation (iTRAQ), a powerful tool to detect the kinds and abundance of proteins, has been widely used in proteomics studies owing to its advantages over two-dimensional (2D) gel electrophoresis [27]. At present, a proteomics analysis of the normal and stem cell-depleted planarian Schmidtea mediterrane, has been performed [28–31], but an investigation of the potential proteome implicated in the initiation of head regeneration in the planarian Dugesia japonica remains unknown.
The aim of this study was to identify a wide range of proteins that are differentially regulated at the early stage of PHR. In this study, iTRAQ combined with LC-MS/MS was employed to quantitatively assess protein expression profiles in the regenerating head fragments at 0 h, 2 h and 6 h after amputation. We found that muscle contraction and ILK signaling might play vital roles in the initiation of PHR through functional enrichment and IPA analysis. The findings of this research provide a molecular basis for further unraveling the mechanism of head regeneration initiation in planarians.
Materials and Methods
Ethics Statement
Planarian samples were collected from the source of Tagang reservoir, Xinxiang, Henan, China. The samples were handled in strict accordance with the guidelines of the Animal Care and Use Committee of Henan Normal University. We confirm that the samples collected were not privately owned or protected in any way and that the field studies did not involve endangered or protected species.
Model preparation of planarian head regeneration
Planarians used in this study belong to the asexual strain of the species Dugesia japonica, and were collected from Tagang reservoir, which is located at the geographic coordinates of longitude (E114° 01'13.48") and latitude (N35°36'4.45"). Animals were cultured in autoclaved tap water in the dark at 20°C. Planarians with a length of 8–10 mm were selected, and starved for at least 1 week prior to use in experiments. A total of 45 planarians mentioned above were randomly assigned to three groups with 15 planarians per group. For regeneration studies, planarians were amputated transversely anterior to the pharynx, and the anterior end was allowed to undergo head regeneration. The regenerating planarians were collected at 0 h, 2 h and 6 h after amputation, and stored at –80°C for further use.
Protein extraction and iTRAQ labeling
Protein extraction was performed using a procedure described previously [32]. Frozen samples were ground in liquid nitrogen to a fine powder and suspended in lysis buffer (30 mM Tris, 7 M urea, 2 M thiourea, 4% CHAPS, 65 mM DTT). The suspension was vortexed at 4℃ for 1 h, and then centrifuged at 20,000 g for 1 h. The protein concentration of each sample was determined using a 2D Quantification kit (GE Healthcare, USA).
A total of 100 μg of each sample was denatured, reduced and alkylated as described in the iTRAQ protocol (Applied Biosystems, USA). Each sample was digested with 0.1 μg/μL trypsin solution at 37°C, overnight. The digested peptides were dried by vacuum centrifugation. Control (0 h), 2 h and 6 h samples were respectively labeled with 114, 115 and 116 iTRAQ tags according to the manufacturer’s protocol (Applied Biosystems, USA). The three samples were pooled and vacuum-dried.
LC-MS/MS, protein identification and data analysis
The pooled sample was separated on the Poly-LC strong cation exchange column (4.6 x 100 mm) on a Nano HPLC System (GE Healthcare) according to the method described in detail previously [33]. In this study, a total of 30 fractions were collected over the gradient, but some were pooled to give a final total of 10 fractions that were desalted using a PepClean C-18 spin column (Sigma, USA), and dried by vacuum centrifugation. Subsequently, the fractionated samples were analyzed by LC-MS/MS based on Q-Exactive mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) [33, 34].
For peptide data analysis, raw mass data were processed using Proteome Discover 1.4 software and searched against the SwissProt database (March 2013) from Uniprot website (http://www.uniprot.org) using Mascot 2.2 (Matrix Science, London, UK). The analysis and search parameters were set as: trypsin as the digestion enzyme with allowance for a maximum of two missed cleavage, Carbamidomethyl (C) and iTRAQplex modification (K and N-terminus) as a fixed modification, Oxidation (M) as a variable modifications, peptide mass tolerance of 20ppm, fragment mass tolerance of 0.1 Da. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium [35] via the PRIDE partner repository with the dataset identifier PXD002100.
In order to measure the false discovery rate (FDR), the peptide mass spectra datasets were used to search a decoy peptide database. The following filters were used in this study, peptide FDR≤0.01 and each protein with at least 2 unique peptides. Expression changes of the identified peptides in the regenerating head fragments were calculated in comparison with the control based on the iTRAQ reporter ion intensities. Based on relative quantification and statistical analysis, 1.4-fold change cutoff was selected to categorize proteins as significantly changed, that is, proteins with iTRAQ ratios > 1.4 were considered to be up-regulated, whereas those with iTRAQ ratios < 0.714 were considered to be down-regulated.
Quantitative real-time PCR analysis of gene expression
In this study, several genes including EIF4A1, EIF4A3, H2B, DDX, CTNNB, CAM, YBX, GST were selected for quantitative real time RT-PCR (qRT-PCR) analysis. The primers were designed with Primer Express 2.0 software according to mRNA sequences of the 8 target genes and internal control ACTB and synthesized by Shanghai Generay Biotech Co., Ltd. The genes and their primer sequences used for qRT-PCR analysis were provided in S1 Table. RNA extraction, cDNA synthesis and qRT-PCR were carried out according to the method described previously [36]. The expression level of gene was calculated using 2-ΔΔCt method. ACTB gene was used as the internal control and the experiments were performed three times. The statistical significance in expression was estimated by t-test, and p < 0.05 was considered to be significant.
Bioinformatics analysis
In order to characterize the expression patterns of the proteins identified in our quantitative proteomics analysis, k-means clustering was utilized to classify the differentially expressed proteins, and Expression Analysis Systematic Explorer (EASE) software on DAVID website was applied to conduct functional enrichment analysis as described in detail previously [37, 38]. EASE analyses were conducted to identify GO categories that were over-represented according to a modified Fisher's exact test. In this study, the GO categories with P-value < 0.05 were considered to be significantly over-represented at the early stage of PHR.
