Abstract
The ability to grow on media containing certain D-amino acids as a sole nitrogen source is widely utilized to differentiate Cryptococcus gattii from C. neoformans. We used the C. neoformans H99 and C. gattii R265 strains to dissect the mechanisms of D-amino acids utilization. We identified three putative D-amino acid oxidase (DAO) genes in both strains and showed that each DAO gene plays different roles in D-amino acid utilization in each strain. Deletion of DAO2 retarded growth of R265 on eleven D-amino acids suggesting its prominent role on D-amino acid assimilation in R265. All three R265 DAO genes contributed to growth on D-Asn and D-Asp. DAO3 was required for growth and detoxification of D-Glu by both R265 and H99. Although growth of H99 on most D-amino acids was poor, deletion of DAO1 or DAO3 further exacerbated it on four D-amino acids. Overexpression of DAO2 or DAO3 enabled H99 to grow robustly on several D-amino acids suggesting that expression levels of the native DAO genes in H99 were insufficient for growth on D-amino acids. Replacing the H99 DAO2 gene with a single copy of the R265 DAO2 gene also enabled its utilization of several D-amino acids. Results of gene and promoter swaps of the DAO2 genes suggested that enzymatic activity of Dao2 in H99 might be lower compared to the R265 strain. A reduction in virulence was only observed when all DAO genes were deleted in R265 but not in H99 indicating a pathobiologically exclusive role of the DAO genes in R265. These results suggest that C. neoformans and C. gattii divergently evolved in D-amino acid utilization influenced by their major ecological niches.
Introduction
Cryptococcus neoformans and Cryptococcus gattii are two closely related basidiomycetous yeasts that cause cryptococcosis in humans and animals [1]. Although these two species share 85–90% genomic identity [2] and cause disease that is treated by the same therapeutic measures, these two species are distinguishable in many ways. The major ecological niche of C. neoformans is pigeon droppings and distributed world-wide. C. neoformans causes meningoencephalitis mainly in HIV infected and other immunocompromised patients [1]. The major ecological niche of C. gattii is higher plants. C. gattii causes diseases more frequently in immunocompetent patients than in immunocompromised individuals and until the 1990s was known to be prevalent only in subtropical to tropical regions [3,4]. However, the outbreak of C. gattii infection reported on Vancouver Island, Canada in early 2000 [5,6] suggested that the species had spread to temperate zones and it is now recognized as an emerging pathogen in the Pacific Northwest [7,8].
The yeast cell morphology of these two species is similar although strains of C. gattii tend to produce more ovoidal to pyriform cells compared to those produced by C. neoformans which are globose with rare pyriform cells [9]. These two species are clearly discernable in the morphology of their sexual spores. However, production of sexual spores is impractical for routine identification. Prior to the availability of molecular tests, laboratories had used serotyping or biochemical tests to differentiate between the two species. The most widely used biochemical tests relied on differences in nutritional growth properties. For instance, creatinine is used as the sole source of nitrogen by both species but the enzyme, creatinine deiminase, responsible for creatinine utilization, is regulated differently between the two species [10]. Creatinine deiminase is repressed by ammonia in C. neoformans but not in C. gattii. This difference is manifested by a change in the color of the diagnostic media as the pH changes [10,11]. C. gattii utilizes glycine as both carbon and nitrogen sources and a majority of C. gattii isolates are resistant to L-canavanine while the majority of C. neoformans strains can utilize glycine only as a nitrogen source but not as a carbon source and are susceptible to L-canavanine [12–14]. As a result, media containing L-canavanine and glycine with bromothymol blue as the color indicator for growth (CGB medium) can efficiently differentiate between these two species [14–16]. Furthermore, most isolates of C. gattii can use D-proline or D-alanine as the sole nitrogen source and can assimilate D-tryptophan to produce pigment whereas only a minor population of C. neoformans can do the same [15,17,18]. The mechanisms responsible for these diagnostic tests, however, have not been investigated. It is known that plants can synthesize D-amino acid derivatives and some D-amino acids such as D-alanine are widely reported in higher plants [19,20]. Furthermore, D-amino acids have been found in the mammalian forebrain and various tissues [21–23]. Because the major ecological niche of C. gattii is plant, the patterns of D-amino acid utilization may bear pathobiological and ecological significance. We, therefore, considered it important to discern the mechanisms underlying the differences between the two species.
We screened for mutants of C. gattii that grew poorly on D-Ala and D-Pro as the sole nitrogen source. One of the isolated mutants contained an insertion at a gene containing a D-amino acid oxidase domain. D-amino acid oxidase (Dao; EC 1.4.3.3) is a flavin adenine dinucleotide (FAD)-containing enzyme that catalyzes the oxidative deamination of D-amino acids to the corresponding α-keto acids and ammonia (http://www.chem.qmul.ac.uk/iubmb/). Dao has high stereoselectivity towards the D-isomers of amino acids and is almost inactive towards the corresponding L-isomers [24]. Dao plays distinct physiological roles in different organisms [25]. For instance, Dao can catabolize D-amino acids and allow bacteria or yeast to grow using D-amino acids as the sole carbon or nitrogen source. Dao also has a regulatory role in the human brain where it controls the levels of the neuromodulator D-serine.
In this study, we found that the two representative genome-sequenced strains, C. neoformans (H99) and C. gattii (R265), each contain three paralogous DAO genes. We characterized the different role of each DAO gene in D-amino acid utilization. Interestingly, DAO genes played a role in pathobiology of C. gattii but not in C. neoformans.
Materials and Methods
Strains and culture conditions
C. neoformans H99 and C. gattii R265 are genome sequenced strains. All other strains derived from C. neoformans H99 and C. gattii R265 are listed S1 Table. Strain Y-1091 of Rhodosporidium toruloides was obtained from USDA Agricultural Research Service. Spot assays were performed as described [26] on YNB media with 2% glucose and 10 mM ammonium sulfate or 10 mM D- or L-amino acids as the sole nitrogen source. For toxicity tests, 100 mM of the indicated D-amino acid was added to YNB medium with 2% glucose and 10mM ammonium sulfate. Plates were incubated at 30°C for 3–14 days and photographed.
Screening and identification of the genes important for growth of R265 on D-amino acid medium
A T-DNA insertion library of R265 was made using Agrobacterium tumefaciens mediated transformation as described previously [27]. To screen for mutants that grow poorly on D-Ala or D-Pro as the sole nitrogen source, about 20,000 individual transformants were replica spotted on yeast nitrogen base media supplemented with 50 μg/ml geneticin, 200 μM cefotaxime and containing either 10 mM ammonium sulfate, D or L-Ala, D or L-Pro as the sole nitrogen source. The plates were incubated at 30°C and growth was monitored for 7 days. Mutants that grew poorly on D-Ala or D-Pro were selected for further analysis. Genomic DNA was isolated and the T-DNA insertion site was mapped in R265 mutants by using a PCR based Vectorette system (Sigma, Woodlands, TX). Genomic sequence flanking the insertion site was subsequently obtained by sequencing the PCR product. BLAST analysis using R-265 database at the Broad institute (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans_b/MultiHome.html) was performed to reveal the loci containing T-DNA insert.
Gene deletion and complementation
The primers used in this study are listed in S2 Table. Genes of interest were disrupted via homologous recombination by biolistic transformation [26,28]. Disruption constructs were created using the overlapping PCR technique by linking the 5’ and 3’ flanking region of the gene of interest to selectable
markers NEO (G418 resistance), NAT (nourseothricin resistance) or HYG (hygromycin resistance) [29]. Homologous integrations were confirmed by PCR and Southern hybridization (data not shown). The deletants were complemented by homologous integration using biolistic transformation. CgDAO2 was reconstituted in Cgdao1ΔCgdao2ΔCgdao3Δ triple deletant with the BLE (phleomycin resistance) marker.
To overexpress the DAO genes, the promoter of GPD (Glyceraldehyde-3-phosphate dehydrogenase) as well as the coding and 3’ untranslated region of each DAO gene were PCR amplified along with the HYG cassette. The PCR products were cloned into BamHI/ApaI site of pCR2.1 using the Clontech In-Fusion HD Cloning Plus kit. After confirmation of the construct by DNA sequencing, the linearized DNA fragment was transformed into indicated strains.
For the promoter and gene swap experiment, the CnDAO2 and CgDAO2 promoters, which contained 867 bp and 827 bp upstream from the first ATG of the coding region, respectively were PCR amplified. To provide the flanking region for homologous integration, the truncated promoter region (R2(f) an H2(f)) was PCR amplified. The final construct was obtained by joining the PCR fragments using the Clontech In-Fusion HD Cloning Plus kit. After confirmation of the construct by DNA sequencing, the linearized DNA fragment was transformed into G418 resistant strains of Cgdao2Δ or Cndao2Δ. The hygromycin resistant transformants were tested for G418 sensitivity. The G418 sensitive transformants, which indicated that homologous integration had occurred at the deleted dao locus, were selected and the integration event was confirmed by Southern blot analysis.
