Introduction
Micro RNAs (miRNAs) constitute a family of small, single-stranded RNAs (ssRNAs) involved in the post-transcriptional regulation of gene expression and in ultimately shaping the transcriptome of a cell. While our views on the biological significance and relevance of miRNA signaling continue to evolve, it is now generally recognized that the principal action of miRNA in all species of plants and animals is to recognize, and then bind, via highly selective hydrogen bonding, to specific complementary ribonucleotide targets in the 3’ prime untranslated region (3’-UTR) of specific messenger RNAs (mRNAs), and in doing so, down-regulate their expression [1–10]. Although miRNAs are considered to be critically important epigenetic regulators of gene expression in eukaryotic development, aging and disease, it is not often appreciated that these ssRNAs: (i) operate ubiquitously and sometimes interactively throughout the plant and animal kingdoms; (ii) are highly selected in their ribonucleotide sequence; (iii) are highly selected in their cell and tissue specificity, (iv) represent an epigenetic signaling system that is evolutionarily ancient; (v) are the smallest yet characterized ribonucleic acid carriers of highly selective genetic regulatory information; (vi) possess highly similar structural and functional features when compared to minimalist plant pathogens known as viroids; (vii) are the most abundant nucleic acids contained in human extracellular fluid (ECF) and cerebrospinal fluid (CSF); and (viii) as major components of the ECF, CSF and blood serum may spread both homeostatic and pathological signaling among neighboring cells and tissues, and perhaps even between individual organisms or species [11–13]. This communication will briefly review some of the more overlooked aspects of the structure, function and mechanism of these fascinating small non-coding RNAs (sncRNAs) in plants and animals, with special emphasis on human central nervous system (CNS) disease, and with specific relevance to Alzheimer disease (AD) wherever possible.
The Unique Sequence Structure, Selectivity, Tissue Specificity and Stability of miRNAs
Elementary ribonucleic acid sequence analysis and bioinformatics predict that a ‘typical’ 22 nucleotide ssRNA that is comprised of 4 different ribonucleotides (adenine, guanine, cytosine and uridine; A,G,C,U) could have over 1013 possible sequence combinations. The experimental observation that there are typically only about 2×103 different miRNAs so far identified in all eukaryotic tissues examined, and that miRNAs are highly developmental stage-, tissue- and cell-specific, even in adjacent cell types suggests an extremely high evolutionary selection pressure to use only specific miRNA sequences that will yield biologically productive miRNA-mRNA interactions [1– 4,14–16]. In fact, extensive studies using miRNA array-, Northern dot blot-, RNA-sequencing, RT-PCR and bioinformatics-based analyses on small ssRNAs suggest that human brain cells probably utilize less than 102 abundant species of miRNA [17–20] and that only a relatively small fraction of these are misregulated in AD and other neurodegenerative disorders with an inflammatory component [17,19,21,22; unpublished observations].
Abundance and Stability of miRNAs
The abundance, complexity and speciation of highly specific miRNAs may vary among human populations in health and disease, and as such, have high potential for the diagnosis of disease [15,16,21]. Like mRNA, miRNAs appear to adopt the same stability rules involving adenine-uridine (AU) rich elements (AREs) in their linear ribonucleotide sequence, and a higher ARE content in miRNAs is generally associated with shorter miRNA half-life. Conversely the absence of AU or UA dinucleotide elements may confer miRNA stability and lengthen miRNA half-life [16,23–27]. Interestingly, while mammalian brain and retinal miRNAs in particular may have in general a relatively short half-life measured in the range of several hours, miRNA half-lives may be considerably extended by miRNA-binding proteins, the adoption of extensive miRNA secondary and tertiary structures, circularization, or by combinations of these RNAse-evading strategies [11,13,16,26,27]. Within miRNA precursors, virtually all of the miRNA sequence base-pairs with complementary sequences in other parts of the same molecule to form double-stranded RNA (dsRNA) structures that are more resistant to degradation than ssRNA alone [27,28] (Figure 1).