In addition, the differentially expressed proteins were analyzed by Ingenuity Pathway Analysis (IPA) version 9.0 (Redwood City, CA, http://www.ingenuity.com) software for predominant canonical pathways [39]. Briefly, a dataset containing these proteins and corresponding extremum of expression values was firstly uploaded into “Dataset Files” of the IPA. Then IPA core analysis was performed. Canonical pathways obtained in this study were identified from the IPA library based on Fisher’s exact test. The significance of the association between the differentially expressed proteins and canonical pathways was measured as described in the literature [40].
Results
Quantitative proteomics analysis revealed tissue proteome alterations at the early stage of planarian head regeneration
To identify proteins associated with the initiation of PHR, iTRAQ was employed to assess proteome changes at 2 h and 6 h following amputation (n = 15), and Q-Exactive mass spectrometer was used to obtain better coverage of tissue proteome. The mass data was searched against the SwissProt database using Mascot 2.2 search engine. The peptide FDR≤0.01 and each protein with at least 2 unique peptides were utilized to filter out the data, and 1595 proteins were indentified (S2 Table). The protein mass distribution mainly concentrated in 10–100 kDa which made up 88.5% of the proteins. The proteins with 2–5 peptides, 6–10 peptides, and above 11 peptides comprised 890, 399 and 306, respectively. Protein sequence coverage with 50–100%, 40–50%,30–40%, 20–30%, 10–20% and under 10% variation accounted for 10.85%, 11.85%, 15.67%, 20.56%, 23.51% and 17.56% coverage, respectively.
Next, we applied a threshold of >1.4-fold to identify proteins that were differentially expressed. A total of 162 proteins meeting the screening criteria were classified as differentially expressed. Among these proteins, 83 proteins were up-regulated and 79 proteins were down-regulated at 2 h and 6 h after amputation compared to control group (S3 Table).
Validation of gene expression by qRT-PCR
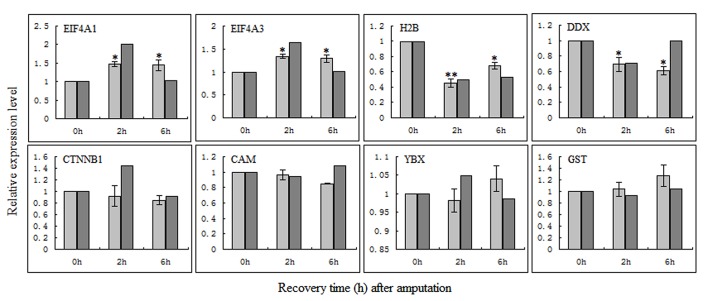
Changes in the protein expression may be due to changes in its mRNA level. In this study, to investigate whether the changes observed in protein expression were the result of transcriptional regulation, we used qRT-PCR to determine mRNA levels of eight selected proteins (EIF4A1, EIF4A3, H2B, DDX, CTNNB1, CAM, YBX, and GST) in the regenerating head fragments at 0 h, 2 h and 6 h after amputation. As shown in Fig 1, in addition to CTNNB1 whose expression trend in protein level is not consistent with that in mRNA level, the changes in mRNA expression of the other seven proteins were basically correlated with their protein expression changes detected by iTRAQ approach. Furthermore, four proteins (EIF4A1, EIF4A3, H2B and DDX) had statistically significant differences, and three proteins (CAM, YBX and GST) had no statistically significant differences, which is consistent with the results from the proteomic analysis.
Fig 1. Validation of the identified proteins at the mRNA level by qRT-PCR analysis.
Light gray histogram indicates mRNA levels detected by qRT-PCR; dark gray histogram indicates protein levels detected by iTRAQ. The mean values are represented in the bar graph. Bars represent standard errors (n = 3); * p<0.05.
Functional enrichment analysis of the differentially expressed proteins at the early stage of planarian head regeneration
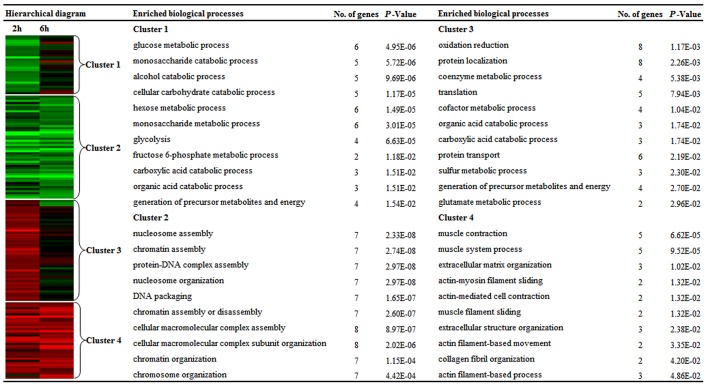
In order to characterize the expression patterns of proteins that exhibited significant change at the early stage of PHR, k-means clustering was utilized to classify the differentially expressed proteins. The results showed that 162 proteins were categorized into 4 clusters: 28 proteins in cluster 1 and 50 proteins in cluster 2 were mainly down-regulated at 2 h and 6 h during PHR, while 48 proteins in cluster 3 and 36 proteins in cluster 4 were primarily up-regulated at 2 h and 6 h during PHR (Fig 2). Details for 162 differentially expressed proteins that grouped into four clusters were shown in S3 Table. Subsequently, EASE analysis was carried out for each cluster to determine whether certain GO categories are over-represented or not. The over-represented GO categories were selected from each cluster according to their corresponding P-value and were shown in Fig 2. A detailed GO annotation of the differentially expressed proteins in each cluster was provided in the S4 Table.
Fig 2. Global protein expression patterns at the early stage of planarian head regeneration.
The “Hierarchical diagram” column represents K-means clustering of total of 162 differentially expressed proteins. Red and green colors denote the expression level higher and lower than the control, respectively. The over-represented functional categories of the four clusters were shown in the “Enriched biological processes” column. Categorical data were analyzed using a modified Fisher's exact test. p < 0.05 was considered to be significant.
