ABSTRACT
Nasal colonization by the human pathogen Staphylococcus aureus is a major risk factor for hospital- and community-acquired infections. A key factor required for nasal colonization is a cell surface-exposed zwitterionic glycopolymer, termed wall teichoic acid (WTA). However, the precise mechanisms that govern WTA-mediated nasal colonization have remained elusive. Here, we report that WTA GlcNAcylation is a pivotal requirement for WTA-dependent attachment of community-acquired methicillin-resistant S. aureus (MRSA) and emerging livestock-associated MRSA to human nasal epithelial cells, even under conditions simulating the nutrient composition and dynamic flow of nasal secretions. Depending on the S. aureus strain, WTA O-GlcNAcylation occurs in either α or β configuration, which have similar capacities to mediate attachment to human nasal epithelial cells, suggesting that many S. aureus strains maintain redundant pathways to ensure appropriate WTA glycosylation. Strikingly, a lack of WTA glycosylation significantly abrogated the ability of MRSA to colonize cotton rat nares in vivo. These results indicate that WTA glycosylation modulates S. aureus nasal colonization and may help to develop new strategies for eradicating S. aureus nasal colonization in the future.
IMPORTANCE
Nasal colonization by the major human pathogen Staphylococcus aureus is a risk factor for severe endogenous infections and contributes to the spread of this microbe in hospitals and the community. Here, we show that wall teichoic acid (WTA) O-GlcNAcylation is a key factor required for S. aureus nasal colonization. These data provide a mechanistic explanation for the capacity of WTA to modulate S. aureus nasal colonization and may stimulate research activities to establish valuable strategies to eradicate S. aureus nasal colonization in high-risk hospitalized patients and in the general community.
INTRODUCTION
Staphylococcus aureus is one of the most important bacterial pathogens in hospital- and community-acquired (CA) infections. S. aureus colonizes the anterior nares of approximately 20 to 30% of the healthy population asymptomatically. Nasal colonization by S. aureus has been found to be a major risk factor for severe endogenous infections and contributes to the transmission and spread of S. aureus in hospitals (1, 2).
S. aureus nasal colonization is a complex and multifactorial process which is understood only incompletely. Both microbial and human host factors appear to mediate and modulate S. aureus nasal colonization. Host factors shaping nasal colonization include, for example, the presence of hemoglobin in nasal secretions (3), altered expression of antimicrobial peptides in human skin (4, 5), nutrient limitation (6), and polymorphisms in immunomodulatory mediators or receptors (7, 8). Equally importantly, S. aureus nasal colonization and stable persistence require bacterial factors which govern efficient attachment to squamous or ciliated epithelial cells located in different areas of the nasal cavity (9, 10). In particular, various S. aureus proteins belonging to the group of “microbial surface components recognizing adhesive matrix molecules” (MSCRAMMs) have been shown to mediate interaction with components of human desquamated nasal epithelial cells (11–13), above all, with cytokeratin-10 and loricrin-binding clumping factor B (ClfB) (14, 15) and iron-regulated surface determinant A (IsdA) (16). isdA and clfB are upregulated during nasal colonization in humans and in vivo models of experimental colonization compared to in vitro growth in broth cultures (17–19). S. aureus mutants lacking ClfB cannot persist in the human nose over a longer time period, underscoring the key role of ClfB in S. aureus nasal long-term colonization (20). However, during early-stage colonization, clfB and isdA are usually expressed only at low levels. In contrast, genes required for the biosynthesis of another cell wall-anchored macromolecule, designated wall teichoic acid (WTA), are upregulated during this period (17, 18). Typically, S. aureus strains, including methicillin-resistant S. aureus (MRSA) and highly virulent clones such as American pandemic clone USA300, synthesize WTA polymers composed of 11 to 40 ribitol-phosphate (RboP) repeating units modified with three tailoring modifications, d-alanine, α-O-N-acetylglucosamine (GlcNAc), and β-O-GlcNAc (10, 21). Interestingly, S. aureus mutants lacking WTA lost their capability to colonize cotton rat nares, while mutants that lost the ability to modify teichoic acids by d-alanylation (WTA and lipoteichoic acid) exhibited reduced colonization. Both types of mutants feature strongly reduced binding capacities to living human primary nasal epithelial cells (22, 23). A recently discovered type F scavenger receptor (SREC-I), present on human nasal epithelial cells, governs S. aureus nasal colonization via charge-dependent interaction with WTA polymers, indicating that the d-alanine modification of WTA contributes to S. aureus nasal colonization (24). However, the role of the WTA aminosugar modification with GlcNAc in nasal colonization, which occurs in two different stereochemical configurations (α- and β-O linkages), remains elusive.
Here, we analyzed the role of S. aureus WTA O-GlcNAcylation during nasal colonization and reveal molecular determinants that contribute to WTA-mediated S. aureus nasal colonization in vitro and in vivo.
RESULTS AND DISCUSSION
S. aureus nasal isolates encode WTA glycosyltransferases.
Two recently discovered enzymes, the α-O-GlcNAc WTA glycosyltransferase TarM and the β-O-GlcNAc glycosyltransferase TarS, can glycosylate S. aureus WTA (25, 26). With only very few exceptions, tarS is present in all sequenced S. aureus genomes. In contrast, tarM is absent in certain clonal complexes (CC), for example, in the emerging CC398, which includes livestock-associated MRSA clones (LA-MRSA CC398). This raises the question of whether TarM-dependent WTA α-O-GlcNAcylation may play a role in S. aureus host tropism (21).
Here, 70 isolates from a collection of genetically diverse nasal S. aureus isolates (49 different spa types associated with at least 15 different sequence types) obtained from healthy volunteers or hospital patients were typed by PCR for the presence or absence of S. aureus WTA-glycosyltransferase-encoding genes. All tested nasal isolates bore tarS, while only a minority (35.7%) of these strains bore both tarM and tarS, indicating that, in principle, nasal S. aureus strains should be capable of glycosylating WTA (Table 1).
Table 1 .
Distribution of WTA glycosyltransferases TarM and TarS in nasal S. aureus isolates
| Parameter | Value for WTA glycosyltransferase-encoding gene(s): |
||
|---|---|---|---|
| tarM | tarS | tarM and tarS | |
| No. of gene-positive nasal isolatesa | 25 | 70 | 25 |
| Rate of gene-positive nasal isolates (%)b | 35.7 | 100 | 35.7 |
A total of 70 S. aureus isolates obtained from healthy carriers or hospital patients was tested. spa typing revealed 49 different spa types associated with at least 15 different sequence types (see also Table S1 in the supplemental material).
Values (%) were calculated by dividing the number of positive isolates by the total number of tested isolates.
Impact of nasal growth conditions on S. aureus WTA glycosylation.