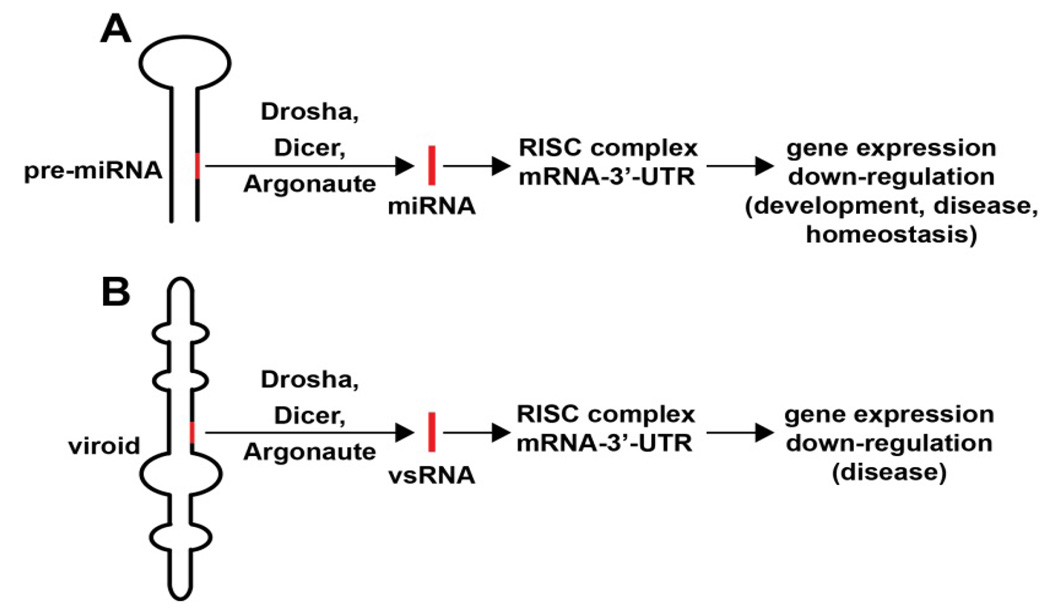
Figure 1. Similarities in miRNA and viroid structure and function.
This highly schematicized figure underscores the remarkable similarities between the structure and function of miRNA and viroids; (A) a typical 75–110 nucleotide primary micro RNA (pri-miRNA) ‘hairpin’ containing an endogenous 21–25 nt miRNA that yields a mature miRNA (red bar) after Drosha- and Dicer-mediated excision and processing; the mature miRNA next associates with a cytoplasmic RNA-induced silencing complex (RISC) and a target mRNA-3’-UTR to degrade and down-regulate expression of that target mRNA, with subsequent effects on the expression of genes involved in homeostasis, development and disease; (B) analogously, a ~246–401 nucleotide closed circular viroid, also containing extensive intra-strand base pairings, dsRNA and stem-loop structures, typically contains a 21–25 nucleotide viroid-specific RNA (vsRNA; red bar) that after host Drosha/Dicer-based processing yields a mature vsRNA; as is the case for miRNAs this vsRNA subsequently targets the RISC and mRNA-3’-UTR complex, down-regulating gene expression to induce progressive developmental and age-related disease in plants [26,27,40,44,72]. In both cases larger miRNA or vsRNA precursors are processed by an RNase III of the family of Dicer-like proteins to generate smaller ‘infectious’ ssRNA species; these sizes are similar to endogenous small interfering RNA (as mature vsRNA or miRNA) to alter the viroid-dependent gene expression in the host plant by viroids, or of miRNA-mRNA processing in animal species including humans [14,26,27,37,42,44,72]. While naked, mature RNAs such as miRNAs and vsRNAs have relatively short half-lives in vitro (for example human neuronal miRNAs appear to be highly labile [27,43], stabilities may be greatly extended by single- or double-stranded RNA-binding proteins, by complex secondary structures, by RNA circularization, by containment in protease- and RNase-resistant vesicles, or by combinations of these and other factors [23,24,26,27,43]. Interestingly, viroids, at about one one-thousandth the size of the smallest known ssRNA virus (see text), are the smallest known self-replicating pathogens of all living species, and both the plant and animal kingdoms have adopted similar ssRNA strategies to store and transmit only the most essential genetic regulatory information in the propagation of either pathological or homeostatic signals. The potential for interaction between various vsRNAs and miRNAs in their hosts, if any, among diverse species of the plant and animal kingdom is currently not known.