Preparation and analysis of RNA
For northern blot analysis, log phase YPD grown cells were washed and transferred to YNB medium containing 10 mM of the different D-amino acids or ammonium sulfate. RNA was extracted from yeast cells using Trizol (Invitrogen, Carlsbad, CA). 32P-labelled DNA probes for northern blots were synthesized from PCR products generated using gene specific primer pairs (S2 Table). The probe for each DAO gene did not cross hybridize amongst each other. For quantitative northern analysis, blots were hybridized with the indicated probe, stripped and hybridized with an ACTIN probe. The blot was exposed to Phosphorimager Screen and quantified with ImageQuant (Molecular Dynamics). Signal of each DAO gene was normalized to that of the ACTIN gene and expressed as the relative amount to H99 or R265. For quantitative RT-PCR, log phase YPD grown cells were washed, transferred to YNB medium and grown for 3 h. Cells were washed and transferred to YNB medium containing 10 mM of the different D-amino acids or ammonium sulfate for 2 h. RNA isolation and quantitative RT-PCR was performed as described [26]. The PCR efficiency and CT determination was performed using the algorithm as described [30]. Data were normalized with ACTIN1 levels and expressed as the amount relative to RNA levels of H99 or R265 grown in ammonium sulfate.
Virulence studies
The animal experiments were carried out with the approval (#A4149-01) and oversight of the Animal Care and Use Committee of the National Institute of Allergy and Infectious Diseases, National Institutes of Health, USA. Infected mice were monitored daily for signs of lethargy, ruffling, and abnormal ambulation. If mice displayed any one of these criteria, they were then monitored twice daily. Mice were sacrificed by gas cylinder CO2 when they became moribund and that day post infection was considered the survival endpoint. Female BALB/c mice (6–8 weeks old) were injected via the lateral tail vein with 0.2 ml suspension of the indicated yeast strains (2.5 x 105 cells/ml in 0.9% NaCl) [31]. Alternatively, BALB/c mice were inoculated with 5,000 yeast cells via intrapharyngeal aspiration [31,32]. Kaplan-Meier analysis of survival was performed using JMP software for Macintosh (SAS Institute, Cary, NC).
Recombinant protein production
PCR was used to amplify each DAO from C. gattii and R. toruloides cDNA. The primers introduced XmnI and BamHI restriction enzyme sequences at the ends of the DAO genes to clone into the pMAL-C5X vector (New England Biolabs, Ipswich, MA) cloning site using the Clontech In-Fusion HD Cloning Plus kit (Clontech, Mountain View, CA). The cloned DAO gene was inserted downstream of MBP so that the recombinant MBP-Dao proteins could be purified by affinity chromatography. Protein extracts were obtained according to the manufacturer’s suggestions. The horseradish peroxidase/o-dianisidine Dao detection assay was used as previously described [33]. Briefly, crude protein extract was added to a reaction mix containing 50 mM KPO4, 0.86 mM o-dianisidine, 100 units horseradish peroxidase, 1.25 uM flavin adenine dinucleotide. After the mixture was heated to 30°C, 100 mM of amino acid was added and the reaction was allowed to progress for 5 minutes. The reaction was stopped by addition of H2SO4 and the activity was assayed by measuring color changes on a spectrophotometer at OD540. The enzyme activity was quantified by comparison to the standard curve generated by pure H2O2.
Results and Discussion
H99 and R265 differ in utilization of D- or L-amino acids as the sole nitrogen source
Although it has been recognized that C. gattii strains can utilize several D-amino acids more efficiently than C. neoformans, a comprehensive comparison in the utilization of the common D-amino acids as the sole nitrogen source has not been carried out. We examined the growth capability of H99 and R265 in each of the common D and L-amino acids as the sole nitrogen source. The growth rate of each strain was determined by monitoring OD600 at several time points for 24 hours. We found that glycine and ammonium sulfate were good nitrogen sources for both R265 and H99 (Table 1). In general, both H99 and R265 grew better in L-amino acids than in D-amino acids. However, R265 grew better than H99 in most D-amino acids. D-Asn was the only D-amino acid that supported better growth of H99 than R265. D-Gln was the only D-amino acid that served as a good nitrogen source for R265. Three D- and L-amino acids (Cys, Thr, and Tyr), one L-amino acid (L-His) and one D-amino acid (D-Ile) were poor nitrogen sources for either of R265 and H99. These growth patterns delineate the differences between H99 and R265 in utilizing the common D- or L-amino acids as the sole nitrogen source.
Table 1. The ability of H99 and R265 to grow in various amino acids as a sole nitrogen source.
| D-amino acid | L-amino acid | |||||||
|---|---|---|---|---|---|---|---|---|
| H99 | R265 | H99 | R265 | |||||
| OD600 a | ranking b | OD600 | ranking | OD600 | ranking | OD600 | ranking | |
| Ala | 0.4 | + | 2.9 | ++ | 4.1 | +++ | 5.4 | +++ |
| Arg | 0.3 | + | 0.8 | + | 5.3 | +++ | 6.4 | +++ |
| Asn | 3.0 | +++ | 1.5 | ++ | 6.1 | +++ | 6.4 | +++ |
| Asp | 0.6 | + | 1.5 | ++ | 6.6 | +++ | 7.6 | +++ |
| Cys | 0.6 | + | 0.8 | + | 0.6 | + | 0.6 | + |
| Gln | 3.2 | +++ | 5.3 | +++ | 4.9 | +++ | 5.2 | +++ |
| Glu | 0.2 | + | 1.3 | ++ | 0.6 | + | 3.5 | +++ |
| His | 0.5 | + | 1.2 | ++ | 0.6 | + | 0.7 | + |
| Ile | 0.5 | + | 0.9 | + | 1.2 | ++ | 1.0 | ++ |
| leu | 0.5 | + | 1.6 | ++ | 1.6 | ++ | 3.2 | +++ |
| Lys | 0.5 | + | 1.1 | ++ | 3.1 | +++ | 3.1 | +++ |
| Met | 0.5 | + | 2.3 | ++ | 4.0 | +++ | 1.4 | ++ |
| Phe | 0.5 | + | 2.5 | ++ | 3.3 | +++ | 1.3 | ++ |
| Pro | 0.3 | + | 2.1 | ++ | 4.6 | +++ | 5.4 | +++ |
| Ser | 0.5 | + | 2.9 | ++ | 2.7 | ++ | 5.0 | +++ |
| Thr | 0.5 | + | 0.9 | + | 0.8 | + | 0.8 | + |
| Trp | 0.5 | + | 1.3 | ++ | 3.1 | +++ | 0.6 | + |
| Tyr | 0.4 | + | 0.3 | + | 0.1 | + | 0.1 | + |
| Val | 0.4 | + | 1.1 | ++ | 0.7 | + | 1.5 | ++ |
| Gly | 4.4 | +++ | 5.7 | +++ | ||||
| NH4SO4 | 4.0 | +++ | 4.2 | +++ | ||||
a. Log phase cells were diluted to OD600 = 0.1 as starting culture. Indicated amino acid was used as a sole nitrogen source in the liquid YNB media. The culture was incubated at 30°C and the OD600 was monitored at several time points for 24h. Only the data from the 24h point are shown. The experiments were repeated twice and the representative data are shown.
b. For comparison, we arbitrary classified the relative growth into three levels: +++ = good N- source, OD600 ≥ 4.0; ++ = moderate N-source, 1.0 < OD600 < 4.0; + = poor N-source, OD600 < 1.0.