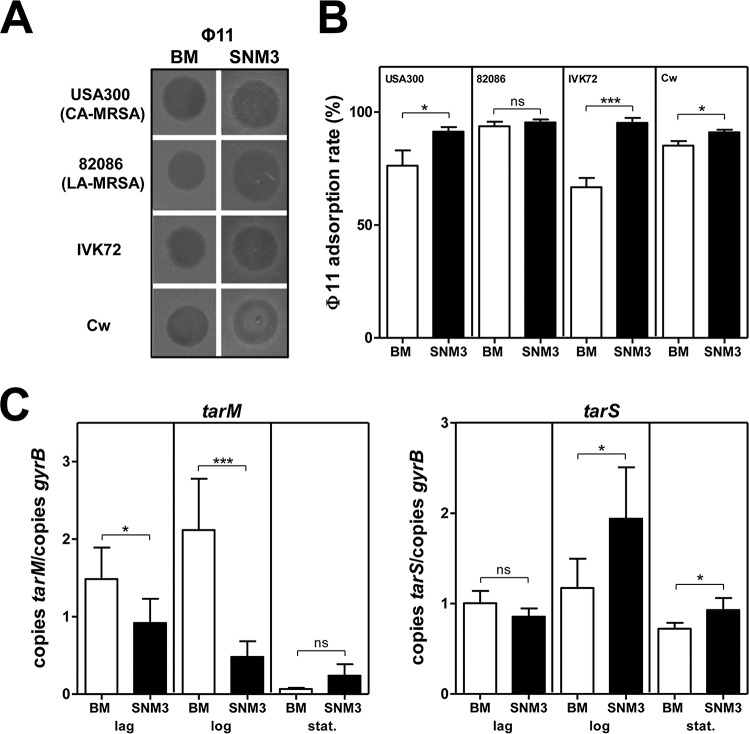
In order to analyze if such nasal strains indeed have glycosylated WTA, their capacity to bind WTA GlcNAc-dependent bacteriophages (phages) such as Φ11 (27) was investigated upon growth in a recently developed synthetic nasal medium (SNM3, containing 0.2 mM bipyridine) providing suitable conditions for simulating the nasal habitat (6). All tested SNM3-grown S. aureus strains were susceptible to phage Φ11, and, interestingly, the phage adsorption rate was increased compared even to that seen with bacteria cultivated in standard laboratory media (Fig. 1A and B). Moreover, even though tarM was downregulated in SNM3, the gene for the TarS β-O-GlcNAc WTA glycosyltransferase was upregulated in community-associated MRSA (CA-MRSA; tarM- and tarS-positive) strain USA300 during the logarithmic- and stationary-growth phases in SNM3 compared to the level seen in bacteria grown in rich B-medium (BM) (Fig. 1C). Thus, WTA polymers are most probably glycosylated under nasal growth conditions but may differ with respect to the type of GlcNAc linkage.
FIG 1 .
Impact of nasal growth conditions on WTA composition and WTA glycosylation. (A and B) S. aureus susceptibility to the WTA GlcNAc-dependent bacteriophage Φ11 (A) along with Φ11 adsorption levels (B). (C) qRT-PCR analysis of WTA glycosyltransferase gene expression levels in S. aureus USA300. mRNA was isolated from lag-, log-, or stationary (stat.)-phase-grown bacteria. For bacteriophage susceptibility and adsorption analysis or analysis of gene expression levels, strains were grown under rich conditions (B-medium [BM]; white columns) or nasal growth conditions (SNM3 [containing 0.2 mM bipyridine]; black columns). Values are given as means and standard deviations (SD; n = 3 to 6). Statistically significant differences calculated using an unpaired two-tailed Student’s t test are indicated as follows: ns (not significant), P > 0.05; *, P < 0.05, **, P < 0.01; ***, P < 0.001.
WTA glycosylation affects interaction of MRSA with epithelial cells.
Next, the impact of WTA glycosylation on nasal colonization and binding to human epithelial cells was clarified by using various S. aureus isolates, including tarM- and tarS-containing CA-MRSA strain USA300 and the RN4220 laboratory strain as well as LA-MRSA CC398 strain 82086 naturally lacking tarM plus corresponding mutants completely lacking WTA glycosylation. Importantly, a lack of WTA glycosylation did not alter expression levels of MSCRAMMs such as ClfB, SdrC, SdrD, IsdA, and SasG, which are known to interact with components of nasal squames (see Fig. S1A in the supplemental material). Accordingly, the ability of S. aureus variants lacking WTA glycosylation to interact with extracellular matrix proteins such as cytokeratin-10, fibronectin, and fibrinogen was equal to that of the wild-type strain, indicating that matrix-protein binding MSCRAMMs are fully functional in the mutants (see Fig. S1B to D).
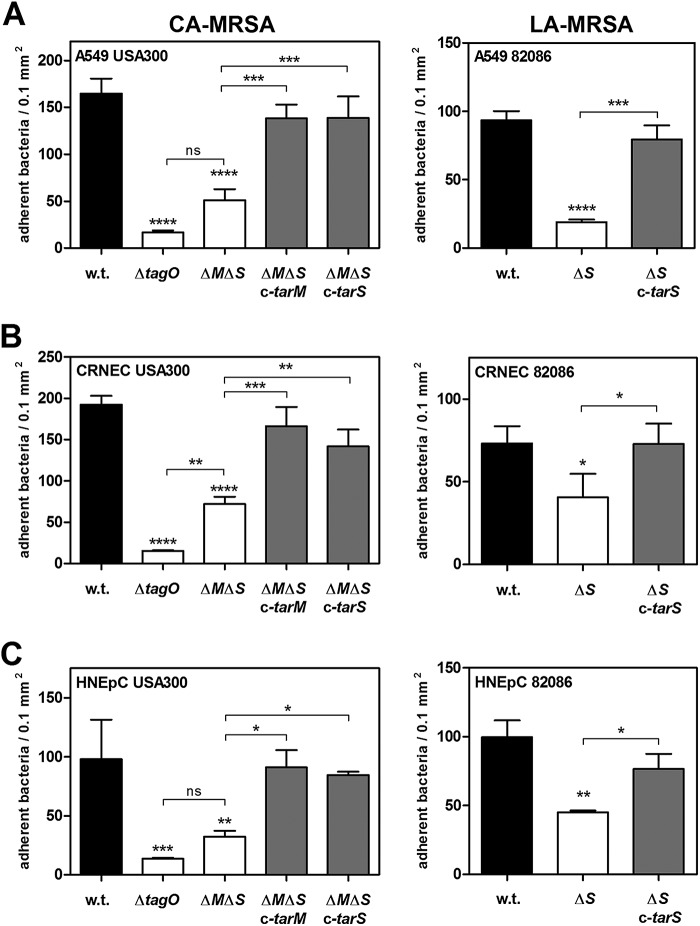
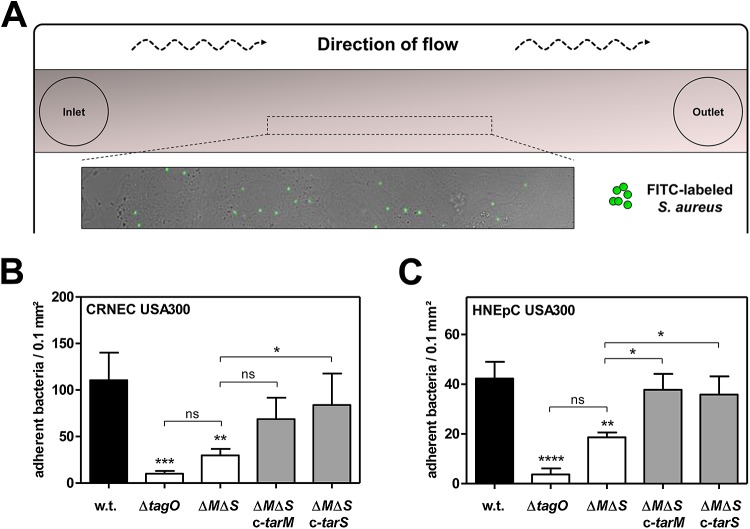
However, the absence of WTA glycosylation caused a strong reduction in the binding capacity of S. aureus to living human airway epithelial cells (A549) and to primary epithelial cells derived from cotton rat (CRNEC) or human nares (HNEpC) (Fig. 2; see also Fig. S2 in the supplemental material). Complementation by either the TarM α-O-GlcNAc WTA glycosyltransferase or the TarS β-O-GlcNAc WTA glycosyltransferase restored the binding capacity of WTA glycosylation-deficient mutants to levels comparable to those seen with the corresponding parental strains, suggesting that WTA glycosylation is required for efficient interaction with nasal epithelial cells but that the stereochemistry of WTA glycosylation is of minor importance for this interaction. Indeed, the binding capacity of S. aureus USA300 mutants, which still glycosylate WTA but lack one of the two glycosyltransferases (TarM or TarS), to A549 cells or to primary nasal epithelial cells was not affected (see Fig. S3). Similar effects were observed