miRNAs Occur Ubiquitously in Plants and Animals
While different miRNAs seem to be abundant in different cells, tissues and plant and animals species, at times primary structures may be very highly conserved as demonstrated by a presence or absence of certain ribonucleotides at specific positions in the miRNA and in the flanking regions of their precursors. Such miRNA sequences may contain fingerprints for conservation across multiple plant and animal species, and these fingerprints represent some of the most highly conserved nucleic acid sequences known [13,14,28,29]. To cite just one recent example, using novel genome-wide computational approaches to detect miRNAs based on both sequence and structure alignment across the plant and animal kingdoms, the miRNA-854 family has been shown to be expressed in Arabidopsis thaliana, Caenorhabditis elegans, Mus musculus and Homo sapiens. Interestingly, across these diverse species, miRNA-854 commonly targets the uridylate binding protein 1b (UBP1b) mRNA 3’-UTR, and UBP1b normally encodes a member of a heterogeneous nuclear RNA binding protein-1 (hnRNP-1) gene family [13,14]. This indicates an evolutionary common origin of miRNA-854 as a regulator of the basal eukaryotic transcription mechanism in both plants and animals for many hundreds of millions of years (Arabidopsis thaliana - Homo sapiens divergence about 1.5 billion years; [13,14,30,31]. While secondary and tertiary structures may also be conserved among multiple miRNAs, and internal stems, loops and mis-paired RNA ‘bulges’ very commonly appear at specifically conserved positions in many pre-miRNA sequences, the actual number of possible miRNA structural configurations may be somewhat limited by the small linear size (~21–25 nucleotides) of the miRNA itself [14,19,28,32] (Figure 1).
miRNAs and Viroids have Extensive Similarity in Structure and Function
miRNAs are the smallest yet identified carriers of highly selective genetic regulatory information in plants and animals. Their ribonucleotide sequences further define and regulate the expression of a relatively discrete subset of cellular mRNAs with which they may interact, thus defining a highly complex and interactive gene regulation network [1–6,33–35]. Across their ~21–25 ribonucleotide sequences miRNAs carry encoded genetic signals which may be transmitted via their unique molecular shape, nucleic acid topology and charge density along their lengths. Interestingly, miRNAs are typically less than about one one-thousandth of the size of a ‘typical virus’; the smallest ssRNA viruses known, in terms of genome size are retroviruses (such as Rous Sarcoma virus with a 3.5×104 nucleotide genome; http://www.ictvonline.org/) [36]. Seven orders, 96 families, 22 subfamilies, 420 genera, and about 2,618 species of viruses have been recently classified by the International Committee on taxonomy of viruses (ICTV), including 5 orders and 47 families of RNA viruses, both double and single stranded [36,37]. Curiously, there is approximately the same number of species of viruses as there are all currently known mammalian miRNAs (numbering about ~2600).
miRNAs and Viroids
Smaller than any known ssRNA viruses are viroids, a family of about 30 plant pathogens consisting of circular ssRNAs ranging in size from 246 to 401 nucleotides. Viroids are of evolutionary, virological and biological interest since they may represent living fossils of pre-cellular evolution in a hypothetical RNA world [13,38–42]. One of the smallest and first discovered viroids is the potato spindle tuber viroid (PSTV), a circular ssRNA which causes infectious disease in potato plants (chiefly Solanum tuberosum), and remains an important agricultural and economic concern throughout the world (Figure 1) [13,38–40]. Viroids represent a class of truly minimalist plant pathogens and it remains controversial whether they are a biological oddity, an evolutionary fossil or a highly evolved plant pathogen [39–42]. Viroids are transcribed by a unidirectional nucleic acid rolling-circle mechanism in the host plant’s chloroplast (for the genus Avsunviroidae) or nuclei (for the genus Pospiviroidae). Relatively recent findings indicate that viroid infection is associated with the appearance of small viroid-specific RNAs (vsRNAs), approximately 21 to 25 nucleotides in size, processed by an RNase III of the family of Dicer-like proteins from a pre-viroid precursor [37] (Figure 1). Abundant in viroid-infected plant species, vsRNAs have sizes similar to naturally occurring endogenous small interfering RNAs and miRNAs, and appear to permanently alter the normal cultivar- and viroid-dependent gene expression patterns in the host [37–42]. Highly analogous to miRNAs, viroid ssRNAs encode no proteins, have no protective protein capsid or coat, do not reverse-transcribe into DNA when they replicate, and are significantly inducible by external stressors, including neurotoxic factors in the environment [18,37–39]. As naked, infectious vsRNA molecules viroid replication requires an endogenous, host-supplied polymerase, initially generated as an internally complementary, double-stranded precursor RNA (previroid) structure from which a mature ssRNA is excised by RNAse III-type Dicer and related riboenzymes (Figure 1). As far as is currently known, miRNAs do not replicate in vivo, as they are probably physically too small to do so, and in vitro require a linker DNA or RNA to efficiently copy themselves using exogenous polymerase systems and replication cofactors. The circularization of pre-viroids, pri-miRNAs, anti-miRNAs or miRNAs, and the formation of complex higher order secondary and tertiary structures, may stabilize them and protect against rapid RNA degradation in cellular environments normally enriched in RNAse and associated depolymerization systems [11,13,41–44].
