Abstract
Purpose
According to current diagnostic criteria, mantle cell lymphoma (MCL) encompasses the usual, aggressive variants and rare, nonnodal cases with monoclonal asymptomatic lymphocytosis, cyclin D1–positive (MALD1). We aimed to understand the biology behind this clinical heterogeneity and to identify markers for adequate identification of MALD1 cases.
Experimental Design
We compared 17 typical MCL cases with a homogeneous group of 13 untreated MALD1 cases (median follow-up, 71 months). We conducted gene expression profiling with functional analysis in five MCL and five MALD1. Results were validated in 12 MCL and 8 MALD1 additional cases by quantitative reverse transcription polymerase chain reaction (qRT-PCR) and in 24 MCL and 13 MALD1 cases by flow cytometry. Classification and regression trees strategy was used to generate an algorithm based on CD38 and CD200 expression by flow cytometry.
Results
We found 171 differentially expressed genes with enrichment of neoplastic behavior and cell proliferation signatures in MCL. Conversely, MALD1 was enriched in gene sets related to immune activation and inflammatory responses. CD38 and CD200 were differentially expressed between MCL and MALD1 and confirmed by flow cytometry (median CD38, 89% vs. 14%; median CD200, 0% vs. 24%, respectively). Assessment of both proteins allowed classifying 85% (11 of 13) of MALD1 cases whereas 15% remained unclassified. SOX11 expression by qRT-PCR was significantly different between MCL and MALD1 groups but did not improve the classification.
Conclusion
We show for the first time that MALD1, in contrast to MCL, is characterized by immune activation and driven by inflammatory cues. Assessment of CD38/CD200 by flow cytometry is useful to distinguish most cases of MALD1 from MCL in the clinical setting. MALD1 should be identified and segregated from the current MCL category to avoid overdiagnosis and unnecessary treatment.
Introduction
Mantle cell lymphoma (MCL) is a lymphoproliferative disorder of mature B cells genetically characterized by the presence of t(11;14)(q13;q32) that juxtaposes the CCND1 proto-oncogene, encoding for cyclin D1 at chromosome 11q13, to the immunoglobulin heavy chain gene, at chromosome 14q32. As a consequence, deregulated expression of the cyclin D1 protein occurs in the mature B-cell compartment (1). While t(11;14)(q13;q32) is considered a primary event in the pathophysiology of MCL, at least in part, by deregulating cell-cycle progression in the target cells (2), this chromosome translocation does not seem to be sufficient for the full transformation of B cells. Different experimental and clinical observations suggest that additional oncogenic events are required for the development of MCL (3, 4).
Patients with MCLs have a poor prognosis with a median overall survival between 3 and 5 years (5). However, cases that meet the World Health Organization (WHO) classification diagnostic criteria for MCL, but lack its aggressive clinical course, have been reported and dubbed as indolent forms of MCL. Such cases are typically characterized by leukemic involvement, with or without splenomegaly and absence of lymphadenopathies, for which they have also been referred to as nonnodal MCL (6–13). In the last years, we and others have proposed that some of these asymptomatic cases may represent a preneoplastic condition that could lead to the development of MCL or instead correspond to other types of lymphoproliferative disorders with the t(11;14)(q13;q32) (8, 13–15).
In an attempt to gain further insight into the heterogeneous behavior of MCL, we have compared a series of classical MCL cases requiring chemotherapy with a homogeneous group of asymptomatic individuals harboring a monoclonal expansion of cyclin D1–positive mature B cells in the peripheral blood that carry the t(11;14)(q13; q32), in the absence of splenomegaly or nodal enlargement due to this process. These cases will be hereafter referred to as MALD1 (monoclonal asymptomatic lymphocytosis, cyclin D1–positive), with the intention of not to prejudge the neoplastic nature of the process. Of note, these asymptomatic cases have ever required or received any treatment after a minimum follow-up of 26 months (median, 71 months). In this study, we have sought for biologic differences between MALD1 and classical MCL, investigating molecular pathways that could be enriched in each of these groups. Finally, we selected differentially expressed genes whose protein products could be easily analyzed by flow cytometry to develop a useful tool for distinguishing MALD1 from classical MCL in routine clinical practice.
Materials and Methods
Patients and samples
Thirty patients studied between 1994 and 2012 who fulfilled the WHO diagnostic criteria for MCL and had available cryopreserved peripheral blood samples were included in this study (1). From them, 17 were diagnosed with MCL requiring chemotherapy at diagnosis and 13 corresponded to individuals with monoclonal expansion of mature B cells in peripheral blood carrying the t(11;14) (q13;q32) and displaying cyclin D1 overexpression (MALD1). Aggressive variants with blastoid or pleomorphic features were excluded from the MCL group. Criteria for inclusion in the MALD1 group were as follows: lack of symptoms, no enlargement of lymphoid organs related to the process (by physical examination or by imaging studies), and follow-up >2 years without requiring any treatment along the whole follow-up period. The decision not to treat was based on a number of features including lack of constitutional symptoms, absence of progressive or bulky disease, and no severe cytopenia or impaired end-organ function as a consequence of the process. In this setting, a watchful waiting approach was adopted. Eight healthy individuals with similar age and gender distribution were also included as controls for microarray analyses. This study was approved by the Institutional Review Board of the participating institutions, where required, and was conducted in accordance with the Declaration of Helsinki.
Histological, immunohistochemical, and immunocytochemical studies
Histologic and immunohistochemical analyses were conducted on formalin-fixed, paraffin-embedded (FFPE) samples. Immunocytochemical studies were conducted on airdried formalin/acetone-fixed peripheral blood smears. Cyclin D1 expression was assessed on histologic sections and/or peripheral blood smears of all cases with t(11;14), regardless of CD5 expression. In each case, 2 primary antibodies were used: clone P2D11F11 (Novocastra) and clone SP4 (Neomarkers Inc.). The immunohistochemical analysis of SOX11 was conducted with an anti-SOX11 polyclonal antibody (Atlas Antibodies), as previously described (9). SOX11 immunostaining on decalcified tissues is not sensitive enough so it was not conducted in bone marrow biopsies. The percentage of Ki-67–positive cells (clone MIB-1; Dako) was recorded for all available cases.
Morphologic and immunophenotypic analysis of B lymphocytes
Peripheral blood and bone marrow smears were stained with May–Grünwald–Giemsa. CD38 and CD200 expression was analyzed in peripheral blood by flow cytometry using a mouse IgG1 anti-human CD38 moAb (HB7, BD Biosciences) and a mouse IgG1 anti-human CD200 moAb (MRCOX-104, BD Biosciences). The expression of both markers was assessed with respect to the mean fluorescence intensity observed in cells labeled with a matched isotypic monoclonal antibody. Lymphocytes were gated in the forward and side scatter plot to avoid the inclusion of debris, monocytes, and doublets. Regarding CD200 expression analysis, and to ensure the exclusion of possible contaminating T and natural killer cells, the resultant cells were then gated to select CD2+ cells, which were electronically excluded (invert gate).
G-banding cytogenetics and FISH
Chromosome analyses were conducted on lymphoid cells from peripheral blood and bone marrow 72-hour 12-O-tetradecanoylphorbol-l3-acetate (TPA)-stimulated cultures following standard procedures (16). When karyotypes presented with 3 or more cytogenetic aberrations, these were classified as complex. FISH analysis was conducted on peripheral blood and bone marrow using the IGH/CCND1 dual-color dual fusion translocation probe and ATM and TP53 locus–specific probes (Abbott Molecular). Cutoff values were 1%, 10%, and 10%, respectively. FISH for IGH/CCND1 was also conducted on FFPE samples (cutoff, 15%; ref. 17).
Analysis of the major translocation cluster (MTC) breakpoint region of IGH/CCND1 and cyclin D1 mRNA isoforms
Analysis of the MTC breakpoint region of IGH/CCND1 was conducted following the BIOMED-2 protocol (18). Positive cases were sequenced by capillary electrophoresis. Sequences were aligned to previously reported MTC region sequences using ClustalW. Identification of the IGHJ segments involved in the translocation and alignment to previously reported sequences were conducted using the IgBlast public server. CCND1 mRNA isoforms were assessed as previously described (19).
