Abstract
Background:
Sebaceoma is a tumor for which the causative oncogenes are not well-understood. Sebaceomas demonstrate some histopathologic features similar to basal cell carcinoma (BCC), such as palisading borders and basaloid cells with additional features, including foamy cytoplasm and indented nuclei.
Aims:
We examine multiple cell-cycle, oncogene, and tumor suppressor gene markers in sebaceomas, to try to find some suitable biological markers for this tumor, and compare with other published studies.
Materials and Methods:
We investigated a panel of immunohistochemical (IHC) stains that are important for cellular signaling, including a cell cycle regulator, tumor suppressor gene, oncogene, hormone receptor, and genomic stability markers in our cohort of sebaceomas. We collected 30 sebaceomas from three separate USA dermatopathology laboratories. The following IHC panel: Epithelial membrane antigen (EMA)/CD227, cytokeratin AE1/AE3, cyclin D1, human breast cancer 1 protein (BRCA-1), C-erb-2, Bcl-2, human androgen receptor (AR), cyclin-dependent kinase inhibitor 1B (p27kip1), p53, topoisomerase II alpha, proliferating cell nuclear antigen, and Ki-67 were tested in our cases.
Results:
EMA/CD227 was positive in the well-differentiated sebaceomas (13/30). Cyclin-dependent kinase inhibitor 1B was positive in tumors with intermediate differentiation (22/30). The less well-differentiated tumors failed to stain with EMA and AR. Most of the tumors with well-differentiated palisaded areas demonstrated positive staining for topoisomerase II alpha, p27kip1, and p53, with positive staining in tumoral basaloid areas (22/30). Numerous tumors were focally positive with multiple markers, indicating a significant degree of variability in the complete group.
Conclusions:
Oncogenes, tumor suppressor genes, cell cycle regulators, and hormone receptors are variably expressed in sebaceomas. Our results suggest that in these tumors, selected marker staining seems to correlate with tumor differentiation; that is, well-differentiated tumors as a group stained with EMA and AR, and palisaded areas demonstrated consistent p53, topoisomerase II alpha and p27kip1 staining. In contrast, less well-differentiated areas stained with a different spectrum of markers.
Keywords: Cell cycle regulators, Sebaceoma, Tumor suppressor genes and oncogenes
Introduction
Sebaceoma is a clinically indolent tumor that should be distinguished from basal cell carcinoma (BCC), squamous cell carcinoma (SCC), and sebaceous carcinoma.[1,2,3,4] Sebaceoma is nosologically difficult to classify vis-a-vis other benign and malignant sebaceous neoplasms. To diagnose a sebaceoma, the tumor must display some degree of sebaceous differentiation within tumor lobules, and additional basaloid differentiation in the periphery of the lobules.[1,2,3,4] The majority of morphological studies classifying these tumors have used hematoxylin and eosin (H and E) data. Occasional studies have utilized immunohistochemical (IHC) markers.[3,4,5] We attempted to evaluate a group of these tumors utilizing a panel of IHC markers, including cytokeratin AE1/AE3, epithelial membrane antigen (EMA), and multiple cell-cycle oncogene and tumor suppressor gene markers. We acknowledge that this study is limited by relatively small number; however, this entity is uncommon and thus our findings will lend some insight to the biologic and/or diagnostic classification of sebaceomas.
Materials and Methods
H and E staining
We studied 30 archival skin biopsies with independent diagnoses of sebaceoma from three dermatopathology laboratories; each biopsy diagnosis was performed by a board certified dermatopathologist in the USA. The study was excepted from Institutional Review Board requirements because the samples were without patient identifiers as well as archival biopsies. The parameters such as age, sex, and race of the subjects for the 30 biopsies were also analyzed. All biopsies were initially fixed in 10% buffered formalin, embedded in paraffin, and cut at 4 micron thicknesses. The tissues were stained with H and E as previously described.[6,7,8] Our study included only lesions that had typical features of basaloid, palisading margins, and the presence of cells with some degree of sebaceous differentiation inside the tumor.[1,2,3,4] Tumors with i) a majority of sebaceous cells demonstrating severe cytologic atypia, or ii) high sebaceous component mitotic rates (focally greater than four mitotic figures per high powered, ×400 microscopic field) were excluded and considered to be sebaceous carcinomas.