Isolation of R265 mutants that grow poorly on D-Pro or D-Ala as the sole nitrogen source
D-Pro and D-Ala are commonly used to distinguish C. gattii from C. neoformans strains [15,17,18]. To characterize the mechanism of D-amino acid metabolism in C. gattii, a T-DNA insertional library of R265 containing about 20,000 individual clones was first constructed. We then screened for mutants that grew poorly on D-Pro or D-Ala but grew normally on L-Pro or L-Ala. Three such mutants were isolated and the genomic location of the T-DNA insertion in each mutant was determined. Two of the mutants had a T-DNA insertion at either the coding region of CNBG_4524 or the flanking region of CNBG_2060, which were annotated by the Broad Institute as myosin-1 and a hypothetical protein, respectively. In spite of several attempts, we were unable to identify the insertion site of the third mutant. Deletion of the CNBG_4524 gene encoding the putative myosin-1 resulted in a growth defect only on D-Pro but not on D-Ala. We, therefore, focused our studies on CNBG_2060. Sequence analysis indicated that CNBG_2060 contained a Glycine/D-amino acid oxidase domain (COG0665) and we designated this gene as DAO2 because a putative D-aspartate oxidase gene had been annotated at the Broad Institute (see below). For convenience, we specified the DAO2 gene from R265 as CgDAO2 to distinguish it from the orthologous gene of H99 (CnDAO2). A BLAST search of the R265 genome using CgDAO2 as a query sequence revealed the presence of two putative DAO paralogs, CNBG_1742 and CNBG_4227, which had been annotated as D-aspartate oxidase and a hypothetical protein respectively by the Broad Institute. We designated CNBG_1742 and CNBG_4227 in R265 as CgDAO1 and CgDAO3, respectively. The sequences of three DAO genes from R265 were used as queries to search for the orthologs in the H99 genome. Three orthologs, CNAG_05802, CNAG_03562, and CNAG_02532 were identified and were designated as CnDAO1, CnDAO2 and CnDAO3, respectively. S1 Fig shows the protein sequence alignment of these Daos.
CgDAO2 is the major DAO in R265
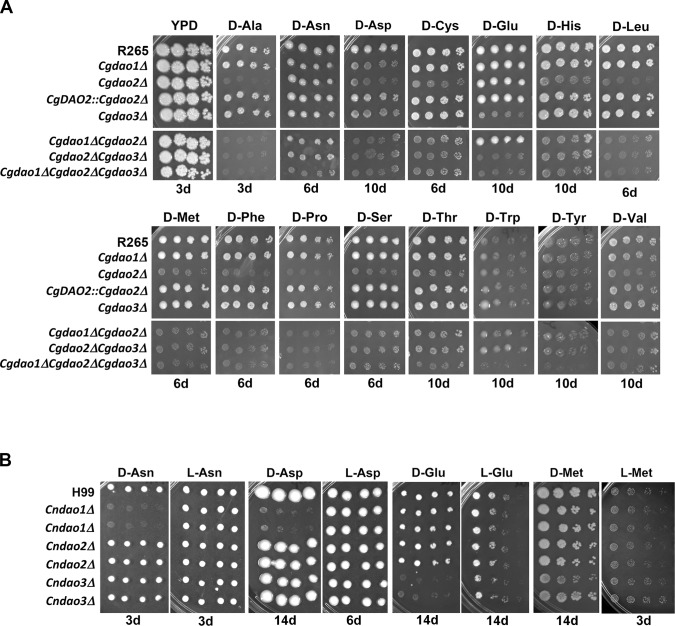
Oxidative deamination of D-amino acids by Dao produces ammonia which can be used as the sole nitrogen source for growth. We deleted each of the three CgDAO genes in R265 and examined its effect on growth on all common D-amino acids. Deletion of individual CgDAO1, CgDAO2 or, CgDAO3 genes did not affect growth on any of the common L-amino acids (Table 2) indicating that DAO genes are not required for L-amino acids utilization. Deletion of CgDAO2 resulted in retarded growth on 11 different D-amino acids and reconstitution of CgDAO2 restored the growth on those D-amino acids (Fig 1A and Table 2) suggesting that CgDAO2 is the major DAO gene required for normal growth of R265 on those 11 D-amino acids. Deletion of CgDAO3 in R265 markedly reduced the growth on D-Glu and slightly affected the growth on D-D-Asp compared to the wild-type. Growth of Cgdao1Δ was also slightly affected in D-Asp. Therefore, D-Asp was the only D-amino acid with which all three single dao deletants showed a phenotype of reduced growth. Although deletion of each CgDAO gene separately did not clearly affect the growth of R265 on D-Asn, D-Trp or D-Tyr, the triple CgDAO gene deletion resulted in severe growth retardation on those three D-amino acids. Interestingly, all R265 dao single deletants grew well on D-Arg, D-Gln, D-Ile and D-Lys (Table 2). These amino acids, except for D-Ile, all contain a side-chain amine group that could be used as a nitrogen source. These observations indicate that DAO genes are important for growth of R265 on all common D-amino acids that lack the side-chain amine group except for D-Ile. It is possible that additional unidentified D-amino acid oxidases or other enzyme(s) in R265 may convert D-Ile to a useable nitrogen source.
Table 2. Summary of growth phenotype associated with deletion of DAO genes.
| D-amino acid | Deletant showing growth defect on D-amino acid a |
|---|---|
| Ala | Cgdao2 |
| Arg | - b |
| Asn | Triple e , Cndao1 |
| Asp | Cgdao1 c , Cgdao2 c , Cgdao3 c , Cndao1 |
| Cys | Cgdao2 |
| Gln | - b |
| Glu | Cgdao3, Cndao3 |
| Gly | NDL d |
| His | Cgdao2 |
| Ile | - b |
| Leu | Cgdao2 |
| Lys | - b |
| Met | Cgdao2, Cndao3 c |
| Phe | Cgdao2 |
| Pro | Cgdao2 |
| Ser | Cgdao2 |
| Thr | Cgdao2 |
| Trp | Triple e |
| Tyr | Triple e |
| Val | Cgdao2 c |
a All dao mutants of R265 and H99 grew normally on L-amino acids.
b No phenotype.
c Growth was only slightly reduced compared to the wild-type.
d No D, L-isomers.
e Triple deletant of R265 displayed growth defect on D-amino acid.
Fig 1. DAO genes are important for growth on D-amino acids.
(A) Phenotype of R265 daoΔ mutants. (B) CnDAO1 and CnDAO3 play different roles for growth on D-amino acids. Three-fold serial dilutions of each indicated strain were spotted on indicated medium and incubated at 30°C. The strains used in the experiments are given on the left. Two independent deletants of each DAO gene from H99 were assayed. Pictures were taken after incubation for 3 days, 6 days, 10 days or 14 days as indicated. The experiments were repeated twice and representative figures are shown.
CnDAO1 and CnDAO3 play different roles in D-amino acids utilization in H99
Although growth of H99 was ignorable compared to R265 on most D-amino acids (Table 1), H99 grew visibly on certain D-amino acids. Therefore, the role of each DAO gene in H99 was also investigated. CnDAO1 was important for the utilization of D-Asn and D-Asp since deletion of CnDAO1 reduced the growth of H99 on these D-amino acids (Fig 1B). The growth of Cndao3Δ was undetectable on D-Glu and was slightly reduced on D-Met compared to the wild type H99 strain. Deletion of CnDAO1 or CnDAO3 did not retard the growth on any of the other D-amino acids. Furthermore, Cndao2Δ behaved as the wild-type H99 on every D-amino acid indicating that unlike CgDAO2 in R265, CnDAO2 was not important for the growth of H99 on any D-amino acid.
DAO3 contributes to adaptation towards D-Glu induced toxicity
D-amino acids have been reported to be toxic to many microorganisms including fungi and bacteria [34–37]. We evaluated the toxicity of six representative amino acids including D-Pro and D-Ala which poorly supported the growth of Cgdao2Δ, D-Trp which affected growth only on the triple deletant, as well as D-Asn, D-Asp and D-Glu, which failed to support growth of Cndao1Δ or Cndao3Δ mutant (Fig 1). The strains were spotted on defined agar media containing 2% glucose and 10 mM ammonium sulfate supplemented with or without 100 mM of the selected D- or L-amino acid. Growth of H99 was not affected by the presence of six D-amino acids tested or their L-enantiomers (Fig 2 and S2 Fig). In contrast, addition of D-Glu or D-Ala slightly reduced the growth of wild-type R265. Furthermore, growth of Cndao3Δ was slightly reduced in the presence of 100 mM D-Glu whereas growth of Cgdao2Δ on D-Ala and Cgdao3Δ on D-Glu was slower compared to the R265 strain (Fig 2). Triple and single deletants of DAO behaved similarly suggesting no additive effects from the deletion of more than one CgDAO gene. Therefore, both CnDAO3 and CgDAO3 influenced growth and detoxification of D-Glu in H99 and R265 respectively. In contrast, only CgDAO2 but not CnDAO2 influenced growth and detoxification of D-Ala.
Fig 2. DAO genes are important for detoxification of D-amino acids.

Three-fold serial dilutions of each strain were spotted onto YNB medium containing 2% glucose and 10 mM ammonium sulfate supplemented with or without 100 mM D- or L-amino acids. Plates were incubated at 30°C for 2 days and photographed. The strains used in the experiments are given on the left.