Mutational studies of immunoglobulin heavy chain V genes
The mutational status of IGHV genes was assessed as previously described (20). Samples in which <2% of base pairs differed from those of the consensus sequence were considered unmutated.
Microarray analyses
Microarray analyses were conducted on RNA isolated from B lymphocytes obtained from peripheral blood by means of immunomagnetic bead selection (B-cell isolation kit II, Miltenyi Biotec GmbH) that yielded purities of ≥90% as assessed by flow cytometry (CD19 APC, clone SJ25C1, BD Biosciences). In cases carrying the t(11;14)(q13;q32) translocation, the percentage of pathologic cells as assessed by flow cytometry was ≥80% of total B lymphocytes.
Microarray expression profiles were obtained using the Affymetrix Human Exon 1.0 ST arrays (Affymetrix). Raw data were normalized to a logarithmic transcript level using the robust multichip average (RMA; ref. 21) and core annotations, obtaining a total of 18,708 transcript clusters. Normalized data were then filtered to avoid noise created by nonexpressed transcript clusters. This filtering was conducted in 2 steps: first, only transcripts with intensity signal >10% of the mean of all intensities of the studied groups were selected; then, those with a variance >80% were considered for further analysis, which lead to 3,479 transcript clusters. Linear models for microarray (LIMMA; ref. 22), a moderated Student t test, was used for detecting differentially expressed genes between groups. Correction for multiple comparisons was conducted using false discovery rate (FDR), and only genes with an adjusted P < 0.05 were considered significant. Hierarchical cluster analysis was also conducted to see how samples and genes aggregated. All data analysis was conducted in R (version 2.15.0) with bioconductor (23) and aroma.affymetrix packages (version 2.7.0; ref. 24). Functional analysis was conducted using the Ingenuity Pathway Analysis (IPA version 8.5; ref. 25) and gene set enrichment analysis (GSEA) computational method (26). The gene sets with P < 0.05 and FDR < 0.2 were considered to be enriched and potentially relevant. The functional heat map was generated by aggregating results of the GSEA and IPA pathway analyses into common categories. The mean of gene expression values belonging to each pathway was taken as the representative value for each sample. Microarray raw data have been deposited in the Gene Expression Omnibus (GEO) database with series accession number GSE45717 (27).
Gene expression analysis by quantitative RT-PCR
cDNAs were prepared from 1 μg RNA using the High-Capacity cDNA Archive Kit (Applied Biosystems). The following genes were analyzed in an ABI Prism 7900HT instrument (Applied Biosystems): CCND1 (Hs00277039_m1), BTLA (Hs00699198_m1), CD200 (Hs01033303_m1), MEF2C (Hs00231149_m1), PON2 (Hs00165563_m1), HMGB3 (Hs00801334_m1), SOX11 (Hs00846583_s1), CD38 (Hs00233552_m1), GAB2 (Hs00373045_m1), LGALS3BP (Hs00174774_m1), MYO6 (Hs00192265_m1), NEO1 (Hs00170143_m1), TXN (Hs00828652_m1), PTK2 (Hs00178587_m1), and ADAMS28 (Hs00248020_m1) along with β-glucuronidase (GUSB; part number 4326320E) using specific commercially available TaqMan gene expression assays. Relative expression levels were calculated with the 2−ΔΔCt method using GUSB as endogenous control.
Generation of the CD38/CD200 classifier
The Classification and Regression Trees (CART) strategy was used to generate a classification algorithm based on CD38 and CD200 expression assessed by flow cytometry. This algorithm was built using R with the Rpart package. Rpart is a nonparametrical 2-step method, which can be easily represented as a decision tree graph. The first step finds the single variable that best splits the cases into 2 groups, on the basis of the minimum Gini index, an estimation based on all possible splits using all available variables. All possible cutoff values for all variables considered in the model are tested to select the split that most decreases the Gini index at the first node. The cases are then assigned to each newly generated node and this process is recursively applied to each of the descendant nodes, until no improvement can be achieved. In the second step, the resulting full tree is trimmed back using a cross-validation procedure (28). The 95% confidence interval (CI) of the proportions of correctly classified patients by the proposed decision tree were computed using methods with continuity correction (29).
Statistical analyses
Comparisons were conducted using the Mann–Whitney U test for continuous and ordinal variables and the Fisher exact test for categorical variables. P < 0.05 from 2-sided tests was considered to indicate statistical significance. The probability of survival was assessed with actuarial life tables, and 95% CIs were calculated. The SPSS 16.0 statistical software package (SPSS) was used for all statistical analyses.
Results
Clinical presentation, laboratory findings, and follow-up of MCL and MALD1 cases
The main clinical and laboratory features at diagnosis are detailed in Table 1 and Supplementary Tables S1 and S2. The reason for referral was different between the 2 groups: while most patients with MCL presented with palpable lymphadenopathies (14 of 17), 12 of 13 MALD1 cases were referred because of abnormal routine peripheral blood tests, including lymphocytosis (7 of 13 cases) and atypical lymphocytes in peripheral blood smears with a normal lymphocyte count (5 of 13 cases). In the remaining case, referral was due to a single submandibular lymphadenopathy related to an inflammatory condition (sarcoid granulomatous reaction). In most MALD1 cases, the morphology of abnormal lymphocytes in blood smears was similar to that of typical chronic lymphocytic leukemia (CLL) and monoclonal B-cell lymphocytosis (MBL) without smudge cells, whereas in MCL cases, the atypical lymphocytes displayed more irregular nuclear contours, as previously reported (30). The absolute lymphocyte count at presentation ranged from 3.1 to 12.2 × 109/L in MALD1 cases and from 0.7 to 69 × 109/L in patients with MCL. MALD1 lymphocyte counts over time have been plotted in Supplementary Fig. S1. All MALD1 individuals and most patients with MCL (69%) showed peripheral blood involvement by flow cytometry, with percentages of abnormal lymphocytes ranging from 11% to 76% in MALD1 cases and from 0% to 86% in patients with MCL. There were no significant differences between MALD1 (5/13) and MCL (6/14) cases regarding CD23 expression, nor in the intensity of surface immunoglobulin expression. Interestingly, the moderate/high intensity of surface immunoglobulin expression in all MALD1 cases (13 of 13) was different to that reported in MBL with typical CLL phenotype, usually negative or weakly positive (31).
Table 1.