miRNAs are Abundant in Systemic Fluids, Including the ECF and CSF of the Human CNS
Emerging studies on human brain extracellular fluid (ECF; a cell-free preparation of the extracellular fluid that encircles human brain cells) and cerebrospinal fluid (CSF; the fluid that surrounds the brain and spinal cord) indicate that miRNAs are the most abundant nucleic acids contained within the circulating fluids of the human CNS [21,45]. Increases in specific miRNAs, confirmed independently using both miRNA arrays (LC Sciences, Houston TX) and reconfirmed using a highly sensitive LED-Northern dot-blot assay, indicate that several NF-κB-sensitive miRNAs detected in ECF and CSF were formerly identified to be up-regulated in brain anatomical regions targeted by AD [15,21,45]. These miRNAs strongly associate with the progressive spreading of inflammatory neurodegeneration [45–48]. The ECF- and CSF-enriched miRNA species include, prominently, miRNA-9, miRNA-34a, miRNA-125b, miRNA-146a and miRNA-155 and others, and their selective enrichment in circulating CSF and ECF in AD suggests that they may be involved in the modulation or proliferation of miRNA-triggered pathogenic signaling throughout the brain and CNS. If abundance is any indication of importance, then these miRNAs may be performing significant pathogenic signaling and disease-transmission functions via the circulation of these CNS fluids. Recent studies further indicate the importance of these and other miRNAs in the regulation of human brain endothelial and epithelial cell-barrier functions [49,50]. A corollary to this is that over-expressed miRNAs may be involved in disease spreading as they pass so easily out of the cell, into adjacent cells, and throughout biological fluids [21,33,45–47,49–54]. Importantly, despite variability in the analysis of AD CSF from many diverse human population samples, increases in the pro-inflammatory miRNA-146a have been reported by multiple independent AD miRNA researchers, and miRNA-146a has also been detected to be significantly up-regulated in transgenic AD murine models, in rare human and rodent prion diseases, epilepsy, and in other neurological disorders associated with progressive inflammatory neurodegeneration [35,46–48,52,54– 56]. Cumulatively these data support the hypothesis that paracrine or endocrine effects of miRNAs originating in stressed constituent cells of the human neurovascular unit may contribute to “spreading events” of AD pathology, and appear to be a unifying characteristic of progressive neurodegenerative disorders [57,58]. As further discussed below; these data additionally suggest that peripheral anti-miRNA strategies may be therapeutically useful in containing the spread of neuropathology not only in AD but in other progressive inflammatory degenerative diseases [25,59].
Interestingly, microRNAs are also abundant in the systemic circulation in humans and may have potential diagnostic value, especially in the earliest stages of AD and other neurological diseases [59,60]. It is not well understood if, like miRNAs, vsRNAs are abundant in the systemic fluids transported by the xylem ‘vessel cell system’ in diseased plants; the high cellulose-, xylan-, and lignin-enriched cell walls located outside of the cell membrane of plant cells may prevent easy access of vsRNA to cytoplasmic compartments [13,38–41].