Expression patterns of DAO genes in eight different D-amino acids
In various fungi, the expression of DAO is induced in the growth media where a D-amino acid is the sole nitrogen source [38–40]. Since growth of R265 on D-Ala and D-Pro required CgDAO2, we examined the expression patterns of CgDAO2 in media containing D-Ala or D-Pro as the sole nitrogen source. The expression of CgDAO2 was detectable in the rich medium YPD, and its expression did not change substantially when the cells were shifted from YPD medium to defined medium containing ammonium sulfate as the sole nitrogen source for 6 h (Fig 3A). However, when R265 was shifted from YPD to D-Ala as the sole nitrogen source, expression of CgDAO2 increased at 3 h after transfer and maintained the increased levels for at least 25 hours. Similarly, expression of CgDAO2 increased slightly at 6 h after shifting from YPD to D-Pro and maintained the increased levels for at least for 25 hours. Therefore, CgDAO2 is required for the growth on D-Ala and D-Pro and its expression levels increase in the presence of these two D-amino acids.
Fig 3. Expression of DAO genes is induced by certain D-amino acids.
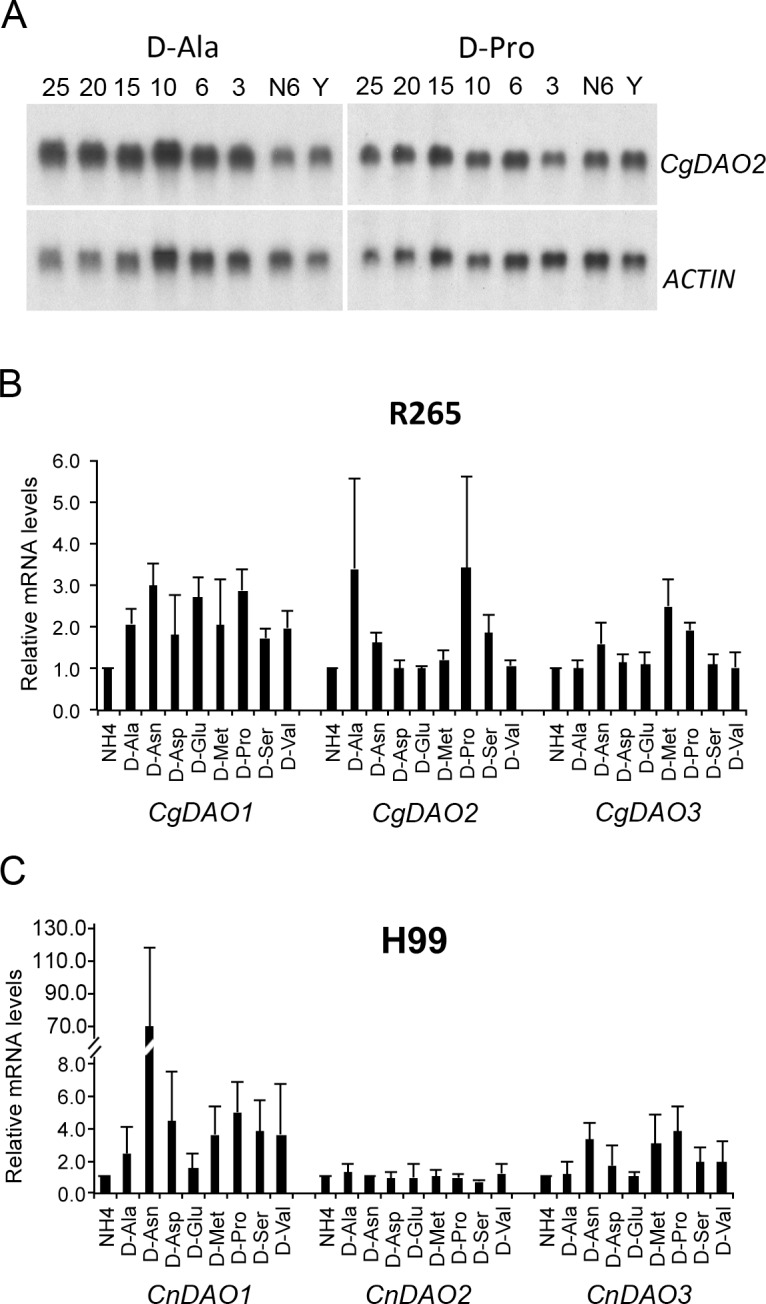
(A) CgDAO2 expression increases in the presence of D-Ala or D-Pro. YPD grown cells (Y) were washed and transferred to YNB medium containing 10 mM D-Ala, D-Pro or ammonium sulfate (N) for the indicated hours. Total RNA (5 μg) was subjected to northern blot analysis using a CgDAO2 probe. Actin served as loading control. (B and C) Expression profiles of R265 and H99 DAO genes in various D-amino acids. YPD grown cells were washed and transferred to YNB medium containing 10mM of the indicated D-amino acids or ammonium sulfate for 2 h. Northern blots were hybridized with the indicated DAO probes. Signals of each DAO gene were normalized to that of the ACTIN gene and expressed as the relative amount to R265 or H99 RNA from the ammonium sulfate grown cultures. The experiments were repeated three times and the error bar represents standard deviation.
We expanded the gene expression analysis of each DAO in both R265 and H99 using northern blot analysis. Eight different D-amino acids were selected based on growth phenotypes; five D-amino acids in which Cgdao2Δ showed a growth deficiency and three D-amino acids in which Cndao1Δ or Cndao3Δ grew poorly. Fig 3B shows that in R265, the expression levels of CgDAO2 increased more than two-fold only in the cells grown on D-Ala or D-Pro while no such change occurred in those grown in other D-amino acids. Although deletion of CgDAO1, CgDAO2 and CgDAO3 each affected the growth of R265 on D-Asp slightly (Fig 1A), none of these genes showed a greater than two-fold increase of the expression levels in D-Asp grown cells. Furthermore, expression levels of CgDAO1 increased more than two-fold in six out of the eight tested D-amino acids while expression levels of CgDAO3 increased more than two-fold in one (D-Met) out of the eight tested D-amino acids.
In H99, expression of CnDAO1 was highly elevated in media containing D-Asn (Fig 3C). Additionally, the expression levels of CnDAO1 in H99 were elevated greater than 2-fold in seven out of the eight tested D-amino acids even though deletion of CnDAO1 only showed growth retardation on D-Asp and D-Asn (Fig 1B and Table 2). Similarly, the expression levels of CnDAO3 increased more than two-fold in D-Asn, D-Met, and D-Pro while deletion of CnDAO3 did not affect the growth of H99 in those D-amino acids. In contrast, the expression of CnDAO3 did not increase in D-Glu even though CnDAO3 was important for the growth on D-Glu or detoxification of D-Glu. Lastly, the expression of CnDAO2 did not change more than two-fold in all eight tested D-amino acids. Since deletion of CnDAO2 had no phenotype in all the tested conditions and expression of CnDAO2 was not inducible by any of the tested D-amino acids, the function of CnDAO2 is not clear for D-amino acid utilization in H99. Taken together, results of the expression profile analysis indicate that the relationship between the growth phenotype of each dao mutant and the expression levels of the corresponding DAO gene in both H99 and R265 is not well-defined. It is known that Gat1 belongs to a conserved family of zinc finger containing transcriptional regulators which activate the transcription of nitrogen catabolite repression sensitive genes when preferred nitrogen sources are absent or limiting [15,41]. It is possible that GAT1 may also control the expression of DAO genes in different D-amino acids. However, no clear relationship was observed between the expression profile of each DAO genes and deletion of GAT1 in H99 or R265. It is possible that other factors may mitigate the expression levels of specific DAO genes in specific D-amino acids.
Certain DAO genes are functionally interchangeable when overexpressed
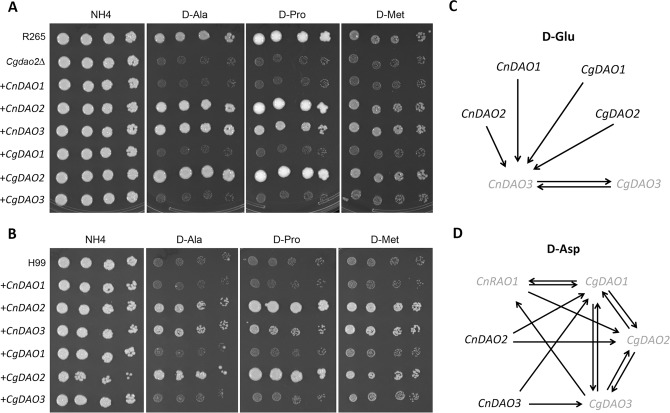
Since the results of phenotypic analysis suggested that each DAO gene functions differently in D-amino acid utilization, we next examined if each DAO gene could functionally substitute other DAO genes by overexpressing individual DAO gene under control of the strong GPD promoter. D-amino acids that affected the growth of single dao-deletion mutants were divided into three groups to test the effect of DAO gene overexpression. The first group included 10 D-amino acids in which Cgdao2Δ grew poorly (Table 2). D-Ala, D-Pro and D-Met were selected as the representatives of this group for the test. D-Glu was the second group since Cgdao3Δ and Cndao3Δ strains both failed to use D-Glu as the nitrogen source. The last group included D-Asp in which the growth of Cgdao1Δ, Cgdao2Δ, Cgdao3Δ and Cndao1Δ was retarded.