Obviously, cluster 1 was mainly enriched with categories of carbohydrate metabolism, and the proteins in this cluster included 6-phosphogluconate dehydrogenase (PGD), phosphofructokinase (PFK), UDP-glucose 6-dehydrogenase (UGDH), and enolase (ENO1). The proteins in cluster 2 were functionally classified into the categories of nucleosome assembly and organization, including members of histone (H1, H2A, H2B, and H4). A higher proportion of proteins in cluster 3 were associated with tanslatation (i.e. members of eukaryotic translation initiation factors and ribosomal proteins), and energy metabolism (i.e. aldehyde dehydrogenase family 5, acyl-coenzyme A oxidase 3, ATP synthase-coupling factor 6, creatine kinase M-type). Muscle contraction-related proteins such as myosins and alpha-actinin-2, extracellular matrix organization-related proteins such as collagens, were predominantly harbored in cluster 4. Some of the over-represented GO categories along with the differentially expressed proteins were summarized in Table 1.
Table 1. Relative expression abundance of the representative proteins in four clusters at the early stage of planarian head regeneration.
| Accession | Protein Name | Gene | MW [kDa] | calc. pI | Coverage | Unique Peptides | 2h* | 6h* |
|---|---|---|---|---|---|---|---|---|
| Carbohydrate and energy metabolism (Cluster 1 and 3) | ||||||||
| F1R2A3 | ATP-dependent 6-phosphofructokinase, platelet type | PFKP | 35.8 | 7.0 | 8.0 | 2 | 0.52 | 1.10 |
| P85968 | 6-phosphogluconate dehydrogenase, decarboxylating | PGD | 53.2 | 7.0 | 29.4 | 8 | 0.56 | 0.94 |
| P30835 | 6-phosphofructokinase, liver type | PFKL | 85.3 | 7.4 | 6.3 | 2 | 0.61 | 1.14 |
| D4A7P8 | 2-oxoglutarate dehydrogenase, mitochondrial | OGDH | 115.2 | 6.9 | 24.1 | 10 | 0.65 | 1.76 |
| O70199 | UDP-glucose 6-dehydrogenase | UGDH | 54.9 | 7.5 | 37.9 | 9 | 0.66 | 1.05 |
| Q64428 | Trifunctional enzyme subunit alpha, mitochondrial | HADHA | 82.6 | 9.1 | 36.6 | 20 | 0.69 | 0.83 |
| P55159 | Serum paraoxonase/arylesterase 1 | PON1 | 39.3 | 5.2 | 23.9 | 7 | 0.70 | 0.81 |
| O02572 | Enolase | ENO1 | 17.1 | 9.0 | 37.9 | 4 | 0.70 | 0.96 |
| P21213 | Histidine ammonia-lyase | HAL | 72.2 | 6.5 | 20.9 | 10 | 0.71 | 0.83 |
| G3V945 | Aldehyde dehydrogenase family 5, subfamily A1 | ALDH5A1 | 56.1 | 8.1 | 24.9 | 10 | 1.32 | 0.70 |
| Q1MTI4 | Triosephosphate isomerase A | TPI1A | 26.8 | 5.0 | 29.8 | 2 | 0.57 | 0.56 |
| Q90XG0 | Triosephosphate isomerase B | TPI1B | 26.8 | 6.9 | 70.6 | 2 | 1.41 | 1.01 |
| F1QXK3 | Acyl-coenzyme A oxidase 3 | ACOX3 | 70.1 | 7.6 | 9.9 | 4 | 1.41 | 1.14 |
| D3ZSC5 | Cystathionine beta-synthase | CBS | 52.4 | 6.7 | 33.5 | 7 | 1.44 | 1.01 |
| A9JRE6 | Dmgdh protein (Fragment) | DMGDH | 97.4 | 7.0 | 25.7 | 3 | 1.44 | 0.95 |
| B0BNG1 | Probable proline dehydrogenase 2 | PRODH2 | 51.1 | 8.5 | 21.1 | 8 | 1.46 | 0.77 |
| Q5SPG8 | Acyl-coenzyme A thioesterase 1 | ACOT1 | 48.6 | 8.3 | 9.8 | 3 | 1.47 | 1.04 |
| Q68FT3 | Pyridine nucleotide-disulfide oxidoreductase domain-containing protein 2 | PYROXD2 | 62.8 | 8.2 | 4.5 | 2 | 1.49 | 0.93 |
| Q6NZW7 | Dihydrolipoamide succinyltransferase | DLST | 48.6 | 8.7 | 11.2 | 2 | 1.49 | 0.98 |
| B1WC61 | Acad9 protein | ACAD9 | 68.8 | 7.7 | 23.2 | 13 | 1.50 | 1.26 |
| Q6PBX8 | NADH dehydrogenase [ubiquinone] flavoprotein 2 | NDUFV2 | 26.6 | 5.6 | 13.9 | 3 | 1.52 | 1.19 |
| P21571 | ATP synthase-coupling factor 6, mitochondrial | ATP5J | 12.5 | 9.4 | 46.3 | 6 | 1.72 | 0.88 |
| Q5BK17 | Iodotyrosine dehalogenase 1 | IYD | 32.8 | 6.6 | 7.7 | 2 | 1.83 | 1.14 |
| P00564 | Creatine kinase M-type | CKM | 43.0 | 7.1 | 21.0 | 5 | 3.01 | 2.89 |
| Nucleosome assembly and organization (Cluster 2) | ||||||||
| P15865 | Histone H1.4 | HIST1H1E | 22.0 | 11.1 | 18.7 | 3 | 0.44 | 0.53 |
| Q7ZUY3 | Histone H2A.x | H2AFX | 15.0 | 10.7 | 39.4 | 3 | 0.38 | 0.53 |
| P0C0S7 | Histone H2A.Z | H2AFZ | 13.5 | 10.6 | 31.3 | 2 | 0.68 | 0.74 |
| A7E2M8 | Histone H2B | HIST2H2BE | 13.6 | 10.4 | 48.4 | 2 | 0.50 | 0.53 |
| A3KPR4 | Histone H4 | HIST1H4A | 11.4 | 11.4 | 53.4 | 3 | 0.61 | 0.71 |
| Q6NYV3 | Histone H1.0 | H1F0 | 21.2 | 10.9 | 9.6 | 2 | 0.24 | 0.28 |