Summary of clinical and pathologic characteristics, as well as clonal diagnostic markers, of the cases included in the present study
| Variable | MCL (n = 17) | MALD1 (n =13) | P |
|---|---|---|---|
| Age ≥ 60 y | 11/17 (65) | 7/13 (54) | 0.711 |
| Males | 7/17 (41) | 7/13 (54) | 0.713 |
| B symptoms | 4/17 (24) | 0/13 (0) | 0.113 |
| Poor performance status (ECOG ≥ 2) Splenomegaly | 3/17 (18) | 0/13 (0) | 0.238 |
| 7/14 (50) | 0/13 (0) | 0.051 | |
| Hemoglobin level, g/L | 117 (67–163) | 155 (94–172) | 0.012 |
| Platelet count, ×109/L | 183 (50–402) | 200 (114–400) | 0.391 |
| Leukocyte count, ×109/L | 8.5 (3.8–87) | 12.1 (7.0–18.9) | 0.286 |
| Lymphocyte count, ×109/La | 3.6 (0.7–69) | 6.5 (3.1–12.2) | 0.187 |
| Albumin < 30 g/L | 1/17 (6) | 0/13 (0) | 1 |
| ESR, mm | 39 (7–140) | 12 (2–31) | 0.001 |
| LDH level ≥ 450 IU/L | 3/17 (19) | 1/13 (8) | 0.613 |
| β2-Microglobulin level ≥ 2.1 mg/L | 12/17 (70) | 1/13 (8) | 0.001 |
| Monoclonal component | 3/17 (18) (IgGL, IgMK) | 2/13 (15) (IgGL, IgMK) | 1 |
| Advanced stage (Ann Arbor III–IV) | 15/17 (88) | 13/13 (100) | 0.492 |
| Extranodal involvement | |||
| ≤1 | 10/17 (59) | 10/13 (77) | 0.440 |
| >1 | 7/17 (41) | 3/13 (23) | |
| Bone marrow infiltration (histology and/or cytology) IPI |
13/16 (81) | 10/10 (100) | 0.262 |
| Low risk | 7/17 (41) | 4/13 (31) | 0.356 |
| Low-intermediate risk | 2/17 (12) | 8/13 (61) | |
| High-intermediate risk | 5/17 (29) | 1/13 (8) | |
| High risk | 3/17 (18) | 0/13 (0) | |
| MIPI | |||
| Low risk | 10/17 (59) | 6/13 (46) | 0.872 |
| Intermediate risk | 1/17 (6) | 6/13 (46) | |
| High risk | 6/17 (35) | 1/13 (8) | |
| Diffuse pattern in secondary lymphoid organs | 7/14 (50) | 0/6 (0) | 0.106 |
| Cyclin D1 expression (IHC and/or ICC) | 17/17 (100) | 13/13 (100) | 1 |
| CD5 expression (IHC and/or FC) | 16/16 (100) | 10/13 (77) | 0.070 |
| SOX11 expression (IHC) | 11/11 (100) | 0/5 (0) | <0.001 |
| Percentage of Ki-67–positive cells in secondary lymphoid organs (IHC) | 20 (10–35) | 5 (3–5) | 0.002 |
| t(11;14) alone (CGC) | 2/8 (25) | 5/13 (39) | 0.183 |
| Complex karyotype (CGC) | 4/15 (50) | 1/13 (8) | 0.333 |
| TP53 deletion (FISH) | 3/14 (21) | 1/13 (8) | 0.596 |
| ATM deletion (FISH) | 4/14 (29) | 0/13 (0) | 0.098 |
| MTC rearrangement (PCR) | 4/17 (24) | 5/13 (39) | 0.443 |
| IGHV hypermutated | 4/13 (31) | 12/12 (100) | <0.001 |
| SOX11 expression (qRT-PCR) | 430 (0–1201) | 10 (0–100) | 0.003 |
NOTE: Results are expressed as cases/total cases (%) for the categorical variables and median (range) for continuous variables. For further details, see Supplementary Tables S1 and S2. Parameters showing statistically significant differences between both groups are shown in bold.
Abbreviations: CGC, conventional G-banding cytogenetics; ECOG, Eastern Cooperative Oncology Group; ESR, erythrocyte sedimentation rate; FC, flow cytometry on peripheral blood; ICC, immunocytochemistry on peripheral blood; IHC, immunohistochemistry on secondary lymphoid organs and/or bone marrow; IPI, international prognostic index; LDH, lactate dehydrogenase; MIPI, MCL international prognostic index.
Normal values for lymphocyte counts in the hospitals submitting cases ranged from 1 × 109/Lto1.5 × 109/L for minimum values and from 3 × 109/L to 5 × 109/L for maximum values.
MCL cases showed lower hemoglobin level (P = 0.012), with higher erythrocyte sedimentation rates (ESR) and β2- microglobulin levels than MALD1 cases (P = 0.001 for both variables). Regarding disease stage and both the IPI and MIPI prognostic indexes, no differences were observed between MCL and MALD1 cases. Of note, 69% and 54% of MALD1 cases were assigned to the IPI and MIPI intermediate- or high-risk groups, respectively. Overall survival of patients with MCL at 2 and 5 years was 88% and 73%, respectively; conversely, all MALD1 individuals remain alive and have not received any treatment with a median follow-up of 71 months (range, 26–186 months).
MALD1 and MCL show distinct histological features
The main histologic and immunohistochemical features of both groups are detailed in Table 1 and Supplementary Table S1. In patients with MCL, the involvement of different organs was apparent in hematoxylin and eosin (HE)- stained slides due to the disruption of normal architecture, and it was confirmed by immunohistochemistry (IHC) and FISH techniques. Tissue samples of secondary lymphoid organs were available from 6 MALD1 cases (upper and lower gastrointestinal tract samples in all 6 cases and lymph nodes in two of them). Noteworthy, the presence of the abnormal lymphoid population would not have been suspected by histology alone, in the absence of previous flow cytometry and/or cytogenetic data. The immunohistochemical analysis of secondary lymphoid organs from MALD1 cases identified cyclin D1–positive small lymphoid cells with predominantly round nuclei in 5 of 6 cases. The cyclin D1–positive cells were mainly located in the lymphoid aggregates lacking germinal centers and in the mantle zones of secondary follicles, without any significant expansion of these areas, thus displaying a so-called in situ histologic pattern. Bone marrow biopsies were available from the aforementioned 6 MALD1 cases. Immunohistochemical analysis revealed the presence of a cyclin D1–positive interstitial lymphoid infiltrate in all of them, which was usually overlooked in HE sections. Only SOX11 and Ki-67 expression were significantly different between MCL and MALD1 groups (P < 0.001 and P = 0.002, respectively). SOX11 was negative in the 5 MALD1 cases in which colonization of secondary lymphoid organs by cyclin D1–positive cells could be proven. On the other hand, 11 of 11 MCL cases showed strong SOX11 nuclear expression, along with a higher percentage of Ki-67–positive cells.
MALD1 shares diagnostic cytogenetic and molecular features with MCL
The main cytogenetic and molecular features are detailed in Table 1 and Supplementary Table S2. All MALD1 and MCL cases displayed a t(11;14)(q13;q32) by G-banding cytogenetics and/or FISH. qRT-PCR analysis confirmed increased CCND1 mRNA expression levels in MALD1 and MCL samples when compared with healthy controls, and these higher levels correlated with cyclin D1 protein overexpression, as detected by IHC. In 0 of 12 MALD1 and 1 of 7 MCL cases, the truncated CCND1 mRNA isoform, associated to shorter survival, was identified (19). Analysis of IGH/CCND1 gene rearrangements at the CCND1 MTC breakpoint region showed a similar percentage of events involving this region in MCL and MALD1 cases. There was a difference in size between the PCR product obtained from MALD1 cases (median size of the PCR product, 269 bp; range, 202–288) and that from patients with MCL (median size, 212 bp; range, 207–213). However, the structural similarities between MALD1 and MCL breakpoints suggest that the mechanisms leading to the IGH/CCND1 translocation in both groups could not differ in their basis (Supplementary Table S3; ref. 32). Finally, all MALD1 cases analyzed showed clonal IGHVDJ gene rearrangements.
MALD1 cells are involved in immune reactions and show evidence of transit through the germinal center
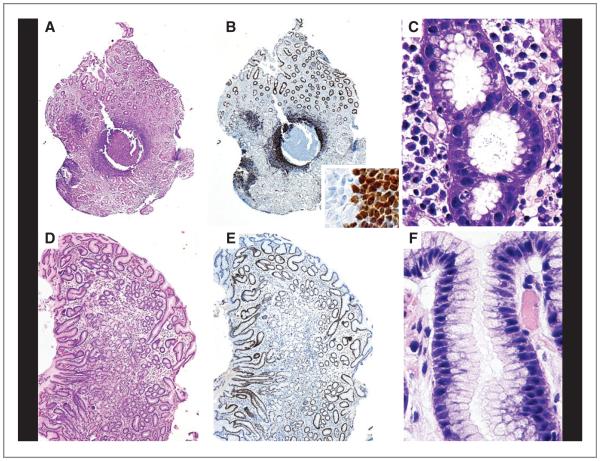
As a part of the routine staging protocol for MCL, upper and lower endoscopy was conducted in most cases (33). A follicular chronic gastritis associated with Helicobacter pylori infection was detected in gastric mucosa biopsies from three MALD1 subjects. Immunohistochemical analysis of these samples revealed numerous cyclin D1–positive lymphoid cells in the follicular mantles of the acquired MALT (Fig. 1A–C). FISH analysis confirmed the presence of t(11;14) (q13; q32) in these cells, and molecular analysis showed the same IGH clonal rearrangements as those found in the other tissues studied (data not shown). Biopsies taken after routine antibiotic treatment for the gastric infection revealed a slight residual atrophy without evidence of bacilli and, more importantly, complete absence of lymphoid infiltrates (Fig. 1D–F). In contrast, in a MCL patient with concurrent H. pylori infection and tumor infiltration, the antibiotic treatment eliminated the H. pylori but not the cyclin D1–positive tumor cells.
