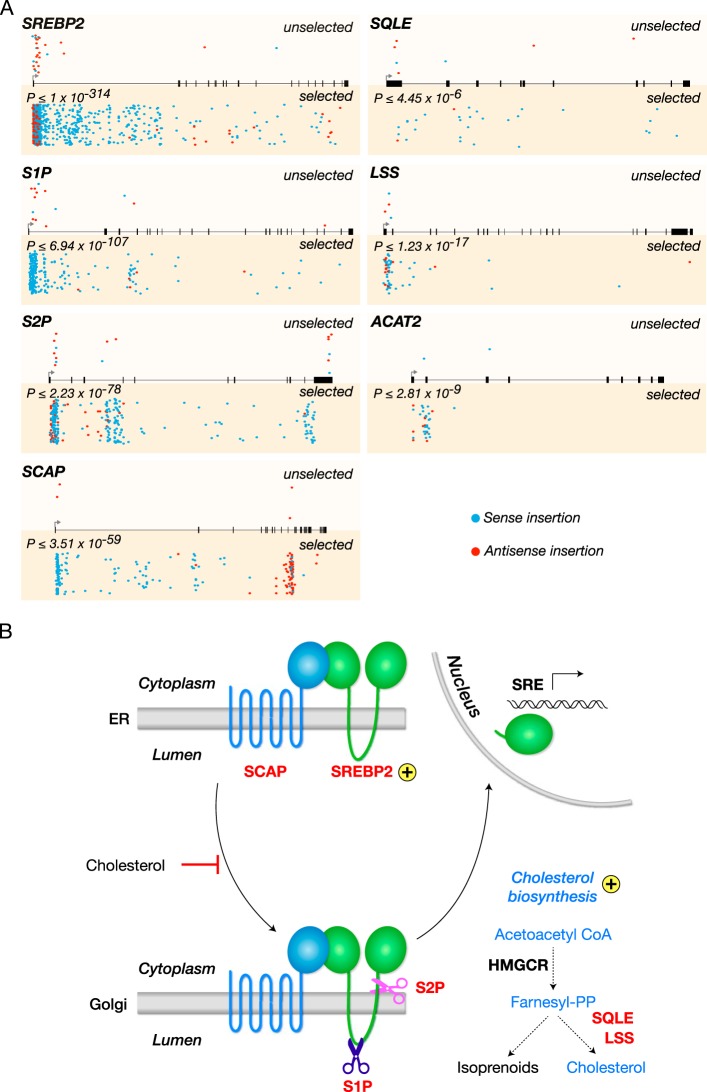
FIG 1 .
A haploid genetic screen reveals multiple genes of the SREBP cholesterol regulatory pathway required for Andes virus entry. (A) Gene trap insertions in genes that are required for Andes virus entry in human HAP1 cells. For every gene, sites of proviral integration are depicted for unselected control cells (top) and cells selected with rVSV-ANDV GP (bottom). P values correspond to the enrichment of disruptive insertions (sense orientation insertions and insertions into exonic regions) in the virus-selected population over levels in the control. Insertion sites in both populations are also scored for orientational bias; integrations in the same transcriptional orientation as the gene (sense) are shown in blue, while those in the opposite orientation (antisense) are depicted in red. All genes with the exception of ACAT2 also show a statistically significant (P ≤ 0.005) bias for sense orientation integrations in the rVSV-ANDV GP-selected cells. (B) Cellular SREBP cholesterol regulatory pathway. Genes identified as hits in the screen are labeled in red. Golgi, Golgi apparatus; PP, pyrophosphate.