IHC staining
The IHC staining was read as positive or negative, in the presence of a negative and a positive control for each marker tested. These readings were performed by an immunodermatologist and were based on the positivity of the stain/mm2 of tumor, at ×200 and ×400 total magnification. Our IHC staining was performed as previously described.[6,7,8] The extent and location of IHC staining for each marker was assessed according to an immunoreactive score (IRS) that evaluated the proportion of cells expressing each marker and the intensity of staining. Staining intensity was graded as 0 (negative), 1 (weak), 2 (moderate), and 3 (strong); and the percentage of positive cells examined was scored as 0 (negative staining), 1 (<10%), 2 (11-50%), 3 (51-80%), and 4 (>80%). The two scores were then multiplied together and the IRS (values from 0-12) was determined: 0 as negative, 1-3 as weak, 4-6 as positive, and 7-12 as strongly positive. No quantitative or semiquantitative results with regard to proliferative markers were done. For IHC, we utilized the following Dako antibodies: Monoclonal mouse anti-human EMA/CD227, monoclonal mouse anti-human p53, monoclonal rabbit anti-human cyclin D1, monoclonal mouse anti-human breast cancer type 1 susceptibility protein (BRCA-1), proliferating cell nuclear antigen (PCNA), monoclonal mouse anti-human Ki-67 antigen, polyclonal rabbit anti-human C-erB-2 oncoprotein, (also called HER2/neu, human EGF receptor 2, and human epidermal growth factor receptor 2), monoclonal mouse anti-human cyclin-dependent kinase inhibitor 1B (p27Kip1), monoclonal mouse anti-human androgen receptor (AR), monoclonal mouse anti-human topoisomerase II alpha, and monoclonal mouse anti-human cytokeratin AE1/AE3. Clone AE1 detects the high molecular weight cytokeratins 10, 14, 15, and 16, and also the low molecular weight cytokeratin 19. Clone AE3 detects the high molecular weight cytokeratins 1, 2, 3, 4, 5, and 6. All tumors were stained with each marker and positive and negative controls were performed for each stain.
Statistical analysis
For statistical analysis, we utilized the nonparametric Mann-Whitney U-test to calculate significant levels for all measurements including the standard deviation (SD). Values of P < 0.05 were considered statistically significant.
Results
From 30 skin biopsies, 17 were from males and 13 from females with a mean age at resection of 60 ± 10 years. Several cellular proliferation markers (c-erb-2, Ki-67, Bcl-2, and Cyclin D1) were focally positive in areas inside the tumors, and also in epidermal areas superjacent to the sebaceomas P < 0.05. The majority of the well-differentiated sebaceomas stained for EMA/CD227 within the tumor, with P < 0.05 (13/30; significantly more staining than in less well-differentiated parts of the tumors). Many tumors were stained by all of the markers; however, EMA was only positive in the well-differentiated ones [Table 1 and Figure 1]. For each antibody staining pattern, please refer to Table 1 for a description of the pattern, as well as respective IRS scores. Basaloid, less well-differentiated areas of the tumors consistently failed to stain with EMA, but were consistently positive with topoisomerase II in areas with a palisading histology (P < 0.05). In these areas, we noted significantly more topoisomerase II staining than in well-differentiated parts of the sebaceomas (24/30). Topoisomerase II was positive in four different patterns among the tumors [Table 1]. PCNA was positive only in 9/30 of the less well-differentiated sebaceomas. In some sebaceomas, we noted mixed patterns, with basaloid cells admixed with areas of sebaceous differentiation and squamous differentiation; the squamous differentiation was highlighted by cytokeratin AE1/AE3 expression. Both the AE1 and AE3 individual clones detect specific high and low molecular weight keratins [Table 1]. In addition, a subset of our sebaceomas displayed a staining overlap with BCCs, based upon the observed positivity of p27kip1, p53, and topoisomerase II alpha markers in the basaloid cells of the sebaceomas [Figure 1]. In comparison to the other sebaceomas in our series, the observed positive staining for these markers was statistically significant, with P < 0.05. Human AR was only positive in well-differentiated sebaceomas [Table 1]. The majority of our biopsies were completely negative for C-erB-2 oncoprotein, BRCA-1, and Ki-67.
Table 1.