In the first group, D-Ala, D-Pro and D-Met, only CgDAO2 but not CgDAO1 or CgDAO3 was able to complement the growth defect of Cgdao2Δ (Fig 4A). In contrast, overexpression of CnDAO2 and CnDAO3 but not CnDAO1 complemented the growth deficiency of Cgdao2Δ on D-Ala, D-Pro and D-Met suggesting that CnDao2 and CnDao3 could oxidize those three D-amino acids when overexpressed in R265.
Fig 4. Some DAO genes are functionally interchangeable when overexpressed.
(A) Overexpression of CnDAO2 and CnDAO3 complements the growth deficiency of Cgdao2Δ on three D-amino acids. Strain Cgdao2Δ was transformed with the overexpression construct of the indicated DAO gene. The resulting strains were spotted onto YNB medium containing ammonium sulfate or the indicated D-amino acids and incubated at 30°C. Pictures were taken after 3 days (NH4) or 5 days (D-Ala, D-Pro and D-Met) of incubation. (B) H99 grows robustly on three D-amino acids when DAO genes are overexpressed. H99 was transformed with the overexpression construct of indicated DAO gene. Spot assays were performed as in (A). (C) Overexpression of every DAO gene complements the growth deficiency of Cndao3Δ. Cndao3Δ and Cgdao3Δ were transformed with each DAO overexpression constructs. The resulting transformants were tested for the ability to grow on D-Glu and the results are summarized. Lighted color letters indicate the original mutants that grow poorly on D-Glu. Dark color letters indicate the original mutants that have no phenotype on D-Glu. Arrow line indicates that overexpression of indicated DAO genes complement the mutant phenotype on D-Glu. (D) Complementation of the growth deficiency of dao mutants on D-Asp by overexpressing DAO genes. The dao mutants showing growth defect on D-Asp were transformed with DAO overexpression constructs. The resulting transformants were tested for the ability to grow on D-D-Asp. Symbols used in the figure are the same as in (C).
Surprisingly, the H99 strain, which grew poorly on D-Ala, D-Pro and D-Met as the sole nitrogen source, grew robustly on these D-amino acids when CgDAO2 was overexpressed in H99 (Fig 4B). Furthermore, overexpression of CnDAO2 and CnDAO3 also enabled H99 to utilize those three D-amino acids. These results suggest that CnDao2 and CnDao3 can convert D-Ala, D-Pro and D-Met into a usable nitrogen source when overexpressed in both H99 and R265. These data also suggest that CnDao1, CgDao1 and CgDao3 lack the ability to oxidize the D-amino acids belonging to the first group.
In the second group, D-Glu, overexpression of each DAO gene from both H99 and R265 enabled growth of Cndao3Δ on D-Glu and the results are summarized in Fig 4C. These data suggest that every DAO gene can functionally substitute for CnDAO3 in H99 and enables the strain to utilize D-Glu as the nitrogen source. In contrast, growth of Cgdao3Δ on D-Glu was restored only by over expression of CnDAO3 and not by any other DAO gene. As mentioned above, growth of R265 and not H99 was affected by the presence of 100 mM D-Glu, thus it is possible that H99 contains additional factors related to D-Glu utilization/tolerance compared to R265. The detailed mechanisms of these differences are yet to be elucidated.
In the third group, D-Asp, the effect of overexpression of each DAO gene was examined in Cgdao1Δ, Cgdao2Δ, Cgdao3Δ and Cndao1Δ strains and the results are summarized in Fig 4D. Overexpression of each DAO gene from either R265 or H99 functionally complemented the growth defect of at least two R265 dao mutants on D-Asp. These data suggest that the Daos from H99 and R265 can catabolize D-Asp when overexpressed in R265. In contrast, CnDAO2 and CnDAO3 were not able to complement the growth defect of Cndao1Δ on D-Asp. In addition, only CgDAO1 and CgDAO3 but not CgDAO2 were able to complement the growth defect of Cndao1Δ. Taken together, our results suggest that every Dao of R265 and H99 has ability to convert all the D-amino acids belonging to the second and third groups into a useable nitrogen source whereas only CgDao2, CnDao2, and CnDao3 have the ability to catabolize the D-amino acids in the first group.
Dao substrate specificity
Our genetic studies suggested the substrate specificity of each Dao. We attempted to biochemically determine the substrate specificity of each Dao using dao mutants of H99 and R265. However, Dao enzymatic activity was undetectable in crude total protein extracts from H99 and R265 while it could be readily detected from a Rhodosporidium toruloides strain known to be a high Dao producer [33,42,43]. Dao activity is known to vary greatly among different organisms [44,45]. It is possible that cryptococcal Daos have very low enzyme activity. To further characterize the Dao protein function, CgDao1, CgDao2 and CgDao3 were individually expressed in E. coli as a recombinant protein fused to maltose-binding protein (MBP). Recombinant Dao of R. toruloides (RtDao) produced by the same vector was used as a positive control.
Table 3 shows the substrate specificity of the recombinant Dao proteins. Eleven D-amino acids that affected the growth of Cgdao1, Cgdao2, or Cgdao3 deletion mutants were examined. The positive control, RtDao, displayed high enzymatic activity towards most D-amino acids except for D-Asp and D-Phe. In contrast, the enzymatic activity of all three recombinant R265 Dao proteins was markedly lower compared to RtDao although the amounts of each recombinant CgDao and RtDao protein produced in E. coli were similar (S3 Fig). Low Dao enzyme activity of the recombinant R265 Dao proteins corroborated our inability to detect the Dao enzymatic activity from crude protein extracts of R265.
Table 3. Substrate specificity of Dao enzymes.
| Protein | RtDao | CgDao1 | CgDao2 | CgDao3 | Concordance with growth phenotypes a |
|---|---|---|---|---|---|
| D-Ala | 41.22 | 0.040 | 0.218* | 0.043 | Yes-dao2Δ |
| D-Asp | 0.97 | 0.916* | 0.053 | 0.049 | Yes b |
| D-Glu | 73.86 | 0.049 | 0.080 | 0.032 | No-dao3Δ |
| D-His | 41.26 | 0.049 | 0.128* | 0.031 | Yes-dao2Δ |
| D-Leu | 15.84 | 0.044 | 0.153* | 0.045 | Yes-dao2Δ |
| D-Met | 27.77 | 0.050 | 0.64* | 0.040 | Yes-dao2Δ |
| D-Phe | 3.97 | 0.046 | 0.043 | 0.031 | No-dao2Δ |
| D-Pro | 39.81 | 0.054 | 0.136* | 0.041 | Yes-dao2Δ |
| D-Ser | 31.13 | 0.062 | 0.129* | 0.044 | Yes-dao2Δ |
| D-Thr | 17.70 | 0.040 | 0.052 | 0.036 | No-dao2Δ |
| D-Val | 50.55 | 0.055 | 1.009* | 0.039 | Yes-dao2Δ |
a Substrate specificity of CgDao enzymes (nM H2O2/ug crude extract) was compared to the growth phenotypes of Cgdao mutants listed in Table 2. “Yes” or “No” designates whether the ability to oxidize D-amino acid is concordant with the growth phenotypes of the specified CgdaoΔ mutant. The experiments were repeated twice and the representative data are shown.
b CgDao1 showed the clearest concordant substrate specificity with Cgdao1Δ growth phenotype.
* Activity is at least two fold higher than the control activity of CgDao using L-amino acid as substrate.