Figure 1.
MALD1 participate in immune reactions. Top, chronic gastritis in a MALD1 case. A, lymphoid follicles formation in the lamina propria (HE; ×20). B, main image: numerous lymphoid cells overexpressing cyclin D1 in the mantle zone (IHC; ×20); inset: boundary between the germinal center and mantle zone (IHC; ×700). C, abundant H. pylori in the lumen of the gastric pits (Giemsa; ×900). Bottom, posttreatment study of the same case. D, mild mucosal atrophy without inflammation (HE; ×40). E, absence of lymphoid cells expressing cyclin D1 (IHC; ×40). F, absence of H. pylori in the lumen of the gastric pits (Giemsa; ×900).
The above findings suggested that despite their clonal nature, MALD1 cells could actively participate or be recruited to antigen-driven immune reactions and would disappear from the involved organ when the antigen is cleared. Thus, we next analyzed whether there was any molecular evidence supporting the involvement of MALD1 cells in immune reactions, by assessing the presence of somatic hypermutation events in the IGHV genes. Analysis of the mutational status of the IGHV genes could be conducted in 12 of 13 MALD1 individuals and in 13 of 17 patients with MCL. All MALD1 cases showed mutated IGHV genes, whereas only 4 of 13 patients with MCL had somatic mutations (P < 0.001; Table 1 and Supplementary Table S2). No significant bias in the usage of VH segments was observed in any of the 2 groups. The occurrence of somatic hypermutation events in IGHV genes all MALD1 cases indicates that MALD1 lymphocytes may have transited through the germinal center. On the other hand, the percentage of somatic hypermutations in the patients with MCL of the present study is 31%, in agreement with previous series (34–36). Remarkably, analysis of the sequences of VH segments in sequential samples of 3 MALD1 individuals showed no evidence of acquired (ongoing) somatic hypermutation events from the time of diagnosis (data not shown). This finding suggests that transit through the germinal center in MALD1 lymphocytes probably occurred before their clonal expansion, an observation that contrasts with the pregerminal center origin postulated for MCL (15, 34).
Gene expression profile of clonal B cells reveals significant biological differences between MALD1 and MCL
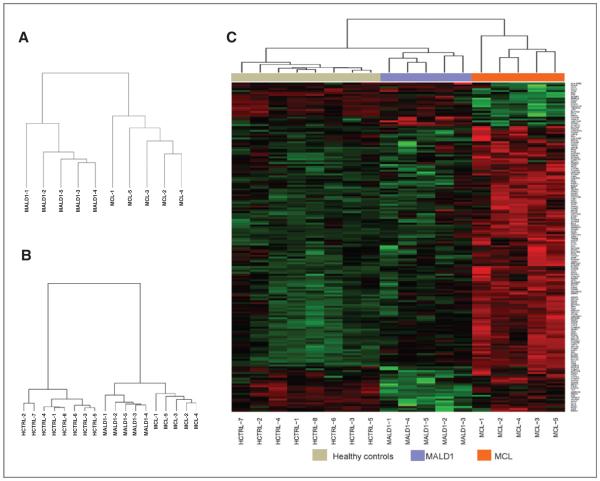
To better understand the basis for possible biologic differences between MCL and MALD1, we analyzed the gene expression profiles of enriched B-cell fractions isolated from peripheral blood of 5 MALD1 subjects, 5 patients with MCL, and 8 healthy individuals. Unsupervised hierarchical clustering of gene expression data revealed robust differences between MALD1 and MCL B cells (Fig. 2A). When compared with normal peripheral blood B cells from healthy individuals, MALD1 and MCL clustered together, indicating that peripheral blood B cells from both groups share significant differences with normal B cells (Fig. 2B). Supervised analysis between MALD1 and MCL samples identified a gene signature composed of 174 differentially expressed transcript clusters (171 genes). This signature clustered together MALD1 and normal B cells (Fig. 2C, Supplementary Table S4). The microarray results were confirmed by qRT-PCR analysis of a subset of top differentially expressed genes (Supplementary Fig. S2). Validation was conducted in the same 10 cases (5 MCL and 5 MALD1) used for microarray analyses. The 20 remaining cases (12 MCL and 8 MALD1) were used as a validation set.
Figure 2.
Gene expression analysis of CD19+ peripheral blood B cells reveals significant differences between MALD1 and MCL. A, unsupervised hierarchical clustering of the gene expression profiles (GEP) from healthy controls (HCTRL), MALD1, and MCL cases. B, unsupervised hierarchical clustering of GEP from MALD1 and MCL cases separates the 2 entities based on distinct gene signatures. C, heatmap showing the results of the supervised analysis using genes differentially expressed between MALD1 and MCL cases. The list of genes appearing in the heatmap is detailed, in order of appearance, in Supplementary Table S4.
The MALD1/MCL gene signature included CD38 [log fold change (logFC) = 1.51; adj. P = 0.024], upregulated in MCL, and CD200 (logFC = 2.04; adj. P = 0.005), upregulated in MALD1, 2 flow cytometric markers commonly used in the study of peripheral blood lymphocytosis. Unlike previous reports (9, 37), SOX11 was not part of the MALD1/MCL gene signature, even though statistically significant differences in SOX11 mRNA expression between MALD1 and MCL were detected by qRT-PCR (Supplementary Fig. S2). This apparent discordance can be explained by the differences between the probe sets included in the Human Exon 1.0 ST array (used in this study), and the 3′IVT array probe sets (HGU133 Plus2 arrays) used in those studies (Supplementary Fig. S3).
Pathway analysis using the IPA tool indicated that the MALD1/MCL gene signature was significantly enriched in genes involved in 2 main biologic processes, namely inflammatory responses in MALD1, and cell growth and proliferation in MCL (Fig. 3, Supplementary Table S5). These differences were confirmed and further refined using the GSEA tool. Many cancer-related signatures (cell cycle and proliferation, Myc deregulation, and self-renewal), as well as gene signatures associated to cyclin D1/Cdk4 activation, were specifically enriched in MCL. In addition, we observed a significant enrichment for DNA damage repair pathways (ATM and global genomic nucleotide excision repair pathways). Both cell cycle and DNA damage repair–related pathways have been previously invoked in the pathogenesis of MCL (28, 29). In contrast, MALD1 gene expression profiles were highly enriched in gene sets related to inflammatory responses, B-cell activation, immunoregulatory interactions, cytokine signaling, and lymphocyte/cell adhesion, and migration, suggesting that inflammatory cues drive the biology of these cells, as also supported by the histologic data (Fig. 1). Thus, gene expression analysis identified significant differences in selected gene expression programs between MCL and MALD1 B cells, in concordance with the clinical and histologic evidence.
Figure 3.
Biologic pathways differentiate MALD1 and MCL cells. Functional heatmap showing relative enrichment of biologic pathways in the 2 groups. This heatmap was generated by aggregating results of the GSEA and IPA analyses in common categories and taking the mean of gene expression in each pathway as the representative value for each sample.
Surface CD38/CD200 expression by flow cytometry is useful to discriminate between MALD1 and MCL
Among other specific surface molecules found to be differentially expressed between MALD1 and MCL, we chose to further validate CD200 and CD38 on peripheral blood samples, given their widespread use in routine phenotypic panels for the study of lymphocytosis (30). Particularly, CD38 was highly expressed in patients with MCL but not in MALD1 cases, an observation further confirmed by qRT-PCR analysis (P = 0.007; Fig. 4A). Flow cytometric data on CD38 surface expression in peripheral blood were available in 37 cases (11 of 17 MCL and 13 of 13 MALD1 from the original series, and 13 additional MCL cases). In agreement with the molecular data, patients with MCL displayed a significantly higher percentage of CD38+ B cells (median, 89%; range, 0%–100%) than that observed among MALD1 individuals (median, 14%; range 0%–35%; P < 0.001; Fig. 4B and C and Supplementary Table S2).