| B8JN00 | Histone H2A | H2AFY2 | 24.3 | 10.2 | 9.8 | 2 | 0.62 | 0.56 |
| Translation (Cluster 3) | ||||||||
| P62755 | 40S ribosomal protein S6 | RPS6 | 28.7 | 10.8 | 24.9 | 2 | 1.41 | 0.83 |
| P62083 | 40S ribosomal protein S7 | RPS7 | 22.1 | 10.1 | 44.9 | 5 | 1.66 | 0.87 |
| P23358 | 60S ribomal protein L12 | RPL12 | 17.8 | 9.4 | 59.4 | 5 | 1.67 | 0.90 |
| H7C5Y5 | 60S ribosomal protein L6 | RPL6 | 33.6 | 10.7 | 29.6 | 7 | 1.38 | 0.61 |
| Q6P3V8 | Eukaryotic translation initiation factor 4A1 | EIF4A1 | 46.1 | 5.5 | 37.0 | 6 | 1.58 | 1.00 |
| A4V6L7 | DEAD box polypeptide 48 protein | EIF4A3 | 43.1 | 7.8 | 7.7 | 2 | 1.65 | 1.01 |
| Protein localization (Cluster 3) | ||||||||
| F1M6Z1 | Apolipoprotein B-100 | APOB | 509.4 | 7.2 | 3.1 | 11 | 1.41 | 1.03 |
| Q5PQK5 | Radixin | RDX | 68.5 | 6.3 | 19.0 | 5 | 1.59 | 0.91 |
| Q3B7E7 | Sorting nexin 3 | SNX3 | 18.8 | 8.6 | 12.4 | 2 | 1.80 | 0.81 |
| Q4KM74 | Vesicle-trafficking protein SEC22b | SEC22B | 24.7 | 8.5 | 20.5 | 4 | 1.55 | 0.92 |
| B2RZD1 | Protein Sec61b | SEC61B | 10.0 | 11.6 | 33.3 | 2 | 1.20 | 0.61 |
| Muscle contraction (Cluster 4) | ||||||||
| Q76BG8 | Fructose-bisphosphate aldolase | ALDOA | 36.1 | 7.9 | 29.9 | 8 | 1.86 | 2.15 |
| G3V885 | Myosin-6 | MYH6 | 223.4 | 5.7 | 30.6 | 3 | 1.11 | 1.41 |
| D3ZCV0 | Protein Actn2 | ACTN2 | 103.8 | 5.5 | 31.0 | 8 | 1.66 | 1.51 |
| F1M789 | Protein Myh13 | MYH13 | 208.8 | 5.6 | 21.5 | 5 | 1.67 | 2.36 |
| F1RBV5 | Uncharacterized protein | MYH4 | 221.6 | 5.8 | 26.3 | 2 | 1.47 | 2.22 |
| B6RK61 | Myosin heavy chain 7B | MYH7B | 221.5 | 5.9 | 9.07 | 3 | 1.45 | 1.29 |
| Q7LZ84 | Myosin heavy chain, cardiac and skeletal muscle | MYH7 | 45.2 | 5.8 | 47.04 | 6 | 1.45 | 1.29 |
| Extracellular matrix organization (Cluster 4) | ||||||||
| F1LNH3 | Protein Col6a2 (Fragment) | COL6A2 | 109.4 | 6.6 | 2.0 | 2 | 1.70 | 1.62 |
| Q9W7R9 | Alpha1 type II collagen | COL2A1 | 135.0 | 7.8 | 5.2 | 6 | 1.85 | 1.38 |
| Q9YIB4 | Collagen alpha-1(I) chain | COL1A1 | 137.5 | 5.6 | 21.0 | 24 | 2.37 | 1.19 |
| P02454 | Collagen alpha-1(I) chain | COL1A1 | 137.9 | 5.9 | 3.4 | 2 | 1.69 | 0.94 |
| Q91145 | Collagen alpha-1(XII) chain | COL12A1 | 101.6 | 5.5 | 5.9 | 5 | 1.74 | 1.35 |
| I3NN31 | Collagen type I alpha 2 | COL1A2 | 127.7 | 8.7 | 5.7 | 6 | 1.68 | 1.25 |
| F1QDL1 | Collagen, type I, alpha 3 | COL1A1B | 136.8 | 5.5 | 1.9 | 2 | 1.42 | 0.98 |
*The iTRAQ ratios for 2 h and 6 h using 0 h as control, and the values marked in bold represent up-regulated or down-regulated expression of proteins.
Modulation of various signal pathways at the early stage of planarian head regeneration
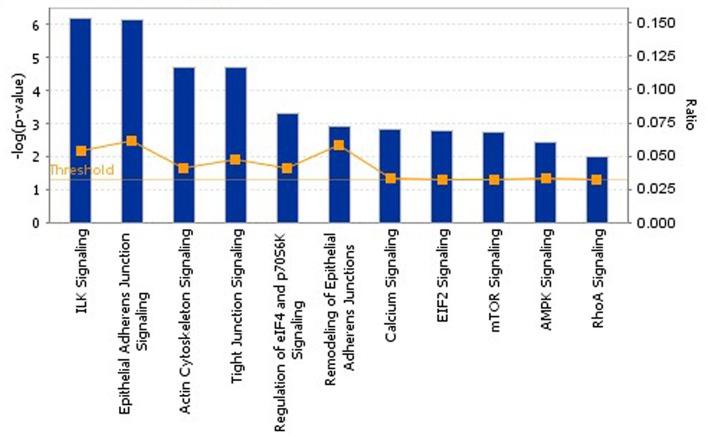
To further clarify which signaling pathways play important roles at the early stage of PHR, pathway analysis was performed to connect the differentially expressed proteins with canonical biological pathways by IPA software. A Benjamini-Hochberg corrected Fischer's exact test was utilized to calculate the p-values associated with a canonical pathway. Additionally, a ratio was used to determine the number of focus molecules to overall molecules in each canonical pathway. Therefore, we analyzed 162 proteins that are differentially expressed at 2 h and 6 h after amputation using IPA, and the significantly enriched canonical pathways are summarized in Fig 3. Among them, “ILK signaling”, “Epithetial Adherens Junction Signaling”, “Actin Cytoskeleton Signaling”, and “Tight Junction Signaling” were highly significantly enriched and involved in cell movement, cell polarity and cell growth. “Regulation of eIF4 and p70S6K Signaling” and “EIF2 Signaling” were highly correlated with protein synthesis through the regulation of translation initiation. “Calcium Signaling” governed the proliferation and differentiation of multipotent stem cells into neuronal fates, and was also found to be regulating planarian regeneration. “mTOR Signaling” plays important roles in regulating cell survival and proliferation. A detailed canonical pathway enrichment of the differentially expressed proteins was provided in S5 Table.