Despite low activity of the three R265 Dao enzymes, CgDao2 showed the broadest substrate specificity. The patterns of CgDao2 and RtDao substrate specificity were similar except that CgDao2 showed relatively lower activity against D-Glu and D-Thr. We noted that substrate specificity of the recombinant CgDaos did not correlate perfectly with the growth phenotypes of each Cgdao mutant (Table 3). For instance, CgDao1 was the only Dao protein which displayed appreciable activity using D-Asp as a substrate and the recombinant CgDao3 showed no appreciable oxidative activity with the substrates tested. Because the recombinant Dao is a MBP fusion protein, it is possible that the MBP fusion caused the observed discrepancy between substrate specificity and growth phenotype. This possibility is supported by the observation that while recombinant RtDao had the highest activity when D-Glu was used as the substrate (Table 3), Dao activity of the native R. toruloides protein was very low against D-Glu [33,43]. The CgDao2 recombinant protein was purified further by removing the MBP with Factor Xa. However, the MBP cleaved CgDao2 lost its ability to oxidize D-amino acids. Since the enzymatic activity of all R265 recombinant Dao proteins was low and our data suggested that the enzymatic activity of CnDao2 was even weaker than CgDao2 (see below), we have not attempted to express any Dao of H99 in E. coli. The precise substrate specificity of each Dao may require further biochemical analysis using highly purified native proteins.
Gene and promoter swap between CgDAO2 and CnDAO2
Since CgDAO2 is the major DAO gene in R265 and overexpression of CnDAO2 complemented the Cgdao2Δ phenotype, we further explored the function of DAO2 genes in both species. Since overexpression of CgDAO2 using the GPD promoter enabled the H99 strain to utilize several D-amino acids, we tested if using the native CgDAO2 promoter could have a similar effect. Insertion of the entire CgDAO2 gene into the Cndao2Δ locus in H99 (Fig 5A upper panel; R2(p)-R2; strain C1720) enabled its growth on D-Ala (Fig 5B). This result indicates that a single copy of the CgDAO2 gene enables H99 to utilize D-Ala. When the CgDAO2 promoter in C1720 was swapped with that of CnDAO2, the resulting strain, C1728 (H2(p)-R2), grew better on D-Ala than H99 but weaker than C1720. The transcriptional levels of CgDAO2 determined by quantitative RT-PCR were higher in C1720 than in C1728 when the cells were grown in ammonium sulfate (Fig 5C). The transcription levels of CgDAO2 were slightly induced in C1720 by D-Ala but not in C1728. These data suggest that the apparent strength of the CgDAO2 promoter relative to that of CnDAO2 may explain the growth difference between C1720 and C1728 on D-Ala.
Fig 5. Gene and promoter swap between CnDAO2 and CgDAO2.
(A) Diagram of the constructs. Cndao2Δ and Cgdao2Δ were transformed with the indicated constructs. The names of the resulting strains are listed on the left. Dotted line indicates the chromosomal regions flanking the deleted locus. Crosses indicate the crossing over event at the homologous regions. The symbols Act(p) = Actin promoter; HYG = hygromycin resistance gene; NEO = neomycin resistance gene; H2 = CnDAO2 without the promoter; R2 = CgDAO2 without the promoter; H2(f) and R2(f) = flanking region of CnDAO2 and CgDAO2 respectively; H2(p) and R2(p) = promoter of CnDAO2 and CgDAO2 respectively. (B) Spot assay of the gene swapped strains. Approximately 600 cells were spotted on D-Ala and the plates were incubated at 30°C for 11 days and photographed. (C) Relative RNA levels. Log phase cells of the indicated strains were transferred to YNB medium containing 10 mM ammonium sulfate or D-Ala for 2 h and the RNA was isolated. The relative mRNA levels were determined by quantitative RT-PCR. Data were normalized with ACTIN levels and expressed as the relative RNA levels of H99 (upper panel) or R265 (lower panel) grown in ammonium sulfate. The experiments were repeated three times and the error bars represent the standard deviation of three technical repeats.
When the promoter of CnDAO2 was replaced with that of CgDAO2, the resulting strain, C1726 (R2(p)-H2), grew slightly better than H99 but poorer than C1728 on D-Ala although CnDAO2 RNA levels in C1726 were higher than the CgDAO2 RNA levels in C1728 on D-Ala (Fig 5C). Since the only difference between these strains was at the CnDAO2 locus, it is possible that the observed growth difference may be due to higher enzymatic activity of CgDao2 in C1728 compared to that of CnDao2 in C1726. The possibility that enzymatic activity of CgDao2 is higher than CnDao2 is supported by the observation that growth of the strains overexpressing CgDAO2 was more robust than those overexpressing CnDAO2 in which both overexpression constructs were under the control of the same GPD promoter (Fig 4A and 4B).
We have shown that overexpression of CnDAO2 was able to complement the Cgdao2Δ phenotype on D-Ala. To examine if the native CnDAO2 promoter could provide a similar effect in the Cgdao2Δ background, the entire CnDAO2 gene was inserted into the Cgdao2Δ locus (Fig 5A bottom panel). The resulting strain, C1722 (H2(p)-H2), failed to restore the growth of Cgdao2Δ on D-Ala (Fig 5B). When the CnDAO2 promoter was replaced by that of CgDAO2, the resulting strain, C1730 (R2(p)-H2), was able to grow on D-Ala although the growth was considerably weaker than R265. Furthermore, when the coding region of CnDAO2 was replaced by that of CgDAO2, growth of the resulting strain, C1724 (H2(p)-R2), on D-Ala was more robust than C1730 although the transcription levels of CgDAO2 were much lower in C1724 than that of the CnDAO2 in C1730 (Fig 5C bottom panel). These results support the notion that the enzymatic activity of CgDao2 may be stronger than CnDao2. These data also indicate that although the promoter strength of CnDAO2 is weaker than CgDAO2 in R265 background and the expression levels of CnDAO2 was not affected by the presence of different D-amino acids (Figs 3C and 5C), CnDAO2 promoter is functional and capable of driving the expression of CgDAO2 in strain C1724 allowing Cgdao2Δ to grow moderately on D-Ala.
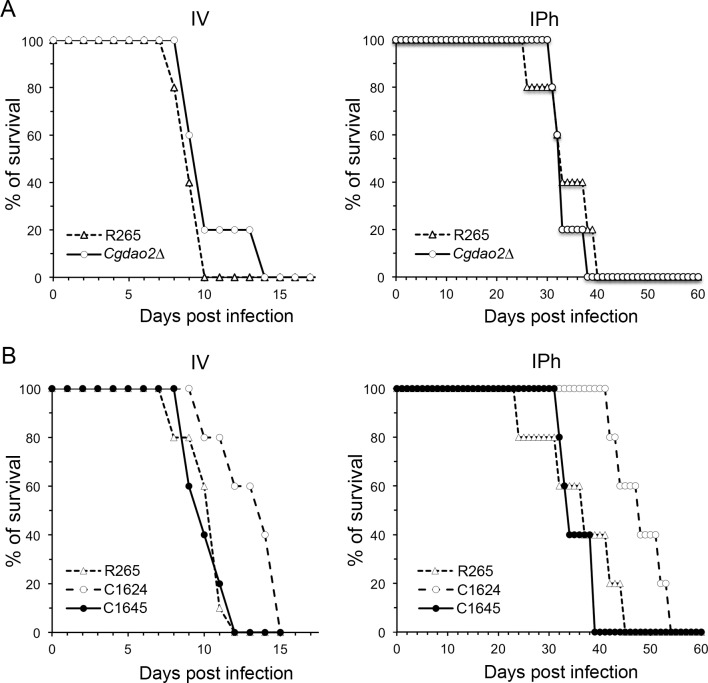
DAO genes play a role in virulence of R265 in murine models
Since CgDAO2 was the major DAO gene in R265, we examined the role of CgDAO2 in virulence using murine models. Deletion of CgDAO2 did not affect virulence in mice infected either by intrapharyngeal aspiration (IPh) or intravenous injection (IV) (Fig 6A). Similarly, neither the deletion of CgDAO1 nor CgDAO3 affected virulence of R265 (data now shown). Interestingly, virulence of the Cgdao1ΔCgdao2ΔCgdao3Δ triple deletant (C1624) was reduced compared to the wild-type in both IPh (p = 0.018, log-rank test) and IV (p = 0.041, log-rank test) models while complementation of CgDAO2 in the triple deletant (C1645) restored the virulence (Fig 6B). This suggests that although the individual DAO gene is not required for the virulence of R265, the three CgDAO genes cooperatively contribute to virulence. On the other hand, the Cndao1ΔCndao3Δ double deletant or the Cndao1ΔCndao2ΔCndao3Δ triple deletant exhibited virulence similar to H99 suggesting that DAO genes in H99 do not play a role in pathogenicity (S4A Fig).
Fig 6. Virulence of the triple dao deletant of R265 is reduced.
(A and B) BALB/c female mice (5 per group) were infected with the indicated strains either by intravenous injection (IV) or intrapharyngeal aspiration (IPh) and the mortality was monitored. C1624: Cgdao1ΔCgdao2ΔCgdao3Δ triple deletant; C1645: Cgdao1Cgdao3Δ double deletant (derived from C1624 by reconstituting CgDAO2). The experiments were repeated twice and the representative data are shown.