Figure 4.
Differences in CD38 and CD200 surface protein and mRNA expression between MALD1 and MCL cases. A, box plots representing the mRNA expression levels of CD38 in the 2 groups, as determined by qRT-PCR and expressed in relative units (Mann–Whitney test). B, box plots depicting the distribution of CD38-positive cells within the clonal peripheral blood B-cell population in the 2 groups (Mann–Whitney test), as assessed by flow cytometry. C, representative examples of flow cytometry analysis of CD38 surface expression in representative MALD1 (top) and MCL (bottom) peripheral blood B cells (CD19+, CD5+, highlighted in red on the right). The mean fluorescence intensity is used as a parameter to plot the levels of CD38 expression (histograms). D, box plot representation of CD200 mRNA total levels in MALD1 and MCL cases and expressed in relative units (Mann–Whitney test). E, box plots depicting the distribution of CD200-positive cells within the clonal peripheral blood B-cell population in the 2 groups (Mann–Whitney test), as assessed by flow cytometry. F, representative examples of flow cytometric analysis of CD200 surface expression in representative MALD1 (top) and MCL (bottom) peripheral blood B cells (highlighted in red) using the mean fluorescence intensity as a parameter (histograms).
Regarding CD200, it was highly expressed in MALD1 cases but not in patients with MCL, and this observation was confirmed by qRT-PCR (P < 0.001; Fig. 4D). CD200 surface expression was available in peripheral blood B-cell lymphocytes of 27 cases (2 of 17 MCL and 12 of 13 MALD1 from the original series, and 13 additional MCL cases; Fig. 4E and F, Supplementary Table S2). In agreement with the qRT-PCR data, all MCL cases showed very low or absent CD200 expression (median, 0%; range, 0%–5%), whereas the majority of MALD1 cases analyzed showed a higher percentage of CD200+ B cells (median, 24%; range, 0%–95%; P < 0.001). These observations suggest that flow cytometric analysis of CD38 and CD200 expression on clonal B cells from peripheral blood samples is useful for the distinction between MCL (CD38+/CD200−) and MALD1 cases (CD38−/CD200+). On the basis of these findings, we designed a classifier to discriminate between both entities in the clinical setting. We generated a CART based on 2 splits, the first one, CD38 ≥ 40%, classifies patients as MCL, whereas the second split, CD200 ≥ 2%, assigns the MALD1 cases (Fig. 5A). We have considered a third non-classified category, encompassing cases with low values for both markers. This algorithm correctly classified 88% (21 of 24; 95% CI, 67%–97%) of the patients with MCL and 85% (11 of 13; 95% CI, 54%–97%) of the MALD1 cases (Fig. 5B and Supplementary Table S6), the rest of the cases remaining as non-classified. Of note, none of the cases was classified in the opposite category. To better characterize the non-classified cases (2 MALD1 and 3 MCL), we analyzed whether SOX11 expression assessed by qRT-PCR could help in classifying these cases. The 2 MALD1 individuals (MALD1-5 and MALD1-10) displayed low levels of SOX11 expression, as expected. On the other hand, in the 2 MCL cases (MCL-1 and MCL-14) with available sample, SOX11 expression was also low and showed mutated IGHV genes. Interestingly, when analyzing the whole series, mutated IGHV cases showed statistically significant higher levels of SOX11 expression than unmutated cases (P = 0.001). These differences were maintained when considering only patients with MCL (P = 0.027).
Figure 5.
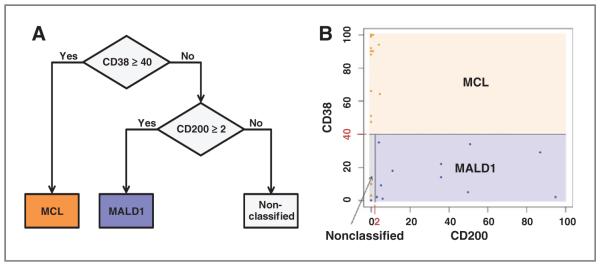
CD38 and CD200 surface expression are helpful to discriminates between MCL and MALD1. A, resulting decision tree graph based on CD38 and CD200 expression assessed by flow cytometry classifies cases in 3 groups: MCL, MALD1, and non-classified. B, graphical distribution of studied cases depicted in blue (MALD1) and orange (MCL). Blue box corresponds to MALD1 assigned group by the tree, orange box corresponds to MCL assigned group, and gray box to the non-classified group.
Discussion
On the basis of current diagnostic criteria, MCL encompasses a spectrum of cases ranging from the usual, aggressive variants to rare, nonnodal cases that remain asymptomatic for many years, even without treatment (6–13, 38). This clinical heterogeneity probably reflects incomplete knowledge of the MCL pathogenesis, with the resulting diagnostic oversimplification. In fact, current prognostic indexes for MCL (IPI, MIPI; refs. 39, 40) are not suitable for MALD1 cases, as they would label some of them as intermediate or even high-risk MCL, mainly due to the presence of leukocytosis and/or to advanced age, despite their benign clinical course (34). It has been speculated that some of the asymptomatic cases represent a preneoplastic condition that would either lead to the development of MCL or remain stable lifelong (7–13, 15, 41, 42). It is even possible that some asymptomatic cases could belong to a different biologic category sharing only some of the diagnostic features of MCL (8, 13, 15). Therefore, it is crucial to gain further insight into the molecular differences between MCL and MALD1 and to find biologic markers that, in addition to clinical presentation, allow identifying MALD1 cases and avoid their overtreatment.
While previous series dealing with this topic are rather heterogeneous (7, 9, 10, 12, 13), we have attempted to characterize a homogeneous group of cases (MALD1) presenting without splenomegaly or nodal enlargement due to the disease and never requiring treatment despite a long follow-up (median, 71 months). This group was compared with a cohort of classical patients with MCL, from which aggressive histologic variants had been excluded to avoid confounding factors related to disease progression. Despite their clonal nature and the presence of t(11;14)(q13;q32), MALD1 cells lack several common features of MCL B lymphocytes and are defined by a paucity of genomic abnormalities, a low proliferative fraction, and their recruitment to active inflammatory/immune foci. Moreover, the normal architecture was preserved in all lymphoid organs analyzed from MALD1 individuals in our series. Characteristically, MALD1 cells have cyclin D1 overexpression and locate only to normal lymphoid structures (i.e., mantle zones), as normal B cells do in immune reactions. In this regard, we have shown for the first time that in those MALD1 cases with H. pylori–associated chronic gastritis, cyclin D1–positive cells disappear from the gastric mucosa after antigen elimination by antibiotic therapy (Fig. 1) whereas these cells persist in MCL cases. This different behavior could reflect that presence of MALD1 cells in the gastric mucosa is dependent on the preservation of their homing compartment (acquired MALT). In agreement with these observations, we have also first shown that MALD1 expression profiles are highly enriched in gene sets related to immune activation and inflammatory responses, indicating that inflammatory cues drive the biology of these cells (Fig. 3, Supplementary Table S5). In contrast, gene expression analysis only detected enrichment of signatures associated with neoplastic behavior and proliferation in MCL cases. Consistent with the above observations, MALD1 B lymphocytes showed features of antigen-experienced cells, namely, the presence of IGHV hypermutations, as well as a replication history (determined by the abundance of IgK-deleting rearrangement excision circles; KREC assay, data not shown; ref. 43), that indicate occasional transit through the germinal center. Notably, the VH segment sequences in consecutive samples of 3 MALD1 individuals showed no evidence of ongoing somatic hypermutation events from the time of diagnosis, strongly suggesting that transit through the germinal center of MALD1 lymphocytes probably occurred before their clonal expansion. These observations contrast with the pregerminal center origin postulated for most MCL (1).