when adherence to primary nasal epithelial cells was monitored under mild shear forces mimicking the dynamic movement of nasal mucus, thereby emphasizing the importance of WTA-GlcNAcylation for interaction with epithelial cells (Fig. 3). However, S. aureus mutants completely lacking WTA polymers (ΔtagO) were also strongly abrogated in binding to epithelial cells compared to the parental strain, which is in agreement with previous reports (22, 23) (Fig. 2 and 3B and C). The more pronounced effect in the ΔtagO mutant than in the glycosylation-deficient mutants may result from the absence of zwitterionic WTA repeating units and their GlcNAcylation, which both appear to be important for interaction with epithelial cell receptors. Together, these findings indicate that backbone glycosylation is an important component of WTA polymers required for efficient interaction with human epithelial cells.
FIG 2 .
WTA glycosylation affects interaction of MRSA with epithelial cells. Data represent binding capacities of S. aureus variants to A549 lung epithelial cells (MOI30) (A), cotton rat nasal epithelial primary cells (CRNEC) (MOI20) (B), or human nasal epithelial primary cells (HNEpC) (MOI30) (C) under static conditions. The binding capacities of various strains encompassing the CA-MRSA USA300 strain and a LA-MRSA CC398 strain (82086) to confluent epithelial cell monolayers were analyzed. The S. aureus wild-type (w.t.) strain and strains lacking WTA (ΔtagO) or WTA glycosylation (ΔtarM ΔtarS [ΔMΔS] or ΔtarS [ΔS], LA-MRSA CC398 strain 82086) and the complemented mutants (ΔMΔS c-tarM, ΔMΔS c-tarS, or ΔS c-tarS, LA-MRSA CC398 strain 82086) are indicated. Values are given as means and standard deviations (SD; n = 3 to 4). Statistically significant differences calculated using one-way analysis of variance (ANOVA) with Bonferroni’s multiple-comparison test are indicated as follows: ns (not significant), P > 0.05; *, P < 0.05, **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
FIG 3 .
WTA glycosylation mediates adherence to primary epithelial cells under dynamic flow conditions. (A) Principle of flow-chamber-based attachment assays mimicking dynamic nasal conditions. (B and C) The binding capacities of S. aureus variants to monolayers of confluent cotton rat nasal epithelial primary cells (CRNEC) (MOI60) (B) or human nasal epithelial primary cells (HNEpC) (MOI60) (C) under conditions of mild shear stress were analyzed. The S. aureus USA300 wild-type (w.t.) strain, strains lacking WTA (ΔtagO) or WTA glycosylation (ΔtarM ΔtarS [ΔMΔS]), and the complemented mutants (ΔMΔS c-tarM and ΔMΔS c-tarS) are indicated. Values are given as means and standard deviations (SD; n = 3 to 4). Statistically significant differences calculated using one-way ANOVA with Bonferroni’s multiple-comparison test are indicated as follows: ns (not significant), P > 0.05; *, P < 0.05, **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
WTA glycosylation affects S. aureus nasal colonization in vivo.
Finally, the impact of WTA glycosylation on S. aureus nasal colonization was analyzed by using a cotton rat in vivo colonization model (22, 28). Cotton rat nares share histological properties with human nares and permit stable long-term nasal colonization with S. aureus (29). Moreover, cotton rats are susceptible to various human respiratory bacterial pathogens, thereby representing an excellent in vivo nasal colonization model (30). Notably, lack of WTA glycosylation significantly abrogated S. aureus nasal colonization in the cotton rat model 3 days after inoculation (Fig. 4). Since absence of WTA glycosylation did not affect S. aureus susceptibility to killing by antimicrobial peptides such as the cathelicidin LL-37 (see Fig. S4 in the supplemental material) or growth in various laboratory media, including the SNM3 synthetic nasal medium (see Fig. S5), the reduced nasal colonization capacity is very likely due to diminished interaction with nasal epithelial cells. Thus, WTA glycosylation affects S. aureus nasal colonization in vivo.
FIG 4 .