Fig 3. The significantly enriched canonical pathways at the early stage of planarian head regeneration by IPA analysis.
Each histogram is a particular canonical pathway. The size of the histogram is correlated with increasing overlap significance (Fisher’s exact test p-value).
Discussion
Planarians have amazing ability to regenerate through the activation of pluripotent stem cells, and present an attractive system for molecular investigation of regeneration initiation. Although some researches have been performed on brain regeneration and head regeneration [8, 24, 41, 42], the mechanisms that initiate PHR remain unclear. In this study, we for the first time employed an iTRAQ-based shotgun proteomic approach to study the initiation of PHR. By this high-throughput quantification method, 162 proteins were found to be significantly altered at 2 h and 6 h after amputation. These proteins were functionally related to carbohydrate and energy metabolism, protein synthesis, nucleosome assembly, muscle contraction and wound healing, and involved in various signal pathways including ILK signaling, calcium signaling, EIF2 signaling and mTOR signaling. Below, the most important proteins related to the initiation of PHR are discussed based on their putative biological functions and signal pathways.
Carbohydrate and energy metabolism
A proteomics study reported that sugar metabolism and energy metabolism was enriched in planarian stem cells, as energy is required for cellular processes including wound healing and regeneration [30]. Consistent with this result, we also found that carbohydrate and energy metabolism was significantly enriched in cluster 1 and 3. Surprisingly, glycolysis was significantly enriched in cluster 1 with the expression pattern of down-regulation at 2 h and slight up-regulation at 6 h after amputation, and the key enzymes for glycolysis including 6-phosphofructokinase (PFK), enolase (ENO1) and triosephosphate isomerase (TPI1) were identified, indicating that glycolysis might be weakened at 2 h, and slightly increased at 6 h of PHR. Our results further support the idea that the amputated tail initially mobilizes the glycogen and lipid reserves during wound healing stage of tail regeneration in lizard [43]. In addition to 2-oxoglutarate dehydrogenase (E1) as one of the key enzymes associated with tricarboxylic acid cycle (TCA cycle) that was down-regulated at 2 h and up-regulated at 6 h after amputation, other key proteins related to TCA cycle and oxidative phosphorylation identified in our study were up-regulated in protein levels, thereby contributing to the maintenance of cellular energy. Furthermore, we found that creatine kinase M-type (CKM) presented a two to three-fold increase in protein levels. CKM, a cytoplasmic enzyme involved in cellular energy homeostasis, reversibly catalyzed the transfer of "energy-rich" phosphate, thus providing the direct energy source for muscle contraction. that TCA cycle and oxidative phosphorylation played important roles for energy supply These results suggested that TCA cycle and oxidative phosphorylation seemed to be preferentially utilized for ATP supply during the first 6 h after amputation while glycolysis appeared to be weakened at 2 h after amputation, which is contrary to the result that glycolysis seemed to be preferentially utilized for ATP supply by surviving hepatocytes in the regenerating livers immediately after partial hepatectomy [44]. The results need to be validated prospectively through other methods.
Nucleosome assembly
Recent studies reported that the expression level of proteins associated with nucleosome assembly responsible for DNA replication and cell division, was significantly altered in the mitotic regenerative neoblasts that drive the nearly unlimited regenerative power of planarians [42, 45]. In this study, seven histones including H1, H2A, H2B and H4 involved in nucleosome assembly were indentified and grouped into in cluster 2. They exhibited expression patterns of down-regulation at 2 h and 6 h after amputation. More importantly, H2B specifically expressed in planarian neoblasts is likely needed for the progression of neoblasts through the S phase of the cell cycle [46]. We found that H2B was down-regulated at both mRNA and protein level at the early stage of PHR. These results suggested that neoblasts at the wound site had not yet undergone cell division within the first 6 h of PHR, which was confirmed by Wenemoser and Reddien [5] who found that the wound-specific signal that induced the first mitotic peak at 6 h after amputation caused an acceleration of G2 rather than S-phase.
Translation
Studies have reported that protein synthesis was enhanced during PHR after amputation [42, 47]. In the present study, translation was significantly enriched in cluster 3 whose expression pattern was up-regulated at the early stage of PHR. 40S ribosomal proteins, 60S ribosomal protein and eukaryotic translation initiation factors responsible for translation were identified as significantly changed. More importantly, our result was in agreement with the result of Solana et al. [46] that eukaryotic initiation factors important for the initiation of protein synthesis were down-regulated after inhibition of neoblast-inducing planarian regeneration by siRNA experiment. Therefore, we speculate that a certain amount of proteins may be needed for synthesis and assembly of various organelles to facilitate proliferation and growth of neoblasts at the early stage of PHR.
Muscle contraction facilitated wound closure during planarian head regeneration
Wound closure is facilitated by muscle contraction [10]. A study by Witchley et al. [48] reported that muscle tissue is a major source of the positional information that guides tissue turnover and regeneration programs. In the present study, muscle contraction and muscle system process were prominent in cluster 4 by EASE analysis. Meanwhile, we found that muscle contraction-related proteins including myosins and alpha-actinin-2, were up-regulated at 2 h and 6 h after amputation. A recent study of planarian Schmidtea mediterranea by Wenemoser et al. [8] identified several early wound response genes including myosins, and myosins were induced between 3 and 12 h following amputation and required for proper patterning during regeneration. Another study also found that myosin heavy chain-A was expressed in the pharynx muscles and pharynx-anchoring muscles during the regeneration process of the pharynx in the planarian Dugesia japonica [49]. These results suggest that muscle contraction plays important roles in planarian regeneration. In addition, migration of planarian epithelial cells to the wound site is essential for wound closure [50]. We found that collagens (COL1A1, COL1A2, COL2A1, COL6A2, COL12A1) were significantly increased in the protein level at the early stage of PHR, and involved in cell migration, differentiation and regeneration [51–53], indicating that collagens are essential for triggering PHR. These results suggest that muscle contraction and muscle-mediated migration maybe participate in the initiation of PHR by activating the expression of the proteins associated with positional information and muscle contraction.