It is well known that single genetic events are not sufficient for malignant transformation (44, 45). Isolated genetic lesions like t(11;14) should not be used as the only criterion to support the diagnosis of lymphoma. In fact, despite being the primary event in the pathogenesis of MCL, additional oncogenic events are known to be required for malignant transformation (3, 46). While the biologic and clinical behavior of MALD1 is far from that expected in neoplastic cells, we have shown that MALD1 lymphocytes carry genetic abnormalities unseen in normal B cells as t(11;14) and clonal expansion. This could eventually prime them for neoplastic transformation in the proper biologic context. Given the exceptionality and limited knowledge of preneoplastic lesions in the lymphoid system, we cannot rule out that MALD1 may act in the long term as a precursor of MCL, similar to what has been reported for MBL and CLL (31, 47–49). In this regard, MALD1 cases would be related to the fraction (20%–40%) of MCL cases with hypermutated IGHV genes, which typically show nonnodal disease at presentation, an indolent clinical course, and a long disease-free survival (35, 36). Finally, according to our results, another possibility that cannot be excluded is that MALD1 cases might instead belong to a distinct disease category, biologically unrelated to MCL.
Several groups have addressed the need for reliable markers to identify cyclin D1–positive cases not requiring early treatment. Among them, SOX11, for which an oncogenic role has been recently proposed, has been the most widely explored (50, 51). Lack of SOX11 expression, alone or in combination with other genes, has been proposed as a candidate marker with conflicting results (9, 13, 52). The limitation of these markers is that, as the majority of asymptomatic cases only show peripheral blood expression, the assessment must be made by qRT-PCR in isolated B cells from peripheral blood. As these techniques are not readily implementable in all laboratories, one of the aims of our study was to identify differentially expressed genes between MALD1 and MCL, providing the basis for a simple routine diagnostic test. Among the differentially expressed and validated genes in our study, we selected CD38 and CD200 because antibodies for their proteins are already used in current diagnostic flow cytometric panels for mature B-cell lymphoproliferative processes (53).
CD38 is an ectoenzyme with an unclear function in B cells (54). CD38 expression is a well-recognized prognostic factor in CLL and it has also been suggested as a marker that partially differentiates nodal from nonnodal MCL (7). In our study, CD38 expression was found to be consistently high in MCL whereas low or absent in MALD1 cases. It has been proposed that CD38 may favor neoplastic B-cell survival by engaging CD31, an adhesion molecule expressed by endothelial cells (55). In MALD1 cases, low CD38 expression may minimize the interaction of MALD1 B cells with CD31-expressing microvessels, thus attenuating clonal B-cell accumulation.
CD200 is an Ig superfamily member frequently upregulated in CLL and has been proposed as a useful marker for the differential diagnosis between CLL and MCL (56, 57). CD200 was found to be consistently low or absent in all MCL but highly expressed in most MALD1 cases. CD200 binds to CD200R, an inhibitory receptor expressed by both lymphoid and myeloid cells. It is tempting to speculate that an increased frequency of inhibitory signals emerging from CD200–CD200R interaction attenuates the proliferation of MALD1 B cells, thus contributing to the asymptomatic behavior of MALD1 cases. Conversely, reduced CD200 expression by conventional MCL may contribute to its more aggressive biologic behavior.
Despite the low prevalence of MALD1 cases, we have generated an algorithm based on CD38 and CD200 expression that correctly assigned most MALD1 and MCL cases; more importantly, such algorithm did not classify any of these cases in the opposite category. SOX11 expression by qRT-PCR did not improve the classification obtained by the algorithm. Our results indicate that a combined analysis of CD38 and CD200 by flow cytometry could be used to discern between MALD1 and MCL. However, as some cases show close values to the cutoff of both markers, these parameters should be considered in the whole clinical and biologic context.
In summary, we have studied a very homogeneous group of untreated asymptomatic individuals with monoclonal cyclin D1–positive lymphocytosis and a long follow-up (MALD1). We have shown for the first time that these cases are characterized by immune activation and driven by inflammatory cues. This is in contrast with the biology of overt MCL and translates into a benign clinical behavior. Although more studies with a larger number of cases are needed, our results suggest that combined flow cytometric assessment of CD38 and CD200 expression provides a simple and useful tool to distinguish most MALD1 cases from patients with MCL in the clinical setting, which may contribute to avoid overdiagnosis and unnecessary treatment of subjects with MALD1.
Supplementary Material
Translational Relevance.
This article shows for the first time that monoclonal asymptomatic lymphocytosis, cyclin D1–positive (MALD1), in contrast to typical mantle cell lymphoma (MCL), is characterized by immune activation and is driven by inflammatory cues. This distinct biology translates into a benign clinical course. It also shows that assessment of CD38 and CD200 by flow cytometry is useful to distinguish most cases of MALD1 from MCL in the clinical setting. According to these results, most of the MALD1 cases can be clearly identified and segregated from the current MCL category avoiding overdiagnosis and unnecessary treatment.
Acknowledgments
The authors thank Laura Pasqualucci, Katia Basso and Paolo Guarneri (Columbia University, New York, NY), Juan Valcárcel (CRG, Barcelona), Francesc Bosch (Hospital Universitari Vall d’Hebron, Barcelona, Spain), and Elías Campo (Hospital Clínic, Barcelona, Spain) for valuable comments and insights, and critical reading of the manuscript; Marta Pulido (IMIM, Barcelona, Spain) for language assistance; Jesús M Hernández-Rivas (Hospital Clínico de Salamanca, Spain), Pilar Giraldo (Hospital Miguel Servet, Zaragoza, Spain), Nicolás González (Hospital Obispo Polanco, Teruel, Spain), and Alicia Smucler (Hospital del Bierzo, Ponferrada, Spain) for samples and clinical data; and Teresa Baró, Federico Rojo, Rosa Navarro, Judith González, Carme Melero, and María Rodríguez-Rivera (Hospital del Mar, Barcelona, Spain) for their expert technical assistance.
Grant Support
This work has been supported, in part, by grants from Instituto de Salud Carlos III RD07/0020/2004, RD09/0076/00036, RD12/0036/0044, (RTICC, FEDER), Generalitat de Catalunya 2009/SGR541, and the "Xarxa de Bancs de Tumors" sponsored by Pla Director d’Oncologia de Catalunya (XBTC).
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
A. Salar is a consultant/advisory board member of Roche. No potential conflicts of interest were disclosed by the other authors.