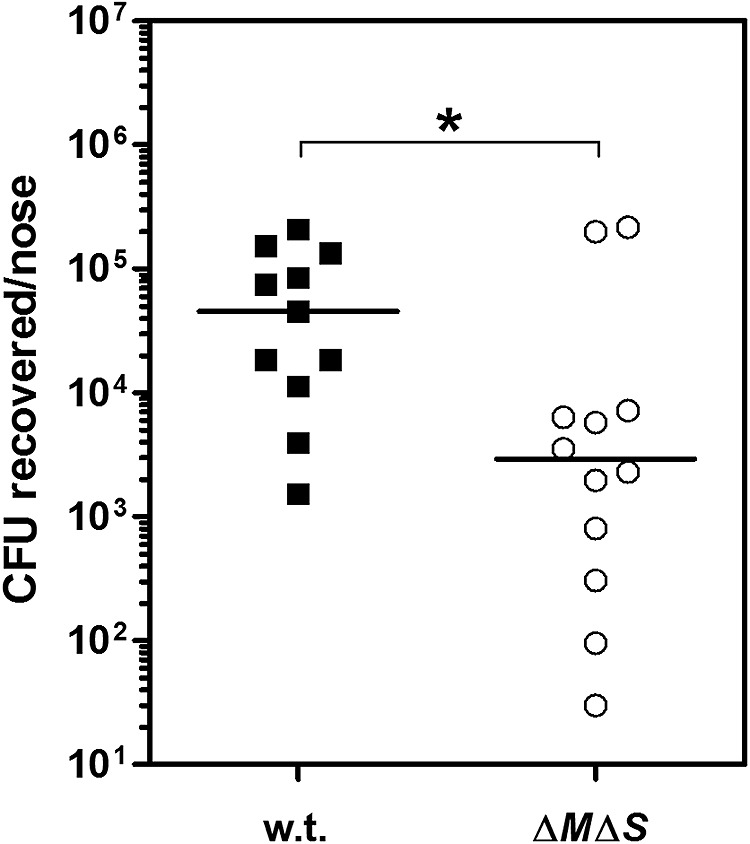
WTA glycosylation affects S. aureus nasal colonization in vivo. Cotton rats were intranasally challenged with the S. aureus USA300 wild-type (w.t.) strain and the USA300 ΔtarM ΔtarS (ΔMΔS) mutant lacking WTA glycosylation. Colonization status was analyzed 3 days after bacterial instillation. Statistically significant differences calculated using a D’Agostino and Pearson omnibus normality test and a subsequent Mann-Whitney test are indicated as follows: *, P < 0.05.
In summary, WTA glycosylation represents a key requirement for efficient S. aureus nasal colonization that most probably operates by modulating interaction with nasal epithelial cells. Since the majority of tested nasal S. aureus isolates encoded only tarS and not tarM, it is tempting to speculate that β-O-GlcNAcylated WTA might be more critical than α-O-GlcNAcylated WTA for S. aureus during nasal colonization. Nevertheless, WTA interaction with epithelial cells does not seem to depend on a specific stereochemistry of GlcNAc. The different types of WTA glycosylation might be more relevant for shaping other host-pathogen interaction processes such as activation of the human complement system via WTA β-O-GlcNAc-specific antibodies or mannose-binding lectin (31). However, WTA-deficient mutants exhibited even more diminished binding capacities to nasal epithelial cells than WTA glycosylation-deficient mutants, suggesting that the zwitterionic WTA repeating units and their GlcNAcylation are probably of the same importance for interaction with host cell receptors. While it has been demonstrated before that the zwitterionic properties of WTA polymers depending on free aminogroups of the d-alanine substituents govern interaction with the human scavenger receptor SREC-I in a charge-dependent manner (24), the identity of the host cell receptor responsible for interaction with WTA glycosyl residues remains unclear. Since SREC-I does not exhibit an obvious carbohydrate recognition domain, other human scavenger receptors have to be considered in the future for WTA glycosylation-dependent binding to epithelial cells. Consequently, antibody-based targeting of these receptors or their bacterial ligands, potentially in combination with nasal elimination of S. aureus by the antibiotic mupirocin (32), might become a suitable strategy for eradicating S. aureus nasal colonization in high-risk hospitalized patients and in the general community.
MATERIALS AND METHODS
Bacterial strains and growth media.
All bacterial strains listed in Table S1 in the supplemental material were grown in lysogeny broth (Becton, Dickinson), B-medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 0.1% K2HPO4, 0.1% glucose), or Mueller-Hinton broth (MHB) or in a recently described synthetic nasal medium (SNM3, containing 0.2 mM bipyridine) (6) at permissive temperatures and were supplemented with the appropriate antibiotics (chloramphenicol [10 µg/ml], streptomycin [250 to 500 µg/ml], and ampicillin [100 µg/ml]). Growth characteristics were monitored at 37°C.
Molecular genetic methods.
For deletion of tarM, tarS, tarM and tarS, or tagO in S. aureus USA300 or in a LA-MRSA CC398 strain (82086), a previously described Escherichia coli-S. aureus shuttle vector, pKOR1, was used (33). For deletion of srtA, an E. coli-S. aureus shuttle vector, pIMAY, was used (34). Gene disruption via pKOR1 or pIMAY was performed via allelic replacement as described before (33, 34). Briefly, knockout plasmids were isolated from appropriate E. coli strains, transformed into electrocompetent S. aureus RN4220 cells, reisolated, and subsequently electroporated in a target strain (USA300 or 82086). Electroporation conditions were described elsewhere (35). Knockout plasmids were integrated onto the genome of target strains at permissive temperatures (for pKOR1, 43°C; for pIMAY, 37°C) and in the presence of chloramphenicol (10 µg/ml). Following plasmid integration, a counterselection step was performed at 30°C using anhydrotetracycline (1 µg/ml). Resulting colonies were patched onto BM agar plates with or without chloramphenicol (10 µg/ml) and screened for plasmid loss. Chloramphenicol-sensitive colonies were screened via PCR to confirm gene deletion. The S. aureus RN4220 variants were described elsewhere (35). For complementation studies, the previously described pRB474 E. coli-S. aureus shuttle vector was used (36).
The presence or absence of S. aureus WTA glycosyltransferase-encoding genes tarM and tarS in nasal S. aureus isolates was verified via PCR using the gene-specific primers listed in Table S2 in the supplemental material. spa typing was performed as described previously (37). Briefly, the spa repeat region was amplified using the published primers listed in Table S2, sequenced, and analyzed using Ridom StaphType 1.5.21 software (37).
Cell culture.
Primary human nasal epithelial cells (HNEpC) were purchased from PromoCell (PromoCell, Heidelberg, Germany) and grown in airway epithelial growth medium (PromoCell) according to the manufacturer’s instructions. Primary cotton rat nasal epithelial cells (CRNEC) were isolated from cotton rat nares and cultivated in Dulbecco’s modified Eagle’s medium-F12 (DMEM-F12) (Gibco-BRL) supplemented with 100 U/ml penicillin, 100 µg/ml streptomycin, 10% heat-inactivated fetal bovine serum, and 2 mM glutamine as previously described (24). A549 lung epithelial cells (American Type Culture Collection [ATCC CCL-185]) were purchased from the ATCC and grown in DMEM-F12 medium (Gibco-BRL) supplemented with 100 U/ml penicillin, 100 µg/ml streptomycin, 10% heat-inactivated fetal bovine serum, and 2 mM glutamine. Epithelial cells were grown at 37°C under 5% CO2.
Interaction of S. aureus with epithelial cells.