Markers for sebaceoma show a spectrum of cell cycle regulators, tumor suppressor genes, and oncogenes
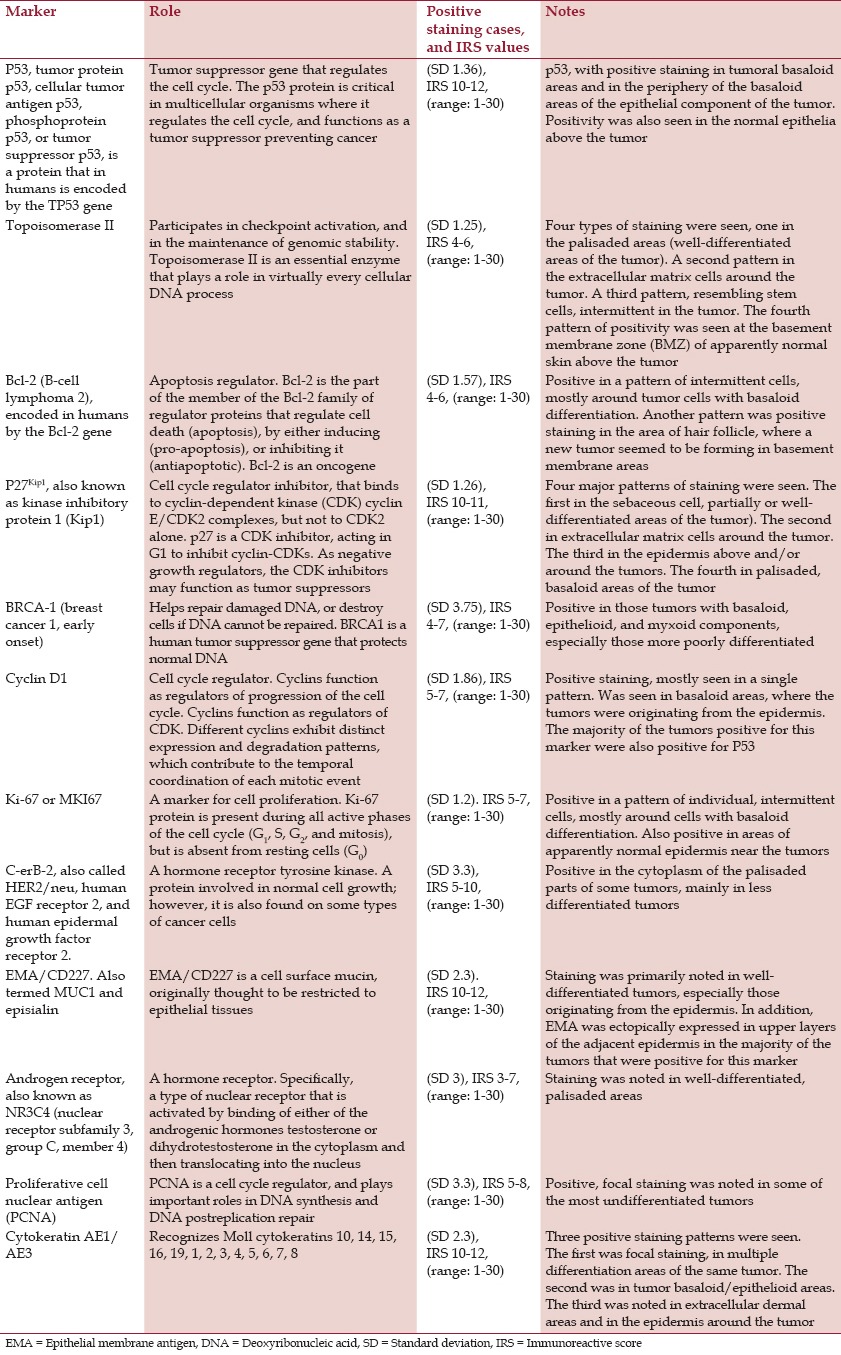
Figure 1.