Authors' Contributions
Conception and design: B. Espinet, A. Ferrer, B. Bellosillo, L. Nonell, J. Lloreta, D. Dominguez-Sola, S. Serrano
Development of methodology: B. Espinet, A. Ferrer, B. Bellosillo, L. Nonell, C. Fernández-Rodríguez, B. Puigdecanet, M. Garcia-Garcia, M. Vela, F. Solé
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): B. Espinet, A. Ferrer, A. Salar, B. Puigdecanet, J. Gimeno, M. Garcia-Garcia, E. Luño, R. Collado, J.T. Navarro, E. de la Banda, P. Abrisqueta, C. Serrano, B. Miñana, L. Florensa, A. Orfao
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): A. Ferrer, B. Bellosillo, L. Nonell, A. Salar, C. Ferńandez-Rodríguez, J. Lloreta, A. Cerutti, L. Florensa, A. Orfao, F. Sanz, F. Solé, D. Dominguez-Sola, S. Serrano
Writing, review, and/or revision of the manuscript: B. Espinet, A. Ferrer, B. Bellosillo, L. Nonell, A. Salar, C. Fernández-Rodríguez, B. Puigdecanet, J. Gimeno, M. Garcia-Garcia, M. Vela, E. Lunño, R. Collado, J.T. Navarro, L. Arenillas, C. Serrano, J. Lloreta, A. Cerutti, L. Florensa, A. Orfao, F. Sanz, F. Solé, D. Dominguez-Sola, S. Serrano
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): B. Espinet, A. Ferrer, L. Nonell, C. Fernández-Rodríguez
Study supervision: B. Espinet, A. Ferrer, B. Bellosillo, F. Solé, D. Dominguez-Sola, S. Serrano
References
- 1.Swerdlow SH, Campo E, Seto M, Muüller-Hermelink HK, Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. World Health Organization classification of tumours of haematopoietic and lymphoid tissues. IARC Press; Lyon, France: 2008. Mantle cell lymphoma; pp. 229–32. [Google Scholar]
- 2.Campo E, Raffeld M, Jaffe ES. Mantle-cell lymphoma. Semin Hematol. 1999;36:115–27. [PubMed] [Google Scholar]
- 3.Hinds PW, Dowdy SF, Eaton EN, Arnold A, Weinberg RA. Function of a human cyclin gene as an oncogene. Proc Natl Acad Sci U S A. 1994;91:709–13. doi: 10.1073/pnas.91.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gladden AB, Woolery R, Aggarwal P, Wasik MA, Diehl JA. Expression of constitutively nuclear cyclin D1 in murine lymphocytes induces B-cell lymphoma. Oncogene. 2006;25:998–1007. doi: 10.1038/sj.onc.1209147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herrmann A, Hoster E, Zwingers T, Brittinger G, Engelhard M, Meusers P, et al. Improvement of overall survival in advanced stage mantle cell lymphoma. J Clin Oncol. 2009;27:511–8. doi: 10.1200/JCO.2008.16.8435. [DOI] [PubMed] [Google Scholar]
- 6.Nodit L, Bahler DW, Jacobs SA, Locker J, Swerdlow SH. Indolent mantle cell lymphoma with nodal involvement and mutated immunoglobulin heavy chain genes. Hum Pathol. 2003;34:1030–4. doi: 10.1053/s0046-8177(03)00410-6. [DOI] [PubMed] [Google Scholar]
- 7.Orchard J, Garand R, Davis Z, Babbage G, Sahota S, Matutes E, et al. A subset of t(11;14) lymphoma with mantle cell features displays mutated IgVH genes and includes patients with good prognosis, nonnodal disease. Blood. 2003;101:4975–81. doi: 10.1182/blood-2002-06-1864. [DOI] [PubMed] [Google Scholar]
- 8.Espinet B, Sole F, Pedro C, Garcia M, Bellosillo B, Salido M, et al. Clonal proliferation of cyclin D1-positive mantle lymphocytes in an asymptomatic patient: an early-stage event in the development or an indolent form of a mantle cell lymphoma? Hum Pathol. 2005;36:1232–7. doi: 10.1016/j.humpath.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez V, Salamero O, Espinet B, Sole F, Royo C, Navarro A, et al. Genomic and gene expression profiling defines indolent forms of mantle cell lymphoma. Cancer Res. 2010;70:1408–18. doi: 10.1158/0008-5472.CAN-09-3419. [DOI] [PubMed] [Google Scholar]
- 10.Ondrejka SL, Lai R, Kumar N, Smith SD, Hsi ED. Indolent mantle cell leukemia: a clinicopathological variant characterized by isolated lymphocytosis, interstitial bone marrow involvement, kappa light chain restriction, and good prognosis. Haematologica. 2011;96:1121–7. doi: 10.3324/haematol.2010.036277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rule SA, Poplar S, Evans PA, O'Connor SJ, Owen RG. Indolent mantle-cell lymphoma: immunoglobulin variable region heavy chain sequence analysis reveals evidence of disease 10 years prior to symptomatic clinical presentation. J Clin Oncol. 2011;29:e437–9. doi: 10.1200/JCO.2010.31.8006. [DOI] [PubMed] [Google Scholar]
- 12.Del Giudice I, Messina M, Chiaretti S, Santangelo S, Tavolaro S, De Propris MS, et al. Behind the scenes of non-nodal MCL: downmodulation of genes involved in actin cytoskeleton organization, cell projection, cell adhesion, tumour invasion, TP53 pathway and mutated status of immunoglobulin heavy chain genes. Br J Haematol. 2012;156:601–11. doi: 10.1111/j.1365-2141.2011.08962.x. [DOI] [PubMed] [Google Scholar]
- 13.Royo C, Navarro A, Clot G, Salaverria I, Gine E, Jares P, et al. Non-nodal type of mantle cell lymphoma is a specific biological and clinical subgroup of the disease. Leukemia. 2012;26:1895–8. doi: 10.1038/leu.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thieblemont C, Nasser V, Felman P, Leroy K, Gazzo S, Callet-Bauchu E, et al. Small lymphocytic lymphoma, marginal zone B-cell lymphoma, and mantle cell lymphoma exhibit distinct gene-expression profiles allowing molecular diagnosis. Blood. 2004;103:2727–37. doi: 10.1182/blood-2003-06-2160. [DOI] [PubMed] [Google Scholar]
- 15.Navarro A, Royo C, Hernandez L, Jares P, Campo E. Molecular pathogenesis of mantle cell lymphoma: new perspectives and challenges with clinical implications. Semin Hematol. 2011;48:155–65. doi: 10.1053/j.seminhematol.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Espinet B, Sole F, Woessner S, Bosch F, Florensa L, Campo E, et al. Translocation (11;14)(q13;q32) and preferential involvement of chromosomes 1, 2, 9, 13, and 17 in mantle cell lymphoma. Cancer Genet Cytogenet. 1999;111:92–8. doi: 10.1016/s0165-4608(98)00228-3. [DOI] [PubMed] [Google Scholar]
- 17.Ventura RA, Martin-Subero JI, Jones M, McParland J, Gesk S, Mason DY, et al. FISH analysis for the detection of lymphoma-associated chromosomal abnormalities in routine paraffin-embedded tissue. J Mol Diagn. 2006;8:141–51. doi: 10.2353/jmoldx.2006.050083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 19.Wiestner A, Tehrani M, Chiorazzi M, Wright G, Gibellini F, Nakayama K, et al. Point mutations and genomic deletions in CCND1 create stable truncated cyclin D1 mRNAs that are associated with increased proliferation rate and shorter survival. Blood. 2007;109:4599–606. doi: 10.1182/blood-2006-08-039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crespo M, Bosch F, Villamor N, Bellosillo B, Colomer D, Rozman M, et al. ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia. N Engl J Med. 2003;348:1764–75. doi: 10.1056/NEJMoa023143. [DOI] [PubMed] [Google Scholar]
- 21.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 22.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 23.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bengtsson H, Simpson K, Bullard J, Hansen K. Aroma.affymetrix: a generic framework in R for analyzing small to very large Affymetrix data sets in bounded memory, Tech Report #745, Department of Statistics, University of California, Berkeley, February 2008. Sep 15, 2013. Available from: http://www.aroma-project.org.
- 25.Ingenuity Pathway Analysis (Ingenuity Systems) 2013 Sep 15; Available from: www.ingenuity.com. [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gene expression Omnibus, a public functional genomics data repository supporting MIAME-compliant data submissions Sep 15, 2013. Available from: http://www.ncbi.nlm.nih.gov/geo.
- 28.Breiman L, Friedman J, Stone CJ, Olshen RA. Chapman & Hall; Boca Raton, Florida: 1984. Classification and Regression Trees. [Google Scholar]
- 29.Website for Statistical Computation The Confidence Interval of a Proportion tool. Sep 15, 2013. Available from: http://www.vassarstats.net/prop1.html.