Since there are many other nitrogen sources other than D-amino acids available in the host, the observed modest pathological effect of triple deletion in R265 could be unrelated to their ability to utilize D-amino acids for growth. However, persistently high concentrations of D-Ser have been reported in the mammalian forebrain [21,22]. We suspected that R265 DAO genes might play a role in brain tropism. We therefore compared the effectiveness of the R265 brain invasion to that of the three different dao mutants. Since R265 cells can efficiently cross the blood-brain barrier and produce severe meningoencephalitis when inoculated intravenously [31], mice were infected via the tail vein and the number of colony forming units in the brain were determined after 3h. No significant difference was found in the brain’s fungal load between the mice infected with the wild-type and those infected with daoΔ mutants including the Cgdao1ΔCgdao2ΔCgdao3Δ triple deletant (S4B Fig). We also compared the brain fungal load of mice challenged by IPh after 7, 14 and 35 days post infection and found no difference between the mice infected with wild-type and those with the triple deletant (S4C Fig). These observations indicate that the dao genes of R265 do not play a role in brain tropism in mice.
Although DAO genes and utilization of D-amino acids have been studied in various fungal species [34–36,38–40,42], this is the first study dealing with a comprehensive analysis of each DAO paralogous gene function in two genetically close pathogenic fungi. Our interest in comparing the mechanism responsible for D-amino acid utilization by the two etiologic agents of cryptococcosis stems from practical reasons. Utilization of certain D-amino acids as the sole nitrogen source has been used as a biochemical tool to distinguish the two species. We chose a representative strain from each species, H99 for C. neoformans and R265 for C. gattii, to determine growth differences in the common D-amino acids. Although three putative D-amino acid oxidases were found in both species, H99 grew poorly in most D-amino acids except for D-Asn and D-Gln. By deleting each DAO gene, we have demonstrated that each DAO gene functions differently with respect to utilization of D-amino acids in H99 and R265 strains.
Several lines of evidence in combination could explain why H99 cannot grow on various D-amino acids. First, overexpression of CnDAO2 and CnDAO3 in H99 enables growth on several D-amino acids suggesting that one of the reasons for the inability of H99 to grow may be due to insufficient expression levels of the CnDAO genes. Second, although expression levels of the DAO2 gene were inducible when the gene was driven by the CgDAO2 promoter in H99 (C1720 and C1726; Fig 5C), the trend of the expression levels was slightly lower in H99 background than in R265 (C1720 vs. R265 and C1726 vs. C1730). These results suggest that H99 may lack some of the components required for an adequate control of DAO gene expression. Lastly, the results of gene and promoter swaps along with overexpression studies further suggest that the enzymatic activity of CnDao2 was lower compared to that of CgDao2.
The major ecological niche of C. gattii is plants [1] and several D-amino acids are known to be present in higher plants [20]. It is possible that the ability to utilize D-amino acids may have been incorporated into C. gattii during the evolutionary process. The virulence difference between triple deletants in R265 and H99 highlights the fact that DAO has evolved differently in these two species. It is not clear, however, if involvement of DAO genes in the pathobiology was a gained function in R265 or a lost function in H99 during evolution.
Supporting Information
Amino acid sequences of DAOs from AhDao (Asterotremella humicola; AB121230), RtDao (Rhodosporidium toruloides; P80324), and TvDao (Trigonopsis variabilis; Q99042), R265 and H99 were compared by CLUSTER W alignment program.
(TIF)
Three-fold serial dilutions of each strain were spotted onto YNB medium containing 2% glucose and 10 mM ammonium sulfate supplemented with or without 100 mM D- or L-amino acids. Plates were incubated at 30°C for 2 days and photographed.
(TIF)
(A) E coli. crude protein extract was isolated, separated in a 4–12% NuPAGE gel and stained with Comassie Blue stain reagent. Twenty ug of Cgdao2 and RtDao and 30 ug of Cgdao1 and Cgdao3 post-induction samples were loaded in wells. Approximately the same amount of pre-induction sample was loaded in wells. (B) Purity of the Dao recombinant proteins after affinity chromatography. Protein was separated by a 4–12% NuPAGE gel and stained with Comassie Blue reagent. Samples 1, 3, and 5 contain crude extract (10ug) and samples 2, 4, and 6 contain purified sample (2ug). Cgdao3 was not shown due to lack of detectable enzymatic activity.
(TIF)
(A) Virulence H99 dao deletants is the same as the wild-type. BALB/c female mice (5 per group) were injected with various strains either by intravenous injection (IV) or intrapharyngeal aspiration (IPh) and the mortality was monitored. (B) Frequency of brain entrance is not reduced in the Cgdao triple deletant. Group of three mice each were infected intravenously with the indicated strains. Brains were isolated 3h post infection and the colony forming units in the brain were determined. Data is expressed as a percentage of the number of the CFU’s in the brain vs. the number of input cells. Bar: mean value of three mice. (C) Fugal burden in the brain of the mice infected by the Cgdao triple deletant is the same as the wild-type. Group of three mice each were infected by intrapharyngeal aspiration. Brains were isolated from the infected mice as indicated time and the colony forming units (CFU) per the brain were determined.
(TIF)
(DOCX)
(DOCX)
Acknowledgments
We thank A. Varma for critical reading of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by funds from the intramural program of the National Institute of Allergy and Infectious Diseases, NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Heitman J, Kozel TR, Kwon-Chung J, Perfect JR, Casadevall A, editors (2011) Cryptococcus: from human pathogen to model yeast. Washington DC: ASM press; 620 p. [Google Scholar]
- 2. Kavanaugh LA, Fraser JA, Dietrich FS (2006) Recent evolution of the human pathogen Cryptococcus neoformans by intervarietal transfer of a 14-gene fragment. Mol Biol Evol 23: 1879–1890. [DOI] [PubMed] [Google Scholar]
- 3. Kwon-Chung KJ, Bennett JE (1992) Medical Mycology. Philadelphia: Lea & Febiger. [Google Scholar]
- 4. Casadevall A, Perfect JR (1998) Cryptococcus neoformans. Washington, DC: ASM press. [Google Scholar]
- 5. Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, Diezmann S, et al. (2005) Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437: 1360–1364. [DOI] [PubMed] [Google Scholar]
- 6. Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, et al. (2004) A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci U S A 101: 17258–17263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Byrnes EJ 3rd, Li W, Lewit Y, Ma H, Voelz K, Ren P, et al. (2010) Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog 6: e1000850 10.1371/journal.ppat.1000850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. MacDougall L, Kidd SE, Galanis E, Mak S, Leslie MJ, Cieslak PR, et al. (2007) Spread of Cryptococcus gattii in British Columbia, Canada, and detection in the Pacific Northwest, USA. Emerg Infect Dis 13: 42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kwon-Chung KJ, Bennett JE, Rhodes JC (1982) Taxonomic studies on Filobasidiella species and their anamorphs. Antonie Van Leeuwenhoek 48: 25–38. [DOI] [PubMed] [Google Scholar]
- 10. Polacheck I, Kwon-Chung KJ (1980) Creatinine metabolism in Cryptococcus neoformans and Cryptococcus bacillisporus . J Bacteriol 142: 15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kwon-Chung KJ, Bennett JE, Theodore TS (1978) Cryptococcus bacillisporus sp. nov.: serotype B-C of Cryptococcus neoformans . International Journal of Systematic Bacteriology 28: 616–620. [Google Scholar]
- 12. Salkin IF, Hurd NJ (1982) New medium for differentiation of Cryptococcus neoformans serotype pairs. J Clin Microbiol 15: 169–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bennett JE, Kwon-Chung KJ, Theodore TS (1978) Biochemical differences between serotypes of Cryptococcus neoformans . Sabouraudia 16: 167–174. [PubMed] [Google Scholar]