Important pathways for priming planarian head regeneration
Studies have indicated that the inhibition of canonical Wnt/β-catenin pathway in planarians promoted head regeneration at the wound site [17, 18, 54]. In the present study, we found that the expression level of β-catenin -1 was decreased in the regenerating head fragments at the mRNA level by qRT-PCR analysis, being agreement with the result that inhibition of β-catenin-1 expression caused planarian to regenerate a head instead of a tail at the wound site. In addition to the canonical Wnt/β-catenin pathway, Ca2+ signaling was found to be significantly changed by IPA analysis, being concordant with the findings that Ca2+-dependent transcriptional regulatory events are required for PHR [55, 56], suggesting that Ca2+ signaling might play an essential role in regulating planarian regeneration. Therefore, we concluded that head regeneration in planarians was promoted by the activation of Ca2+ signaling and the inhibition of Wnt/β-catenin pathway.
Furthermore, this study made a discovery that several signaling pathways significantly enriched by IPA analysis were associated with the migration of cells into the wound site, such as ILK signaling, epithetial adherens junction signaling, actin cytoskeleton signaling, and tight junction signaling, indicating that these pathways were likely to participate in regulating the initiation of PHR. Among them, ILK signaling was the most significantly enriched pathway, and ACTN2 and myosins related to this signaling pathway were up-regulated at the early stage of PHR by iTRAQ analysis. Delightedly, studies have found that the activation of ILK after skin wounding is critical for tissue repair and wound healing [22, 57], which confirmed the speculation that ILK signaling might trigger PHR by promoting wound healing. In addition, regulation of eIF4 and EIF2 signaling and EIF2 signaling significantly enriched by IPA analysis were highly correlated with protein synthesis through the regulation of translation initiation. Meanwhile, we also found that eukaryotic translation initiation factors and ribosomal proteins were up-regulated at the early stage of PHR, suggesting that regulation of eIF4 and EIF2 signaling and EIF2 signaling were essential for priming PHR. Recently it has been shown that amputation triggers two peaks in neoblast mitoses during planarian regeneration [5]. The first mitotic peak is a body-wide response to injury and necessary for blastema formation, and this process requires mTOR signaling [58]. In this study, we found that mTOR signaling was significantly enriched at the early stage of PHR. The results further confirmed a key role for mTOR signaling in priming PHR.
Conclusions
The proteomic technology iTRAQ was employed to analyze changes in the expression of proteins in the initiation of PHR. A total of 162 proteins differentially expressed at 2 h and 6h after amputation were identified. On the basis of GO annotation, these proteins were involved in several functional categories including carbohydrate and energy metabolism, translation, muscle contraction, extracellular matrix organization, etc. Meanwhile, we make a discovery that various signaling pathways, especially ILK, calcium, EIF2 and mTOR signaling, were involved in the initiation of PHR by IPA analysis. In conclusion, we for the first time found that muscle contraction and ILK signaling played important roles in the initiation of PHR through global analysis of proteome dynamics. The findings of this research improve our understanding of the molecular mechanisms underlying head regeneration initiation in planarians.
Supporting Information
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
Acknowledgments
We want to thank Shanghai Applied Protein Technology co.ltd for iTRAQ test. We would also like to thank Dr. Cuifang Chang for her kindly providing us with bioinformatics method.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by National Basic Research 973 Pre-Research Program of China (No. 2012CB722304), National Nature Science Foundation of China (No. 31201093), Major Scientific and Technological Projects of Henan (No. 111100910600), Natural Science Foundation of Henan (No. 112300413204 and No. 122300413207). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Gentile L, Cebria F, Bartscherer K (2011) The planarian flatworm: an in vivo model for stem cell biology and nervous system regeneration. Dis Model Mech 4: 12–19. 10.1242/dmm.006692 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wagner DE, Wang IE, Reddien PW (2011) Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332: 811–816. 10.1126/science.1203983 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rink JC (2013) Stem cell systems and regeneration in planaria. Dev Genes Evol 223: 67–84. 10.1007/s00427-012-0426-4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eisenhoffer GT, Kang H, Sanchez Alvarado A (2008) Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell 3: 327–339. 10.1016/j.stem.2008.07.002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wenemoser D, Reddien PW (2010) Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev Biol 344: 979–991. 10.1016/j.ydbio.2010.06.017 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tanaka EM, Reddien PW (2011) The cellular basis for animal regeneration. Dev Cell 21: 172–185. 10.1016/j.devcel.2011.06.016 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pellettieri J, Fitzgerald P, Watanabe S, Mancuso J, Green DR, Sanchez Alvarado A (2010) Cell death and tissue remodeling in planarian regeneration. Dev Biol 338: 76–85. 10.1016/j.ydbio.2009.09.015 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wenemoser D, Lapan SW, Wilkinson AW, Bell GW, Reddien PW (2012) A molecular wound response program associated with regeneration initiation in planarians. Genes Dev 26: 988–1002. 10.1101/gad.187377.112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reddien PW, Sanchez Alvarado A (2004) Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol 20: 725–757. 10.1146/annurev.cellbio.20.010403.095114 . [DOI] [PubMed] [Google Scholar]