Adherence of S. aureus to epithelial cells was analyzed as described elsewhere (22). Briefly, approximately 5 × 104 A549, 2.5 × 104 CRNEC, or 2.5 × 104 HNEpC cells were seeded to 24-well culture plates and grown at 37°C under 5% CO2 in appropriate cell culture media. Bacterial overnight cultures grown in B-medium at 37°C were diluted in MHB and grown at 37°C to the mid-logarithmic-growth phase, washed three times with phosphate-buffered saline (PBS), and labeled with fluorescein isothiocyanate (FITC; 0.1 mg/ml) for 1 h at 37°C. FITC-labeled bacteria were washed three times with PBS and resuspended in RPMI medium (Sigma, St. Louis, MO, USA). Bacterial cell numbers were adjusted using a Neubauer chamber. Confluent epithelial cell monolayers were washed twice with RPMI medium and inoculated with FITC-labeled bacteria for 1 h at 37°C under 5% CO2. Parameters of multiplicities of infection (MOI) were chosen in a strain-dependent manner and are individually given in the corresponding figure legends. Subsequently, epithelial cell monolayers were washed twice with RPMI and subsequently with PBS. Cells were fixed at room temperature for 10 min using 3.5% paraformaldehyde (PFA)-PBS. PFA was removed, and wells were coated with 1 ml PBS. No morphological changes were observed after this procedure. Adherent bacteria per 0.1 mm2 were counted (in at least 6 random fields of 0.1 mm2) using a fluorescence microscope.
Adherence under shear stress conditions was monitored in chamber slides (μ-Slide VI0.4; ibidi, Martinsried, Germany) as described before (24) with minor modifications. Briefly, approximately 2 × 104 HNEpC or 2 × 104 CRNEC cells were seeded to chamber slides and cultivated as described before. Confluent epithelial cell monolayers were washed twice with RPMI medium. Subsequently, peristaltic pumps (Amersham) were used for infection (20 min) with FITC-labeled bacteria at a flow rate of 1 ml/h, mimicking mild shear stress conditions at 0.3 dynes/cm2. MOI parameters are given in the corresponding figure legends. Cell monolayers were washed in two consecutive steps using RPMI medium and PBS and fixed as described before. Adherent bacteria per 0.1 mm2 were counted (in at least 8 random fields of 0.1 mm2) using a fluorescence microscope.
Adherence to extracellular matrix proteins.
Adherence to extracellular matrix proteins was analyzed as described elsewhere (23) with minor modifications. Briefly, 96-well microtiter plates were coated with keratin (50 µg/well), fibrinogen (2.5 µg/well), or fibronectin (15 µg/well) (all obtained from Sigma, St. Louis, MO, USA) mixed in sodium carbonate buffer (50 mM, pH 9.6) overnight at 4°C. Next, wells were blocked by using 100 µl 2% bovine serum albumin (BSA)–PBS for 2 h at room temperature. Wells were washed with 200 µl PBS and incubated with 100 µl bacteria (5 × 109 CFU/ml) for 1.5 h at 37°C. Wells were washed three times with 200 µl PBS. Bacteria were stained using 100 µl crystal violet (2 mg/ml in PBS) for 2 min at room temperature. Subsequently, wells were washed five times with 400 µl PBS. A 100 µl volume of (70%) ethanol was added to each well, and the absorbance at 600 nm (A600) was determined.
Experiments with phages.
Phage Φ11 susceptibility of S. aureus was analyzed as described before (35). When bacteria were grown under nasal growth conditions using SNM3 (containing 0.2 mM bipyridine), phage susceptibility was analyzed on solid SNM3 agar overlaid with SNM3 soft agar (0.4% agar). Phage Φ11 adsorption to various S. aureus isolates grown in rich medium (B-medium) or in SNM3 (containing 0.2 mM bipyridine) was analyzed as described previously (35). Briefly, phage Φ11 was propagated on the S. aureus RN4220 wild-type strain and used for adsorption experiments. The MOI was set to 0.1, and percent phage adsorption to test strains was calculated by determining the number of PFU of unbound phages in supernatants and subtracting that value from the total number of input PFU as a ratio to the total number of input PFU.
RNA isolation and preparation.
B-medium overnight cultures were diluted in fresh media (B-medium or SNM3 [containing 0.2 mM bipyridine]) and grown at 37°C. Bacteria were harvested in the lag-, log-, or stationary-growth phase and subsequently resolved in 1 ml TRIzol (Invitrogen/Life Technologies, Karlsruhe, Germany). Bacteria were mechanically disrupted with 0.5 ml zirconia-silica beads (Carl-Roth, Karlsruhe, Germany) (0.1 mm in diameter) using a FastPrep 24 homogenizer (MP Biomedicals) (2 cycles of 20 s at a speed of 6.5 m/s). Samples were stored at −80°C or subsequently used for further preparation. Chloroform (200 μl) was added, and samples were thoroughly mixed for 60 s and incubated for 3 min at room temperature. Samples were centrifuged (12,000 × g, 15 min, 4°C), and the supernatant was mixed with 500 µl isopropanol. Samples were incubated for 10 min at room temperature and centrifuged (12,000 × g, 30 min, 4°C). The pellet was washed with 500 µl (70%) ethanol, and the sample was centrifuged at 7,500 × g for 5 min (4°C). The pellet was air dried and dissolved in 50 µl nuclease-free water. After incubation at 55°C for 10 min, the sample was mixed well for 4 min and 5 µg RNA was digested with DNase I (Roche) and stored at −80°C.
qRT-PCR.
The primers used for quantitative real-time PCR (qRT-PCR) are listed in Table S2 in the supplemental material. RNA was transcribed into cDNA, and qRT-PCR was performed according to the instructions of the manufacturer using Brilliant II SYBR green 1-Step master mix (Agilent). Relative quantifications were analyzed using Roche’s LightCycler 480 II system. Transcription levels of target genes analyzed in this study were normalized against the expression of the gyrB housekeeping gene (38).
S. aureus killing by antimicrobial peptides.
Killing by antimicrobial peptides was analyzed as described before (22) with minor modifications. Briefly, S. aureus overnight cultures were diluted in MHB and grown to the mid-logarithmic-growth phase. Cells were washed 3 times in potassium phosphate buffer (PPB) (pH 7.5) containing 0.005% human serum albumin (HSA) (PPB-HSA). Approximately 2.0 × 105 bacteria (in PPB-BSA) were treated with 10 µg/ml LL-37 (Sigma, St. Louis, MO, USA). Killing was analyzed over a time period of 180 min. Each reaction was stopped by addition of 250 µl ice-cold PPB-BSA. Serial dilutions were plated on appropriate agar plates to analyze killing by determination of CFU, and the results are given as a ratio to the total number of input CFU.
Cotton rat nasal colonization model.