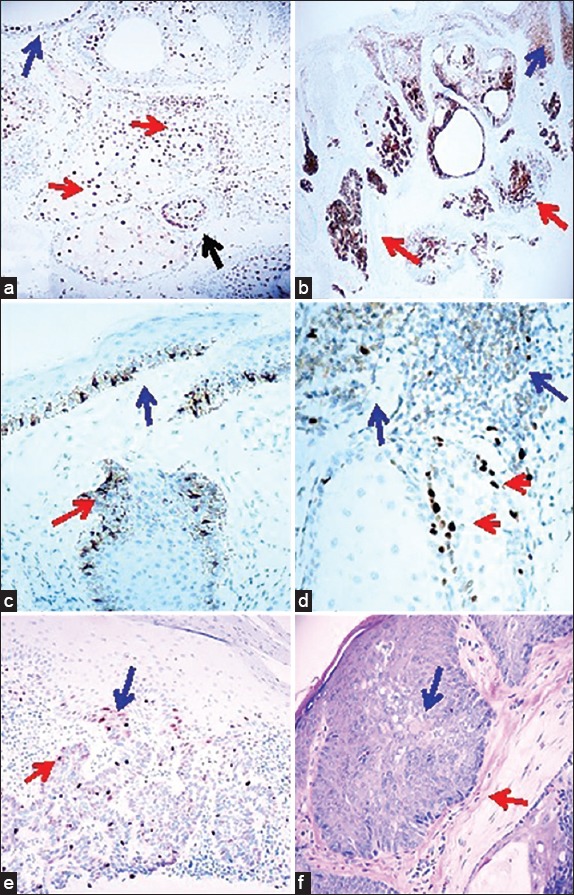
Immunohistochemical staining panel on sebaceoma. a. IHC, with P27Kip positive staining inside a tumor (brown staining; red arrows), and also in the adjacent epidermis above the tumor (brown staining; blue arrow) (100×) (the black arrows is positive around the tumor. b. IHC, demonstrating EMA/CD227 positive staining in a tumor (brown staining; red arrows) and also in the suprajacent epidermis (brown staining; blue arrow) (40×). c. IHC, demonstrating PCNA in the normal tissue above the tumor and in the bulb origin from the epidermis, positive staining inside a tumor palisade areas (brown staining; red arrow), and in the suprajacent epidermis (brown staining; blue arrow) (200×). d. IHC staining with Topoisomerase II, positive inside a tumor in palisaded areas (red arrow) and in surrounding peritumoral areas (brown staining; blue arrow) (400×). e. Cyclin D1 IHC staining, positive in palisaded areas of a tumor (brown staining; red arrow) as well as in an area where the tumor seems to be originating from the epidermis (brown staining; blue arrow) f. H&E staining of a tumor demonstrates focal features of BCC, with the palisaded areas (red arrow) and sebaceous differentiation areas (blue arrow).
Discussion
The H and E histologic diagnosis of sebaceoma is challenging and IHC markers have been previously utilized to assist in the diagnosis.[1,2,3,9,10] Our results similarly identified high variability among sebaceomas with regard to their expression of markers of i) cell cycle regulators, ii) oncogenes, iii) tumor suppressor genes, and iv) related proteins and hormones receptors, suggesting an immunologic spectrum of tumors defined as sebaceomas.
We identified expression of the majority of these markers in our cohort of sebaceomas. The expression of many of the markers was observed within the tumors and interestingly, also in the surrounding tumoral microenvironment including the overlying epidermis. Several tumors were demonstrated with variegated expression in foci within the tumor, and also surrounding the tumors. These two findings may be indicative of a significant immunologic variability among sebaceomas, as well as the possibility that the tumors contribute variably to immunologic changes in their surrounding microenvironment. This microenvironment expression data may prove useful, since tumoral microenvironment alterations are becoming increasingly important in the setting of personalized therapy. We found diverse degrees and patterns of positivity for specific markers across multiple cases, reinforcing these concepts. Our study has suggested that not all sebaceoma cells are identically altered, accounting for the variability of staining in each tumor. Pertinently, aneuploidy (an atypical number of chromosomes) is a trait common to most solid tumors.[11]
PCNA expression is found within many essential cellular processes, such as deoxyribonuclease acid (DNA) replication, repair of DNA damage, chromatin structure maintenance, chromosome segregation, and cell-cycle progression. Overexpression of cyclin D1 expression has been observed frequently in a variety of tumors, such those described here.[12] p27Kip is known to be involved in regulation of cell proliferation, and may contribute to the tumor genesis seen in sebaceomas. Moreover, studies in several tumor types indicate that p27Kip expression levels have both prognostic and therapeutic implications in several tumors.[12] We are conducting further studies with this marker; our preliminary results show that p27Kip is a proliferation marker that tends to be positive where proliferating sebaceous cells are present.
BRCA-1 is a human gene that produces a tumor suppressor protein. The BRCA-1 protein helps repair damaged DNA, and therefore plays a role in ensuring the stability of the cell's genetic material. When the BRCA-1 gene is mutated or altered, such that its protein product is not made or does not function correctly, damaged DNA may not be repaired properly. As a result, cells are more likely to develop additional genetic alterations that can lead to cancer. For the markers we explored, is difficult to know if the body is already responding to the tumor via tumor suppressor genes, and thus attempting to neutralize any genomic instability.[10,12] Our exploratory study is limited by its sample size; however, our study demonstrates that sebaceomas express different IHC markers depending on their degree of differentiation. We conclude that sebaceomas display abnormal expression of several oncogenes, tumor suppressor genes, cell cycle regulators, and hormone receptors depending of the degree of differentiation. Further molecular characterizations and correlations with the degree of differentiation of the tumors are warranted. In this context, we are correlating our data with further research on BCCs and SCCs, with the aim of determining a diagnosis, and further specific information on tumor differentiation based on the altered expression of oncogenes, cell cycle regulators, and tumor suppressor factors.