- 10. Handberg-Thorsager M, Fernandez E, Salo E (2008) Stem cells and regeneration in planarians. Front Biosci 13: 6374–6394. . [DOI] [PubMed] [Google Scholar]
- 11. Tasaki J, Shibata N, Sakurai T, Agata K, Umesono Y (2011) Role of c-Jun N-terminal kinase activation in blastema formation during planarian regeneration. Dev Growth Differ 53: 389–400. 10.1111/j.1440-169X.2011.01254.x . [DOI] [PubMed] [Google Scholar]
- 12. Tasaki J, Shibata N, Nishimura O, Itomi K, Tabata Y, Son F, et al. (2011) ERK signaling controls blastema cell differentiation during planarian regeneration. Development 138: 2417–2427. 10.1242/dev.060764 . [DOI] [PubMed] [Google Scholar]
- 13. Almuedo-Castillo M, Sureda-Gomez M, Adell T (2012) Wnt signaling in planarians: new answers to old questions. Int J Dev Biol 56: 53–65. 10.1387/ijdb.113451ma . [DOI] [PubMed] [Google Scholar]
- 14. Rink JC, Gurley KA, Elliott SA, Sanchez Alvarado A (2009) Planarian Hh signaling regulates regeneration polarity and links Hh pathway evolution to cilia. Science 326: 1406–1410. 10.1126/science.1178712 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yazawa S, Umesono Y, Hayashi T, Tarui H, Agata K (2009) Planarian Hedgehog/Patched establishes anterior-posterior polarity by regulating Wnt signaling. Proc Natl Acad Sci U S A 106: 22329–22334. 10.1073/pnas.0907464106 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reddien PW, Bermange AL, Kicza AM, Sanchez Alvarado A (2007) BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development 134: 4043–4051. 10.1242/dev.007138 . [DOI] [PubMed] [Google Scholar]
- 17. Gurley KA, Rink JC, Sanchez Alvarado A (2008) Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science 319: 323–327. 10.1126/science.1150029 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Petersen CP, Reddien PW (2008) Smed-betacatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science 319: 327–330. 10.1126/science.1149943 . [DOI] [PubMed] [Google Scholar]
- 19. Petersen CP, Reddien PW (2011) Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science 332: 852–855. 10.1126/science.1202143 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Umesono Y, Tasaki J, Nishimura Y, Hrouda M, Kawaguchi E, Yazawa S, et al. (2013) The molecular logic for planarian regeneration along the anterior-posterior axis. Nature 500: 73–76. 10.1038/nature12359 . [DOI] [PubMed] [Google Scholar]
- 21. Shafiei MS, Rockey DC (2012) The function of integrin-linked kinase in normal and activated stellate cells: implications for fibrogenesis in wound healing. Lab Invest 92: 305–316. 10.1038/labinvest.2011.155 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Serrano I, Diez-Marques ML, Rodriguez-Puyol M, Herrero-Fresneda I, Raimundo Garcia del M, Dedhar S, et al. (2012) Integrin-linked kinase (ILK) modulates wound healing through regulation of hepatocyte growth factor (HGF). Exp Cell Res 318: 2470–2481. 10.1016/j.yexcr.2012.08.001 . [DOI] [PubMed] [Google Scholar]
- 23. Ho E, Dagnino L (2012) Emerging role of ILK and ELMO2 in the integration of adhesion and migration pathways. Cell Adh Migr 6: 168–172. 10.4161/cam.20399 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Agata K, Umesono Y (2008) Brain regeneration from pluripotent stem cells in planarian. Philos Trans R Soc Lond B Biol Sci 363: 2071–2078. 10.1098/rstb.2008.2260 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qin YF, Fang HM, Tian QN, Bao ZX, Lu P, Zhao JM, et al. (2011) Transcriptome profiling and digital gene expression by deep-sequencing in normal/regenerative tissues of planarian Dugesia japonica. Genomics 97: 364–371. 10.1016/j.ygeno.2011.02.002 . [DOI] [PubMed] [Google Scholar]
- 26. Nishimura O, Hirao Y, Tarui H, Agata K (2012) Comparative transcriptome analysis between planarian Dugesia japonica and other platyhelminth species. BMC Genomics 13: 289 10.1186/1471-2164-13-289 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, et al. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 3: 1154–1169. 10.1074/mcp.M400129-MCP200 . [DOI] [PubMed] [Google Scholar]
- 28. Adamidi C, Wang Y, Gruen D, Mastrobuoni G, You X, Tolle D, et al. (2011) De novo assembly and validation of planaria transcriptome by massive parallel sequencing and shotgun proteomics. Genome Res 21: 1193–1200. 10.1101/gr.113779.110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boser A, Drexler HC, Reuter H, Schmitz H, Wu G, Scholer HR, et al. (2013) SILAC proteomics of planarians identifies Ncoa5 as a conserved component of pluripotent stem cells. Cell Rep 5: 1142–1155. 10.1016/j.celrep.2013.10.035 . [DOI] [PubMed] [Google Scholar]
- 30. Fernandez-Taboada E, Rodriguez-Esteban G, Salo E, Abril JF (2011) A proteomics approach to decipher the molecular nature of planarian stem cells. BMC Genomics 12: 133 10.1186/1471-2164-12-133 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Onal P, Grun D, Adamidi C, Rybak A, Solana J, Mastrobuoni G, et al. (2012) Gene expression of pluripotency determinants is conserved between mammalian and planarian stem cells. Embo j 31: 2755–2769. 10.1038/emboj.2012.110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geng X, Xu T, Niu Z, Zhou X, Zhao L, Xie Z, et al. (2014) Differential proteome analysis of the cell differentiation regulated by BCC, CRH, CXCR4, GnRH, GPCR, IL1 signaling pathways in Chinese fire-bellied newt limb regeneration. Differentiation. 10.1016/j.diff.2014.10.002 . [DOI] [PubMed]
- 33. Hsieh HC, Chen YT, Li JM, Chou TY, Chang MF, Huang SC, et al. (2009) Protein profilings in mouse liver regeneration after partial hepatectomy using iTRAQ technology. J Proteome Res 8: 1004–1013. 10.1021/pr800696m . [DOI] [PubMed] [Google Scholar]
- 34. Nicholson J, Neelagandan K, Huart AS, Ball K, Molloy MP, Hupp T (2012) An iTRAQ proteomics screen reveals the effects of the MDM2 binding ligand Nutlin-3 on cellular proteostasis. J Proteome Res 11: 5464–5478. 10.1021/pr300698d . [DOI] [PubMed] [Google Scholar]