For analysis of S. aureus nasal colonization in vivo, a cotton rat model was used (22). Cotton rats continuously received drinking water containing 2.5 mg/ml streptomycin to reduce natural nasal flora 3 days before the experiment. The cotton rats were anesthetized and instilled intranasally with 10 µl of 1 × 107 CFU of the S. aureus USA300 wild type or USA300 ΔtarM ΔtarS. Three days after bacterial instillation, animals were euthanized and nares were removed surgically. Nares were subjected to vortex mixing for 30 s in 1 ml PBS containing 0.5% Tween 20. Serial dilutions were plated on appropriate agar plates to calculate nasal colonization data.
Ethics statement.
All animal experiments were performed in strict accordance with the German regulations of the Gesellschaft für Versuchstierkunde/Society for Laboratory Animal Science (GV-SOLAS) and the European Health Law of the Federation of Laboratory Animal Science Associations (FELASA). The protocol was approved by the Regierungspräsidium Tübingen, Tübingen, Germany (permit no. T1/10).
Statistical analysis.
Statistical analysis was performed by using GraphPad Prism (GraphPad Software, Inc., La Jolla, USA; version 5.04). Statistically significant differences were calculated by using appropriate statistical methods as indicated. P values of ≤0.05 were considered significant.
SUPPLEMENTAL MATERIAL
MSCRAMMs are functional in S. aureus variants lacking WTA glycosylation. (A) qRT-PCR analysis of S. aureus surface protein gene expression levels. mRNA was isolated from lag-, log-, or stationary (stat.)-phase-grown bacteria (B-medium [BM]), and gene expression in the S. aureus USA300 wild-type (w.t.) strain (black columns) and in the USA300 ΔtarM ΔtarS mutant lacking WTA glycosylation (white columns) was analyzed. Values are given as means and standard deviations (SD; n = 3). Statistically significant differences calculated using an unpaired two-tailed Student’s t test are indicated as follows: ns (not significant), P > 0.05. (B, C, and D) Binding capacities of S. aureus USA300 variants to extracellular matrix proteins (cytokeratin-10 [B], fibrinogen [C], and fibronectin [D]). The S. aureus USA300 wild-type (w.t.) strain and strains lacking WTA glycosylation (ΔtarM ΔtarS [ΔMΔS]) or surface proteins (ΔsrtA) are indicated. Values are given as means and standard deviations (SD; n = 3). Statistically significant differences calculated using one-way ANOVA with Bonferroni’s multiple-comparison test are indicated as follows: ns (not significant), P > 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Download
S. aureus RN4220 requires WTA glycosylation for interaction with epithelial cells. Data represent binding capacities of S. aureus RN4220 variants to A549 lung epithelial cells (MOI15) (A) or human nasal epithelial primary cells (HNEpC) (MOI30) (B) under static conditions. The binding capacities of S. aureus RN4220 variants to confluent epithelial cell monolayers were analyzed. The S. aureus wild-type (w.t.) strain, strains lacking WTA (ΔtagO) or WTA glycosylation (ΔtarM ΔtarS [ΔMΔS]), and the complemented mutants (ΔMΔS c-tarM, ΔMΔS c-tarS) are indicated. Values are given as means and standard deviations (SD; n = 3 to 7). Statistically significant differences calculated using one-way ANOVA with Bonferroni’s multiple-comparison test are indicated as follows: ns (not significant), P > 0.05; *, P < 0.05, **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Download
Stereochemical linkage of S. aureus WTA glycosylation is dispensable for interaction with epithelial cells. Data represent the binding capacities of S. aureus USA300 variants to A549 lung epithelial cells (MOI30) (A), cotton rat nasal epithelial primary cells (CRNEC) (MOI20) (B), or human nasal epithelial primary cells (HNEpC) (MOI30) (C) under static conditions. The binding capacities to confluent epithelial cell monolayers were analyzed. The S. aureus USA300 wild-type (w.t.) strain and strains lacking the α-O-GlcNAc WTA transferase TarM (ΔtarM [ΔM]) or the β-O-GlcNAc WTA transferase TarS (ΔtarS [ΔS]) are indicated. Values are given as means and standard deviations (SD; n = 3). Statistically significant differences calculated using one-way ANOVA with Bonferroni’s multiple-comparison test are indicated as follows: ns (not significant), P > 0.05. Download
Impact of WTA glycosylation on killing by the human cationic antimicrobial peptide LL-37. Data represent S. aureus killing kinetics by cathelicidin LL-37. The killing of the S. aureus USA300 wild-type (w.t.) strain (black line) and the USA300 ΔtarM ΔtarS mutant lacking WTA glycosylation (blue line) was analyzed. Values are given as means and standard deviations (SD; n = 3). No differences were observed. Download
Lack of WTA glycosylation is dispensable for growth of S. aureus. Data represent the growth characteristics of the S. aureus USA300 wild-type (w.t.) strain (black line) and the USA300 ΔtarM ΔtarS mutant lacking WTA glycosylation (blue line). Growth at 37°C was monitored in rich B-medium (A), in minimal medium (MHB) (B), or in the synthetic nasal medium SNM3 containing 0.2 mM bipyridine (C). Values are given as means and standard deviations (SD; n = 3). No differences were observed. Download
Bacterial strains and phages used in this study.
Oligonucleotides used in this study.
ACKNOWLEDGMENTS
We thank Bernhard Krismer and Matthias Marschal for providing nasal S. aureus isolates, Berit Schulte for help with spa typing, and Alexander Zipperer for help with cotton rats.
This work was supported by grants from the German Research Council (TRR34, TRR156, and SFB766) and from the the German Center for Infection Research (DZIF) to A.P. and C.W.
Footnotes
Citation Winstel V, Kühner P, Salomon F, Larsen J, Skov R, Hoffmann W, Peschel A, Weidenmaier C. 2015. Wall teichoic acid glycosylation governs Staphylococcus aureus nasal colonization. mBio 6(4):e00632-15. doi:10.1128/mBio.00632-15.