- 30.Pittaluga S, Verhoef G, Criel A, Maes A, Nuyts J, Boogaerts M, et al. Prognostic significance of bone marrow trephine and peripheral blood smears in 55 patients with mantle cell lymphoma. Leuk Lymphoma. 1996;21:115–25. doi: 10.3109/10428199609067588. [DOI] [PubMed] [Google Scholar]
- 31.Rawstron AC, Green MJ, Kuzmicki A, Kennedy B, Fenton JA, Evans PA, et al. Monoclonal B lymphocytes with the characteristics of "indolent" chronic lymphocytic leukemia are present in 3.5% of adults with normal blood counts. Blood. 2002;100:635–9. doi: 10.1182/blood.v100.2.635. [DOI] [PubMed] [Google Scholar]
- 32.Welzel N, Le T, Marculescu R, Mitterbauer G, Chott A, Pott C, et al. Templated nucleotide addition and immunoglobulin JH-gene utilization in t(11;14) junctions: implications for the mechanism of translocation and the origin of mantle cell lymphoma. Cancer Res. 2001;61:1629–36. [PubMed] [Google Scholar]
- 33.Salar A, Juanpere N, Bellosillo B, Domingo-Domenech E, Espinet B, Seoane A, et al. Gastrointestinal involvement in mantle cell lymphoma: a prospective clinic, endoscopic, and pathologic study. Am J Surg Pathol. 2006;30:1274–80. doi: 10.1097/01.pas.0000208899.15859.cb. [DOI] [PubMed] [Google Scholar]
- 34.Kuppers R, Klein U, Hansmann ML, Rajewsky K. Cellular origin of human B-cell lymphomas. N Engl J Med. 1999;341:1520–9. doi: 10.1056/NEJM199911113412007. [DOI] [PubMed] [Google Scholar]
- 35.Hadzidimitriou A, Agathangelidis A, Darzentas N, Murray F, fau-Larue MH, Pedersen LB, et al. Is there a role for antigen selection in mantle cell lymphoma? Immunogenetic support from a series of 807 cases. Blood. 2011;118:3088–95. doi: 10.1182/blood-2011-03-343434. [DOI] [PubMed] [Google Scholar]
- 36.Navarro A, Clot G, Royo C, Jares P, Hadzidimitriou A, Agathangelidis A, et al. Molecular subsets of mantle cell lymphoma defined by the IGHV mutational status and SOX11 expression have distinct biologic and clinical features. Cancer Res. 2012;72:5307–16. doi: 10.1158/0008-5472.CAN-12-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ek S, Dictor M, Jerkeman M, Jirstrom K, Borrebaeck CA. Nuclear expression of the non B-cell lineage Sox11 transcription factor identifies mantle cell lymphoma. Blood. 2008;111:800–5. doi: 10.1182/blood-2007-06-093401. [DOI] [PubMed] [Google Scholar]
- 38.Thieblemont C, Houlgatte R, Felman P, Traverse-glehen A, Baseggio L, Rolland D, et al. Indolent Mantle Cell Lymphoma (MCL): a retrospective detailed clinical and morphological analysis of 21 patients, with histological, cytological, cytogenetic, interphase genetic, immunoglobulin gene, and gene expression profiling analysis. ASH Annual Meeting Abstracts.2008. p. 1780. [Google Scholar]
- 39.A predictive model for aggressive non-Hodgkin's lymphoma The International Non-Hodgkin's Lymphoma Prognostic Factors Project. N Engl J Med. 1993;329:987–94. doi: 10.1056/NEJM199309303291402. [DOI] [PubMed] [Google Scholar]
- 40.Hoster E, Dreyling M, Klapper W, Gisselbrecht C, van Hoof A, Kluin-Nelemans HC, et al. A new prognostic index (MIPI) for patients with advanced-stage mantle cell lymphoma. Blood. 2008;111:558–65. doi: 10.1182/blood-2007-06-095331. [DOI] [PubMed] [Google Scholar]
- 41.Martin P, Chadburn A, Christos P, Weil K, Furman RR, Ruan J, et al. Outcome of deferred initial therapy in mantle-cell lymphoma. J Clin Oncol. 2009;27:1209–13. doi: 10.1200/JCO.2008.19.6121. [DOI] [PubMed] [Google Scholar]
- 42.Eve HE, Furtado MV, Hamon MD, Rule SA. Time to treatment does not influence overall survival in newly diagnosed mantle-cell lymphoma. J Clin Oncol. 2009;27:e189–90. doi: 10.1200/JCO.2009.23.9731. [DOI] [PubMed] [Google Scholar]
- 43.van Zelm MC, Szczepanski T, van der BM, van Dongen JJ. Replication history of B lymphocytes reveals homeostatic proliferation and extensive antigen-induced B cell expansion. J Exp Med. 2007;204:645–55. doi: 10.1084/jem.20060964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirt C, Schuler F, Dolken L, Schmidt CA, Dolken G. Low prevalence of circulating t(11;14)(q13;q32)-positive cells in the peripheral blood of healthy individuals as detected by real-time quantitative PCR. Blood. 2004;104:904–5. doi: 10.1182/blood-2004-02-0738. [DOI] [PubMed] [Google Scholar]
- 45.Nieto WG, Almeida J, Romero A, Teodosio C, Lopez A, Henriques AF, et al. Increased frequency (12%) of circulating chronic lymphocytic leukemia-like B-cell clones in healthy subjects using a highly sensitive multicolor flow cytometry approach. Blood. 2009;114:33–7. doi: 10.1182/blood-2009-01-197368. [DOI] [PubMed] [Google Scholar]
- 46.Jares P, Colomer D, Campo E. Genetic and molecular pathogenesis of mantle cell lymphoma: perspectives for new targeted therapeutics. Nat Rev Cancer. 2007;7:750–62. doi: 10.1038/nrc2230. [DOI] [PubMed] [Google Scholar]
- 47.Landgren O, Albitar M, Ma W, Abbasi F, Hayes RB, Ghia P, et al. B-cell clones as early markers for chronic lymphocytic leukemia. N Engl J Med. 2009;360:659–67. doi: 10.1056/NEJMoa0806122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 49.Nieto WG, Teodosio C, Lopez A, Rodriguez-Caballero A, Romero A, Barcena P, et al. Non-CLL-like monoclonal B-cell lymphocytosis in the general population: prevalence and phenotypic/genetic characteristics. Cytometry B Clin Cytom. 2010;78(Suppl 1):S24–34. doi: 10.1002/cyto.b.20543. [DOI] [PubMed] [Google Scholar]
- 50.Vegliante MC, Palomero J, Perez-Galan P, Roue G, Castellano G, Navarro A, et al. SOX11 regulates PAX5 expression and blocks terminal B-cell differentiation in aggressive mantle cell lymphoma. Blood. 2013;121:2175–85. doi: 10.1182/blood-2012-06-438937. [DOI] [PubMed] [Google Scholar]
- 51.Ferrando AA. SOX11 is a mantle cell lymphoma oncogene. Blood. 2013;121:2169–70. doi: 10.1182/blood-2013-02-480418. [DOI] [PubMed] [Google Scholar]
- 52.Nygren L, Baumgartner WS, Klimkowska M, Christensson B, Kimby E, Sander B. Prognostic role of SOX11 in a population-based cohort of mantle cell lymphoma. Blood. 2012;119:4215–23. doi: 10.1182/blood-2011-12-400580. [DOI] [PubMed] [Google Scholar]
- 53.van Dongen JJ, Lhermitte L, Bottcher S, Almeida J, van der Velden VH, Flores-Montero J, et al. EuroFlow antibody panels for standardized n-dimensional flow cytometric immunophenotyping of normal, reactive and malignant leukocytes. Leukemia. 2012;26:1908–75. doi: 10.1038/leu.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malavasi F, Deaglio S, Damle R, Cutrona G, Ferrarini M, Chiorazzi N. CD38 and chronic lymphocytic leukemia: a decade later. Blood. 2011;118:3470–8. doi: 10.1182/blood-2011-06-275610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zucchetto A, Benedetti D, Tripodo C, Bomben R, Dal Bo M, Marconi D, et al. CD38/CD31, the CCL3 and CCL4 chemokines, and CD49d/vascular cell adhesion molecule-1 are interchained by sequential events sustaining chronic lymphocytic leukemia cell survival. Cancer Res. 2009;69:4001–9. doi: 10.1158/0008-5472.CAN-08-4173. [DOI] [PubMed] [Google Scholar]
- 56.Dorfman DM, Shahsafaei A. CD200 (OX-2 membrane glycoprotein) expression in b cell-derived neoplasms. Am J Clin Pathol. 2010;134:726–33. doi: 10.1309/AJCP38XRRUGSQOVC. [DOI] [PubMed] [Google Scholar]
- 57.Palumbo GA, Parrinello N, Fargione G, Cardillo K, Chiarenza A, Berretta S, et al. CD200 expression may help in differential diagnosis between mantle cell lymphoma and B-cell chronic lymphocytic leukemia. Leuk Res. 2009;33:1212–6. doi: 10.1016/j.leukres.2009.01.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.