miRNA and the Spreading of CNS Disease
Alzheimer’s disease (AD), as the prototypical example of a progressive human neurological disease whose most significant risk factor is aging, appears to originate in the limbic system of the human brain and subsequently spreads radially from the hippocampal CA1 and superior temporal lobe regions into more distal lobes of the brain, including, eventually, the frontal and parietal poles and primary visual cortex [8,25,48,60,61]. Hence, a classical neuropathological feature of AD is this progressive and propagating nature of AD inflammatory neuropathology, including the age-related deposition of Aβ42 peptides as insoluble senile plaque deposits [62–66]. The mechanism of AD spreading throughout these anatomical regions of the human brain involved in cognition and memory are not well understood. The small size of miRNAs, the recent identification of miRNA protective proteins, the circularization of miRNAs into complex secondary and tertiary structures, and the fact that miRNAs may concentrate into small membrane-encased secretory vesicles suggest that miRNAs may represent novel transmissible factors for paracrine, endocrine and related forms of intercellular and inter-tissue communication and potential disease spreading amongst CNS [67–69; unpublished observations]. Interestingly, miRNAs are secreted into the surrounding cell culture growth medium when human primary neuronal-glial cocultures are stressed with cytokines (such as IL-1β and TNFα) and other AD-relevant stressors [70,71]. When control neuronal-glial co-cultures are subsequently treated with this surrounding ‘conditioned growth medium’, these brain cells take on characteristics of the stressed cells from which the surrounding ‘conditioned growth medium’ was derived [71; unpublished observations]. Understanding the mechanism for the potential pathological proliferation of AD by miRNAs may of course expose unique opportunities for the development of novel diagnostic techniques, including anti-miRNA-based therapeutic strategies, not only for AD but also for related neurodegenerative disorders such as Parkinson’s disease, Down’s syndrome (trisomy 21), fronto-temporal dementia, prion disease, and other progressive neurological diseases with an inflammatory component.
Concluding Remarks
In summary, at least 7 independent lines of research currently support the contention that miRNAs may be involved in the pathological spreading of AD, including the following empirical observations: (i) their similarity in structure, function and pathological mechanism to viroids, the smallest infectious agent known, which spread degenerative disease in plants; (ii) that the growth medium of stressed primary human brain cells contain elevated pro-inflammatory miRNA levels; (iii) that these same potentially pathogenic miRNAs are abundant in AD ECF and CSF; (iv) that the pathogenic miRNAs found to be elevated in AD brain tissues are also abundant in AD ECF and CSF; (v) that miRNA-containing growth medium from stressed primary human brain cells can in vitro down-regulate pro-inflammatory regulatory glycoproteins and receptors such as complement factor H (CFH) and the triggering receptor in myeloid cells (TREM2; CFH and TREM2 are known to be down-regulated in AD); (vi) that when individual miRNAs of the same type found to be up-regulated in conditioned growth medium are added to normal human brain cells in primary culture, AD-type changes rapidly ensue; and (vii) that anti-miRNA (AM) strategies can quench the phenomenon of miRNA-mediated inflammatory spreading in stressed primary human brain cells [21,33,52,53,60,62,64,70–73].
What is also noteworthy is that miRNAs may be highly mobile not only between cells and tissues but between species, carrying genetic regulatory information outside of the cells and tissues from which they were initially generated [11,13,21,33,73]. While several studies need to be independently replicated, the very recent observations of (1) plant-derived miRNAs being enriched in humans who consume these plants in their diet; and (2) human microbiome-derived sncRNA and miRNA, and plant miRNA translocation across endothelial barriers, between cells and tissues, and even between individual species, indicates that human neurobiology may be significantly impacted by the actions of microbiome-mediated or externally-derived sncRNA or miRNA trafficking, and the integration of a cell, tissue or even an entire organism into its local environment [11–13,21,50,71,73; unpublished].
Summary
Our concepts of the structure and function of sncRNA, including miRNA and vsRNA and their regulatory mechanisms in development and aging, and how they fit into the fascinating realm of dysfunctional RNA metabolism in diseases of plants and animals, and more specifically human neurological diseases such as AD, continues to evolve. Similarities in the molecular-genetics of miRNA and viroid generation, structure and function suggest that some very basal and conserved mechanisms of disease processes have been preserved across the evolution of multiple and diverse plant and animal species. Indeed, pathogenically up-regulated miRNAs and sncRNAs can be considered as an ancient, epigenetic system used to down-regulate specific mRNAs and their expression, and this elegant regulatory system has been maintained across a plant and animal species divergence that occurred some 1.5 billion years ago [13,14,30,31,69]. Significantly up-regulated miRNAs in neurodegenerative disorders such as AD may help explain the large number of brain gene mRNAs, essential for homeostatic brain activity, that are observed to be progressively down-regulated in AD susceptible anatomical regions as AD advances [62,70,74–77]. In the case of AD, and perhaps other progressive age-related neurological diseases, miRNA- and/or anti-miRNA-directed therapeutics represent an obvious choice for future pharmacological treatment strategies, and these would enable the diseased CNS to regain more normalized and homeostatic gene expression functions.