REFERENCES
- 1.Von Eiff C, Becker K, Machka K, Stammer H, Peters G. 2001. Nasal carriage as a source of Staphylococcus aureus bacteremia. N Engl J Med 344:11–16. doi: 10.1056/NEJM200101043440102. [DOI] [PubMed] [Google Scholar]
- 2.Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, Nouwen JL. 2005. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis 5:751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 3.Pynnonen M, Stephenson RE, Schwartz K, Hernandez M, Boles BR. 2011. Hemoglobin promotes Staphylococcus aureus nasal colonization. PLoS Pathog 7:e1002104. doi: 10.1371/journal.ppat.1002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nurjadi D, Herrmann E, Hinderberger I, Zanger P. 2013. Impaired beta-defensin expression in human skin links DEFB1 promoter polymorphisms with persistent Staphylococcus aureus nasal carriage. J Infect Dis 207:666–674. doi: 10.1093/infdis/jis735. [DOI] [PubMed] [Google Scholar]
- 5.Zanger P, Nurjadi D, Vath B, Kremsner PG. 2011. Persistent nasal carriage of Staphylococcus aureus is associated with deficient induction of human beta-defensin 3 after sterile wounding of healthy skin in vivo. Infect Immun 79:2658–2662. doi: 10.1128/IAI.00101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krismer B, Liebeke M, Janek D, Nega M, Rautenberg M, Hornig G, Unger C, Weidenmaier C, Lalk M, Peschel A. 2014. Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. PLoS Pathog 10:e1003862. doi: 10.1371/journal.ppat.1003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van den Akker EL, Nouwen JL, Melles DC, van Rossum EF, Koper JW, Uitterlinden AG, Hofman A, Verbrugh HA, Pols HA, Lamberts SW, van Belkum A. 2006. Staphylococcus aureus nasal carriage is associated with glucocorticoid receptor gene polymorphisms. J Infect Dis 194:814–818. doi: 10.1086/506367. [DOI] [PubMed] [Google Scholar]
- 8.Emonts M, Uitterlinden AG, Nouwen JL, Kardys I, Maat MP, Melles DC, Witteman J, Jong PT, Verbrugh HA, Hofman A, Hermans PW, Belkum A. 2008. Host polymorphisms in interleukin 4, complement factor H, and C-reactive protein associated with nasal carriage of Staphylococcus aureus and occurrence of boils. J Infect Dis 197:1244–1253. doi: 10.1086/533501. [DOI] [PubMed] [Google Scholar]
- 9.Foster TJ, Geoghegan JA, Ganesh VK, Höök M. 2014. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol 12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weidenmaier C, Peschel A. 2008. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol 6:276–287. doi: 10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- 11.Corrigan RM, Rigby D, Handley P, Foster TJ. 2007. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 153:2435–2446. doi: 10.1099/mic.0.2007/006676-0. [DOI] [PubMed] [Google Scholar]
- 12.Roche FM, Meehan M, Foster TJ. 2003. The Staphylococcus aureus surface protein SasG and its homologues promote bacterial adherence to human desquamated nasal epithelial cells. Microbiology 149:2759–2767. doi: 10.1099/mic.0.26412-0. [DOI] [PubMed] [Google Scholar]
- 13.Corrigan RM, Miajlovic H, Foster TJ. 2009. Surface proteins that promote adherence of Staphylococcus aureus to human desquamated nasal epithelial cells. BMC Microbiol 9:22. doi: 10.1186/1471-2180-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mulcahy ME, Geoghegan JA, Monk IR, O’Keeffe KM, Walsh EJ, Foster TJ, McLoughlin RM. 2012. Nasal colonisation by Staphylococcus aureus depends upon clumping factor B binding to the squamous epithelial cell envelope protein loricrin. PLoS Pathog 8:e1003092. doi: 10.1371/journal.ppat.1003092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Brien LM, Walsh EJ, Massey RC, Peacock SJ, Foster TJ. 2002. Staphylococcus aureus clumping factor B (ClfB) promotes adherence to human type I cytokeratin 10: implications for nasal colonization. Cell Microbiol 4:759–770. doi: 10.1046/j.1462-5822.2002.00231.x. [DOI] [PubMed] [Google Scholar]
- 16.Clarke SR, Brummell KJ, Horsburgh MJ, McDowell PW, Mohamad SA, Stapleton MR, Acevedo J, Read RC, Day NP, Peacock SJ, Mond JJ, Kokai-Kun JF, Foster SJ. 2006. Identification of in vivo-expressed antigens of Staphylococcus aureus and their use in vaccinations for protection against nasal carriage. J Infect Dis 193:1098–1108. doi: 10.1086/501471. [DOI] [PubMed] [Google Scholar]
- 17.Burian M, Rautenberg M, Kohler T, Fritz M, Krismer B, Unger C, Hoffmann WH, Peschel A, Wolz C, Goerke C. 2010. Temporal expression of adhesion factors and activity of global regulators during establishment of Staphylococcus aureus nasal colonization. J Infect Dis 201:1414–1421. doi: 10.1086/651619. [DOI] [PubMed] [Google Scholar]
- 18.Burian M, Wolz C, Goerke C. 2010. Regulatory adaptation of Staphylococcus aureus during nasal colonization of humans. PLoS One 5:e10040. doi: 10.1371/journal.pone.0010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkins A, Diep BA, Mai TT, Vo NH, Warrener P, Suzich J, Stover CK, Sellman BR. 2015. Differential expression and roles of Staphylococcus aureus virulence determinants during colonization and disease. mBio 6:e02272-14. doi: 10.1128/mBio.02272-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wertheim HF, Walsh E, Choudhurry R, Melles DC, Boelens HA, Miajlovic H, Verbrugh HA, Foster T, van Belkum A. 2008. Key role for clumping factor B in Staphylococcus aureus nasal colonization of humans. PLoS Med 5:e17. doi: 10.1371/journal.pmed.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winstel V, Xia G, Peschel A. 2014. Pathways and roles of wall teichoic acid glycosylation in Staphylococcus aureus. Int J Med Microbiol 304:215–221. doi: 10.1016/j.ijmm.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 22.Weidenmaier C, Kokai-Kun JF, Kristian SA, Chanturiya T, Kalbacher H, Gross M, Nicholson G, Neumeister B, Mond JJ, Peschel A. 2004. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat Med 10:243–245. doi: 10.1038/nm991. [DOI] [PubMed] [Google Scholar]
- 23.Weidenmaier C, Kokai-Kun JF, Kulauzovic E, Kohler T, Thumm G, Stoll H, Götz F, Peschel A. 2008. Differential roles of sortase-anchored surface proteins and wall teichoic acid in Staphylococcus aureus nasal colonization. Int J Med Microbiol 298:505–513. doi: 10.1016/j.ijmm.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 24.Baur S, Rautenberg M, Faulstich M, Grau T, Severin Y, Unger C, Hoffmann WH, Rudel T, Autenrieth IB, Weidenmaier C. 2014. A nasal epithelial receptor for Staphylococcus aureus WTA governs adhesion to epithelial cells and modulates nasal colonization. PLoS Pathog 10:e1004089. doi: 10.1371/journal.ppat.1004089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown S, Xia G, Luhachack LG, Campbell J, Meredith TC, Chen C, Winstel V, Gekeler C, Irazoqui JE, Peschel A, Walker S. 2012. Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc Natl Acad Sci U S A 109:18909–18914. doi: 10.1073/pnas.1209126109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia G, Maier L, Sanchez-Carballo P, Li M, Otto M, Holst O, Peschel A. 2010. Glycosylation of wall teichoic acid in Staphylococcus aureus by TarM. J Biol Chem 285:13405–13415. doi: 10.1074/jbc.M109.096172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia G, Corrigan RM, Winstel V, Goerke C, Gründling A, Peschel A. 2011. Wall teichoic acid-dependent adsorption of staphylococcal siphovirus and myovirus. J Bacteriol 193:4006–4009. doi: 10.1128/JB.01412-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kokai-Kun JF. 2008. The cotton rat as a model for Staphylococcus aureus nasal colonization in humans: cotton rat S. aureus nasal colonization model. Methods Mol Biol 431:241–254. [DOI] [PubMed] [Google Scholar]