- 35. Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Rios D, et al. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol 32: 223–226. 10.1038/nbt.2839 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dong Z, Yuwen Y, Wang Q, Chen G, Liu D (2012) Eight genes expression patterns during visual system regeneration in Dugesia japonica. Gene Expr Patterns 12: 1–6. 10.1016/j.gep.2011.08.001 . [DOI] [PubMed] [Google Scholar]
- 37. Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. 10.1038/nprot.2008.211 . [DOI] [PubMed] [Google Scholar]
- 38. Wang G, Xu C, Zhi J, Hao Y, Zhang L, Chang C (2012) Gene expression profiles reveal significant differences between rat liver cancer and liver regeneration. Gene 504: 41–52. 10.1016/j.gene.2012.04.086 . [DOI] [PubMed] [Google Scholar]
- 39. Kramer A, Green J, Pollard J Jr., Tugendreich S (2014) Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30: 523–530. 10.1093/bioinformatics/btt703 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kulkarni YM, Suarez V, Klinke DJ 2nd, (2010) Inferring predominant pathways in cellular models of breast cancer using limited sample proteomic profiling. BMC Cancer 10: 291 10.1186/1471-2407-10-291 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fraguas S, Barberan S, Iglesias M, Rodriguez-Esteban G, Cebria F (2014) egr-4, a target of EGFR signaling, is required for the formation of the brain primordia and head regeneration in planarians. Development 141: 1835–1847. 10.1242/dev.101345 . [DOI] [PubMed] [Google Scholar]
- 42. Sandmann T, Vogg MC, Owlarn S, Boutros M, Bartscherer K (2011) The head-regeneration transcriptome of the planarian Schmidtea mediterranea. Genome Biol 12: R76 10.1186/gb-2011-12-8-r76 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Alibardi L (2014) Histochemical, Biochemical and Cell Biological aspects of tail regeneration in lizard, an amniote model for studies on tissue regeneration. Prog Histochem Cytochem 48: 143–244. 10.1016/j.proghi.2013.12.001 . [DOI] [PubMed] [Google Scholar]
- 44. Sun Y, Deng X, Li W, Yan Y, Wei H, Jiang Y, et al. (2007) Liver proteome analysis of adaptive response in rat immediately after partial hepatectomy. Proteomics 7: 4398–4407. 10.1002/pmic.200600913 . [DOI] [PubMed] [Google Scholar]
- 45. Guo T, Peters AH, Newmark PA (2006) A Bruno-like gene is required for stem cell maintenance in planarians. Dev Cell 11: 159–169. 10.1016/j.devcel.2006.06.004 . [DOI] [PubMed] [Google Scholar]
- 46. Solana J, Kao D, Mihaylova Y, Jaber-Hijazi F, Malla S, Wilson R, et al. (2012) Defining the molecular profile of planarian pluripotent stem cells using a combinatorial RNAseq, RNA interference and irradiation approach. Genome Biol 13: R19 10.1186/gb-2012-13-3-r19 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen X, Xu C (2015) Protein expression profiling in head fragments during planarian regeneration after amputation. Dev Genes Evol 225: 79–93. 10.1007/s00427-015-0494-3 . [DOI] [PubMed] [Google Scholar]
- 48. Witchley JN, Mayer M, Wagner DE, Owen JH, Reddien PW (2013) Muscle cells provide instructions for planarian regeneration. Cell Rep 4: 633–641. 10.1016/j.celrep.2013.07.022 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sakai T, Kato K, Watanabe K, Orii H (2002) Planarian pharynx regeneration revealed by the expression of myosin heavy chain-A. Int J Dev Biol 46: 329–332. . [PubMed] [Google Scholar]
- 50. Pascolini R, Tei S, Vagnetti D, Bondi C (1984) Epidermal cell migration during wound healing in Dugesia lugubris. Observations based on scanning electron microscopy and treatment with cytochalasin. Cell Tissue Res 236: 345–349. . [DOI] [PubMed] [Google Scholar]
- 51. Wolf K, Alexander S, Schacht V, Coussens LM, von Andrian UH, van Rheenen J, et al. (2009) Collagen-based cell migration models in vitro and in vivo. Semin Cell Dev Biol 20: 931–941. 10.1016/j.semcdb.2009.08.005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Alexander MS, Casar JC, Motohashi N, Myers JA, Eisenberg I, Gonzalez RT, et al. (2011) Regulation of DMD pathology by an ankyrin-encoded miRNA. Skelet Muscle 1: 27 10.1186/2044-5040-1-27 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rankin CR, Hilgarth RS, Leoni G, Kwon M, Den Beste KA, Parkos CA, et al. (2013) Annexin A2 regulates beta1 integrin internalization and intestinal epithelial cell migration. J Biol Chem 288: 15229–15239. 10.1074/jbc.M112.440909 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Iglesias M, Almuedo-Castillo M, Aboobaker AA, Salo E (2011) Early planarian brain regeneration is independent of blastema polarity mediated by the Wnt/beta-catenin pathway. Dev Biol 358: 68–78. 10.1016/j.ydbio.2011.07.013 . [DOI] [PubMed] [Google Scholar]
- 55. Nogi T, Zhang D, Chan JD, Marchant JS (2009) A novel biological activity of praziquantel requiring voltage-operated Ca2+ channel beta subunits: subversion of flatworm regenerative polarity. PLoS Negl Trop Dis 3: e464 10.1371/journal.pntd.0000464 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Beane WS, Morokuma J, Adams DS, Levin M (2011) A chemical genetics approach reveals H,K-ATPase-mediated membrane voltage is required for planarian head regeneration. Chem Biol 18: 77–89. 10.1016/j.chembiol.2010.11.012 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li JF, Duan HF, Wu CT, Zhang DJ, Deng Y, Yin HL, et al. (2013) HGF accelerates wound healing by promoting the dedifferentiation of epidermal cells through beta1-integrin/ILK pathway. Biomed Res Int 2013: 470418 10.1155/2013/470418 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gonzalez-Estevez C, Felix DA, Smith MD, Paps J, Morley SJ, James V, et al. (2012) SMG-1 and mTORC1 act antagonistically to regulate response to injury and growth in planarians. PLoS Genet 8: e1002619 10.1371/journal.pgen.1002619 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.