- 14. Kwon-Chung KJ, Polacheck I, Bennett JE (1982) Improved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotypes B and C). J Clin Microbiol 15: 535–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ngamskulrungroj P, Chang Y, Roh J, Kwon-Chung KJ (2012) Differences in nitrogen metabolism between Cryptococcus neoformans and C. gattii, the two etiologic agents of cryptococcosis. PLoS One 7: e34258 10.1371/journal.pone.0034258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nishikawa MM, Sant'Anna OD, Lazera MS, Wanke B (1996) Use of D-proline assimilation and CGB medium for screening Brazilian Cryptococcus neoformans isolates. J Med Vet Mycol 34: 365–366. [PubMed] [Google Scholar]
- 17. Dufait R, Velho R, De Vroey C (1987) Rapid identification of the two varieties of Cryptococcus neoformans by D-proline assimilation. Mykosen 30: 483 [PubMed] [Google Scholar]
- 18. Mukamurangwa P, Raes-Wuytack C, De Vroey C (1995) Cryptococcus neoformans var. gattii can be separated from var. neoformans by its ability to assimilate D-tryptophan. J Med Vet Mycol 33: 419–420. [PubMed] [Google Scholar]
- 19. Friedman M (1999) Chemistry, nutrition, and microbiology of D-amino acids. J Agric Food Chem 47: 3457–3479. [DOI] [PubMed] [Google Scholar]
- 20. Robinson T (1976) D-amino acids in higher plants. Life Sci 19: 1097–1102. [DOI] [PubMed] [Google Scholar]
- 21. Hashimoto A, Nishikawa T, Oka T, Takahashi K, Hayashi T (1992) Determination of free amino acid enantiomers in rat brain and serum by high-performance liquid chromatography after derivatization with N-tert.-butyloxycarbonyl-L-cysteine and o-phthaldialdehyde. J Chromatogr 582: 41–48. [DOI] [PubMed] [Google Scholar]
- 22. Schell MJ, Molliver ME, Snyder SH (1995) D-serine, an endogenous synaptic modulator: localization to astrocytes and glutamate-stimulated release. Proc Natl Acad Sci U S A 92: 3948–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. D'Aniello A, D'Onofrio G, Pischetola M, D'Aniello G, Vetere A, Petrucelli L, et al. (1993) Biological role of D-amino acid oxidase and D-aspartate oxidase. Effects of D-amino acids. J Biol Chem 268: 26941–26949. [PubMed] [Google Scholar]
- 24. Khoronenkova SV, Tishkov VI (2008) D-amino acid oxidase: physiological role and applications. Biochemistry (Mosc) 73: 1511–1518. [DOI] [PubMed] [Google Scholar]
- 25. Pollegioni L, Piubelli L, Sacchi S, Pilone MS, Molla G (2007) Physiological functions of D-amino acid oxidases: from yeast to humans. Cell Mol Life Sci 64: 1373–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chang YC, Khanal Lamichhane A, Garraffo HM, Walter PJ, Leerkes M, Kwon-Chung KJ (2014) Molecular mechanisms of hypoxic responses via unique roles of Ras1, Cdc24 and Ptp3 in a human fungal pathogen Cryptococcus neoformans . PLoS Genet 10: e1004292 10.1371/journal.pgen.1004292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McClelland CM, Chang YC, Kwon-Chung KJ (2005) High frequency transformation of Cryptococcus neoformans and Cryptococcus gattii by Agrobacterium tumefaciens . Fungal Genet Biol 42: 904–913. [DOI] [PubMed] [Google Scholar]
- 28. Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR (1993) Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol 175: 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuwayama H, Obara S, Morio T, Katoh M, Urushihara H, Tanaka Y (2002) PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res 30: E2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao S, Fernald RD (2005) Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol 12: 1047–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ngamskulrungroj P, Chang Y, Sionov E, Kwon-Chung KJ (2012) The Primary Target Organ of Cryptococcus gattii Is Different from That of Cryptococcus neoformans in a Murine Model. Mbio 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rao GV, Tinkle S, Weissman DN, Antonini JM, Kashon ML, Salmen R, et al. (2003) Efficacy of a technique for exposing the mouse lung to particles aspirated from the pharynx. J Toxicol Environ Health A 66: 1441–1452. [DOI] [PubMed] [Google Scholar]
- 33. Gabler M, Hensel M, Fischer L (2000) Detection and substrate selectivity of new microbial D-amino acid oxidases. Enzyme Microb Technol 27: 605–611. [DOI] [PubMed] [Google Scholar]
- 34. Takahashi S, Kakuichi T, Fujii K, Kera Y, Yamada RH (2005) Physiological role of D-aspartate oxidase in the assimilation and detoxification of D-aspartate in the yeast Cryptococcus humicola . Yeast 22: 1203–1212. [DOI] [PubMed] [Google Scholar]
- 35. Soutourina J, Plateau P, Blanquet S (2000) Metabolism of D-aminoacyl-tRNAs in Escherichia coli and Saccharomyces cerevisiae cells. J Biol Chem 275: 32535–32542. [DOI] [PubMed] [Google Scholar]
- 36. Ohnishi E, Macleod H, Horowitz NH (1962) Mutants of Neurospora deficient in D-amino acid oxidase. J Biol Chem 237: 138–142. [PubMed] [Google Scholar]
- 37. Champney WS, Jensen RA (1970) Molecular events in the growth inhibition of Bacillus subtilis by D-tyrosine. J Bacteriol 104: 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Horner R, Wagner F, Fischer L (1996) Induction of the d-Amino Acid Oxidase from Trigonopsis variabilis . Appl Environ Microbiol 62: 2106–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Molla G, Motteran L, Piubelli L, Pilone MS, Pollegioni L (2003) Regulation of D-amino acid oxidase expression in the yeast Rhodotorula gracilis . Yeast 20: 1061–1069. [DOI] [PubMed] [Google Scholar]
- 40. Takahashi S, Okada H, Abe K, Kera Y (2012) D-amino acid-induced expression of D-amino acid oxidase in the yeast Schizosaccharomyces pombe . Curr Microbiol 65: 764–769. 10.1007/s00284-012-0227-z [DOI] [PubMed] [Google Scholar]
- 41. Kmetzsch L, Staats CC, Simon E, Fonseca FL, Oliveira DL, Joffe LS, et al. (2011) The GATA-type transcriptional activator Gat1 regulates nitrogen uptake and metabolism in the human pathogen Cryptococcus neoformans . Fungal Genet Biol 48: 192–199. 10.1016/j.fgb.2010.07.011 [DOI] [PubMed] [Google Scholar]
- 42. Molla G, Vegezzi C, Pilone MS, Pollegioni L (1998) Overexpression in Escherichia coli of a recombinant chimeric Rhodotorula gracilis d-amino acid oxidase. Protein Expr Purif 14: 289–294. [DOI] [PubMed] [Google Scholar]
- 43. Lee YH, Chu WS (1996) D-Amino acid oxidase activity from Rhodosporiduim toruloides . Letters in Applied Microbiology 23: 283–286. [Google Scholar]
- 44. Saleem A, Moharram AM, Fathy N (2012) Production and optimization of D-amino acid oxidase which is involved in the biosynthesis of beta-lactam antibiotics. African Journal of Microbiology Research 6: 4365–4376. [Google Scholar]
- 45. Tishkov VI, Khoronenkova SV (2005) D-Amino acid oxidase: structure, catalytic mechanism, and practical application. Biochemistry (Mosc) 70: 40–54. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid sequences of DAOs from AhDao (Asterotremella humicola; AB121230), RtDao (Rhodosporidium toruloides; P80324), and TvDao (Trigonopsis variabilis; Q99042), R265 and H99 were compared by CLUSTER W alignment program.
(TIF)
Three-fold serial dilutions of each strain were spotted onto YNB medium containing 2% glucose and 10 mM ammonium sulfate supplemented with or without 100 mM D- or L-amino acids. Plates were incubated at 30°C for 2 days and photographed.
(TIF)
(A) E coli. crude protein extract was isolated, separated in a 4–12% NuPAGE gel and stained with Comassie Blue stain reagent. Twenty ug of Cgdao2 and RtDao and 30 ug of Cgdao1 and Cgdao3 post-induction samples were loaded in wells. Approximately the same amount of pre-induction sample was loaded in wells. (B) Purity of the Dao recombinant proteins after affinity chromatography. Protein was separated by a 4–12% NuPAGE gel and stained with Comassie Blue reagent. Samples 1, 3, and 5 contain crude extract (10ug) and samples 2, 4, and 6 contain purified sample (2ug). Cgdao3 was not shown due to lack of detectable enzymatic activity.
(TIF)
(A) Virulence H99 dao deletants is the same as the wild-type. BALB/c female mice (5 per group) were injected with various strains either by intravenous injection (IV) or intrapharyngeal aspiration (IPh) and the mortality was monitored. (B) Frequency of brain entrance is not reduced in the Cgdao triple deletant. Group of three mice each were infected intravenously with the indicated strains. Brains were isolated 3h post infection and the colony forming units in the brain were determined. Data is expressed as a percentage of the number of the CFU’s in the brain vs. the number of input cells. Bar: mean value of three mice. (C) Fugal burden in the brain of the mice infected by the Cgdao triple deletant is the same as the wild-type. Group of three mice each were infected by intrapharyngeal aspiration. Brains were isolated from the infected mice as indicated time and the colony forming units (CFU) per the brain were determined.
(TIF)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.