- 29.Prince GA, Jenson AB, Horswood RL, Camargo E, Chanock RM. 1978. The pathogenesis of respiratory syncytial virus infection in cotton rats. Am J Pathol 93:771–791. [PMC free article] [PubMed] [Google Scholar]
- 30.Niewiesk S, Prince G. 2002. Diversifying animal models: the use of hispid cotton rats (Sigmodon hispidus) in infectious diseases. Lab Anim 36:357–372. doi: 10.1258/002367702320389026. [DOI] [PubMed] [Google Scholar]
- 31.Kurokawa K, Jung DJ, An JH, Fuchs K, Jeon YJ, Kim NH, Li X, Tateishi K, Park JA, Xia G, Matsushita M, Takahashi K, Park HJ, Peschel A, Lee BL. 2013. Glycoepitopes of staphylococcal wall teichoic acid govern complement-mediated opsonophagocytosis via human serum antibody and mannose-binding lectin. J Biol Chem 288:30956–30968. doi: 10.1074/jbc.M113.509893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bode LG, Kluytmans JA, Wertheim HF, Bogaers D, Vandenbroucke-Grauls CM, Roosendaal R, Troelstra A, Box AT, Voss A, van der Tweel I, van Belkum A, Verbrugh HA, Vos MC. 2010. Preventing surgical-site infections in nasal carriers of Staphylococcus aureus. N Engl J Med 362:9–17. doi: 10.1056/NEJMoa0808939. [DOI] [PubMed] [Google Scholar]
- 33.Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Monk IR, Shah IM, Xu M, Tan MW, Foster TJ. 2012. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 3:e00277-11. doi: 10.1128/mBio.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winstel V, Liang C, Sanchez-Carballo P, Steglich M, Munar M, Broker BM, Penades JR, Nubel U, Holst O, Dandekar T, Peschel A, Xia G. 2013. Wall teichoic acid structure governs horizontal gene transfer between major bacterial pathogens. Nat Commun 4:2345. doi: 10.1038/ncomms3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brückner R. 1992. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene 122:187–192. doi: 10.1016/0378-1119(92)90048-T. [DOI] [PubMed] [Google Scholar]
- 37.Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, Vogel U. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol 41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goerke C, Fluckiger U, Steinhuber A, Bisanzio V, Ulrich M, Bischoff M, Patti JM, Wolz C. 2005. Role of Staphylococcus aureus global regulators sae and sigmaB in virulence gene expression during device-related infection. Infect Immun 73:3415–3421. doi: 10.1128/IAI.73.6.3415-3421.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MSCRAMMs are functional in S. aureus variants lacking WTA glycosylation. (A) qRT-PCR analysis of S. aureus surface protein gene expression levels. mRNA was isolated from lag-, log-, or stationary (stat.)-phase-grown bacteria (B-medium [BM]), and gene expression in the S. aureus USA300 wild-type (w.t.) strain (black columns) and in the USA300 ΔtarM ΔtarS mutant lacking WTA glycosylation (white columns) was analyzed. Values are given as means and standard deviations (SD; n = 3). Statistically significant differences calculated using an unpaired two-tailed Student’s t test are indicated as follows: ns (not significant), P > 0.05. (B, C, and D) Binding capacities of S. aureus USA300 variants to extracellular matrix proteins (cytokeratin-10 [B], fibrinogen [C], and fibronectin [D]). The S. aureus USA300 wild-type (w.t.) strain and strains lacking WTA glycosylation (ΔtarM ΔtarS [ΔMΔS]) or surface proteins (ΔsrtA) are indicated. Values are given as means and standard deviations (SD; n = 3). Statistically significant differences calculated using one-way ANOVA with Bonferroni’s multiple-comparison test are indicated as follows: ns (not significant), P > 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Download
S. aureus RN4220 requires WTA glycosylation for interaction with epithelial cells. Data represent binding capacities of S. aureus RN4220 variants to A549 lung epithelial cells (MOI15) (A) or human nasal epithelial primary cells (HNEpC) (MOI30) (B) under static conditions. The binding capacities of S. aureus RN4220 variants to confluent epithelial cell monolayers were analyzed. The S. aureus wild-type (w.t.) strain, strains lacking WTA (ΔtagO) or WTA glycosylation (ΔtarM ΔtarS [ΔMΔS]), and the complemented mutants (ΔMΔS c-tarM, ΔMΔS c-tarS) are indicated. Values are given as means and standard deviations (SD; n = 3 to 7). Statistically significant differences calculated using one-way ANOVA with Bonferroni’s multiple-comparison test are indicated as follows: ns (not significant), P > 0.05; *, P < 0.05, **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Download
Stereochemical linkage of S. aureus WTA glycosylation is dispensable for interaction with epithelial cells. Data represent the binding capacities of S. aureus USA300 variants to A549 lung epithelial cells (MOI30) (A), cotton rat nasal epithelial primary cells (CRNEC) (MOI20) (B), or human nasal epithelial primary cells (HNEpC) (MOI30) (C) under static conditions. The binding capacities to confluent epithelial cell monolayers were analyzed. The S. aureus USA300 wild-type (w.t.) strain and strains lacking the α-O-GlcNAc WTA transferase TarM (ΔtarM [ΔM]) or the β-O-GlcNAc WTA transferase TarS (ΔtarS [ΔS]) are indicated. Values are given as means and standard deviations (SD; n = 3). Statistically significant differences calculated using one-way ANOVA with Bonferroni’s multiple-comparison test are indicated as follows: ns (not significant), P > 0.05. Download
Impact of WTA glycosylation on killing by the human cationic antimicrobial peptide LL-37. Data represent S. aureus killing kinetics by cathelicidin LL-37. The killing of the S. aureus USA300 wild-type (w.t.) strain (black line) and the USA300 ΔtarM ΔtarS mutant lacking WTA glycosylation (blue line) was analyzed. Values are given as means and standard deviations (SD; n = 3). No differences were observed. Download
Lack of WTA glycosylation is dispensable for growth of S. aureus. Data represent the growth characteristics of the S. aureus USA300 wild-type (w.t.) strain (black line) and the USA300 ΔtarM ΔtarS mutant lacking WTA glycosylation (blue line). Growth at 37°C was monitored in rich B-medium (A), in minimal medium (MHB) (B), or in the synthetic nasal medium SNM3 containing 0.2 mM bipyridine (C). Values are given as means and standard deviations (SD; n = 3). No differences were observed. Download
Bacterial strains and phages used in this study.
Oligonucleotides used in this study.