Further suggestive studies may include comparison of sebaceoma with sebaceous adenoma and with sebaceous carcinoma and the presence of oncogenes, tumor suppressor genes, and cell cycle regulators.
Acknowledgments
Mr. Jonathan S Jones at Georgia Dermatopathology Associates provided excellent technical assistance. Bruce R Smoller, MD, Chair of Pathology and Laboratory Medicine, University of Rochester Medical Center, Rochester, New York provided helpful advice and comments.
Footnotes
Source of Support: Nil.
Conflict of Interest: None declared.
References
- 1.Misago N, Suse T, Uemura T, Narisawa Y. Basal cell carcinoma with sebaceous differentiation. Am J Dermatopathol. 2004;26:298–303. doi: 10.1097/00000372-200408000-00006. [DOI] [PubMed] [Google Scholar]
- 2.Misago N, Mihara I, Ansai S, Narisawa Y. Sebaceoma and related neoplasms with sebaceous differentiation: A clinicopathologic study of 30 cases. Am J Dermatopathol. 2002;24:294–304. doi: 10.1097/00000372-200208000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Bayer-Garner IB, Givens V, Smoller B. Immunohistochemical staining for androgen receptors: A sensitive marker of sebaceous differentiation. Am J Dermatopathol. 1999;21:426–31. doi: 10.1097/00000372-199910000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Asadi-Amoli F, Khoshnevis F, Haeri H, Jahanzad I, Pazira R, Shahsiah R. Comparative examination of androgen receptor reactivity for differential diagnosis of sebaceous carcinoma from squamous cell and basal cell carcinoma. Am J Clin Pathol. 2010;134:22–6. doi: 10.1309/AJCP89LYTPNVOBAP. [DOI] [PubMed] [Google Scholar]
- 5.Sinard JH. Immunohistochemical distinction of ocular sebaceous carcinoma from basal cell and squamous cell carcinoma. Arch Ophthalmol. 1999;117:776–83. doi: 10.1001/archopht.117.6.776. [DOI] [PubMed] [Google Scholar]
- 6.Abreu-Velez AM, Googe PB, Howard MS. Immune reactivity in psoriatic Munro-saboureau microabscesses, stratum corneum and blood vessels. N Am J Med Sci. 2012;4:257–65. doi: 10.4103/1947-2714.97204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abreu-Velez AM, Howard MS, Yi H, Gao W, Hashimoto T, Grossniklaus HE. Neural system antigens are recognized by autoantibodies from patients affected by a new variant of endemic pemphigus foliaceus in Colombia. J Clin Immunol. 2011;31:356–68. doi: 10.1007/s10875-010-9495-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abreu-Velez AM, Howard MS, Hashimoto T, Grossniklaus HE. Human eyelid meibomian glands and tarsal muscle are recognized by autoantibodies from patients affected by a new variant of endemic pemphigus foliaceus in El-Bagre, Colombia, South America. J Am Acad Dermatol. 2010;62:437–47. doi: 10.1016/j.jaad.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heyderman E, Graham RM, Chapman DV, Richardson TC, McKee PH. Epithelial markers in primary skin cancer: An immunoperoxidase study of the distribution of epithelial membrane antigen (EMA) and carcinoembryonic antigen (CEA) in 65 primary skin carcinomas. Histopathology. 1984;8:423–34. doi: 10.1111/j.1365-2559.1984.tb02354.x. [DOI] [PubMed] [Google Scholar]
- 10.Hatta N, Hirano T, Kimura T, Hashimoto K, Mehregan DR, Ansai S, et al. Molecular diagnosis of basal cell carcinoma and other basaloid cell neoplasms of the skin by the quantification of Gli1 transcript levels. J Cutan Pathol. 2005;32:131–6. doi: 10.1111/j.0303-6987.2005.00264.x. [DOI] [PubMed] [Google Scholar]
- 11.Suijkerbuijk SJ, Kops GJ. Preventing aneuploidy: The contribution of mitotic checkpoint proteins. Biochim Biophys Acta. 2008;1786:24–31. doi: 10.1016/j.bbcan.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Orr B, Compton DA. A double-edged sword: How oncogenes and tumor suppressor genes can contribute to chromosomal instability. Front Oncol. 2013;3:164. doi: 10.3389/fonc.2013.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]