Acknowledgements
Sincere thanks are extended to Drs. PN Alexandrov, B Bhattacharjee, F Culicchia, C Eicken, C Hebel and Y. Zhao for short post-mortem interval (PMI) human brain tissues or extracts, miRNA array work and initial data interpretation, and to D Guillot and J Lockwood for expert technical assistance. Additional thanks are extended to the physicians and neuropathologists who have provided high quality, short post-mortem interval human brain tissues for study. Additional human control and AD brain tissues were provided by the Memory Impairments and Neurological Disorders (MIND) Institute and the University of California, Irvine Alzheimer’s Disease Research Center (UCI-ADRC; NIA P50 AG16573). Research on miRNA in the Lukiw laboratory involving the innate-immune response in AD, amyloidogenesis and neuro-inflamamtion was supported through COBRE III Pilot Project NIH/NIGMS Grant P30-GM103340, an unrestricted grant from Research to Prevent Blindness (RPB), the Louisiana Biotechnology Research Network (LBRN), NIH Grant NEI EY006311 and NIA Grant AG038834.
Footnotes
This research was presented in part at the Alzheimer Association International Conference 2013 (AAIC 2013) held in Boston MA USA 13–18 July 2013.
References
- 1.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 2.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 3.Tuschl T. RNA interference and small interfering RNAs. Chembiochem. 2001;2:239–245. doi: 10.1002/1439-7633(20010401)2:4<239::AID-CBIC239>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 5.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, et al. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 7.Mehler MF, Mattick JS. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol Rev. 2007;87:799–823. doi: 10.1152/physrev.00036.2006. [DOI] [PubMed] [Google Scholar]
- 8.Lukiw WJ, Zhao Y, Cui JG. An NF-kappaB-sensitive micro RNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J Biol Chem. 2008;283:31315–31322. doi: 10.1074/jbc.M805371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Witkos TM, Koscianska E, Krzyzosiak WJ. Practical Aspects of microRNA Target Prediction. Curr Mol Med. 2011;11:93–109. doi: 10.2174/156652411794859250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perkel JM. Assume nothing: the tale of circular RNA. Biotechniques. 2013;55:55–57. doi: 10.2144/000114061. [DOI] [PubMed] [Google Scholar]
- 12.Sarkies P, Miska EA. Molecular biology. Is there social RNA? Science. 2013;341:467–468. doi: 10.1126/science.1243175. [DOI] [PubMed] [Google Scholar]
- 13.Hill JM, Zhao Y, Bhattacharjee S, Lukiw WJ. miRNAs and viroids utilize common strategies in genetic signal transfer. Front Mol Neurosci. 2014;7:10. doi: 10.3389/fnmol.2014.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arteaga-Vázquez M, Caballero-Pérez J, Vielle-Calzada JP. A family of microRNAs present in plants and animals. Plant Cell. 2006;18:3355–3369. doi: 10.1105/tpc.106.044420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lukiw WJ. NF-κ B-regulated micro RNAs (miRNAs) in primary human brain cells. Exp Neurol. 2012;235:484–490. doi: 10.1016/j.expneurol.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukiw WJ. Variability in micro RNA (miRNA) abundance, speciation and complexity amongst different human populations and potential relevance to Alzheimer’s disease (AD) Front Cell Neurosci. 2013;7:133. doi: 10.3389/fncel.2013.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burmistrova OA, Goltsov AY, Abramova LI, Kaleda VG, Orlova VA, et al. MicroRNA in schizophrenia: genetic and expression analysis of miR-130b (22q11) Biochemistry (Mosc) 2007;72:578–582. doi: 10.1134/s0006297907050161. [DOI] [PubMed] [Google Scholar]
- 18.Lukiw WJ, Pogue AI. Induction of specific micro RNA (miRNA) species by ROS-generating metal sulfates in primary human brain cells. J Inorg Biochem. 2007;101:1265–1269. doi: 10.1016/j.jinorgbio.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukiw WJ. Evolution and complexity of micro RNA in the human brain. Front Genet. 2012;3:166. doi: 10.3389/fgene.2012.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuva-Aydemir Y, Simkin A, Gascon E, Gao FB. MicroRNA-9: functional evolution of a conserved small regulatory RNA. RNA Biol. 2011;8:557–564. doi: 10.4161/rna.8.4.16019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexandrov PN, Dua P, Hill JM, Bhattacharjee S, Zhao Y, et al. microRNA (miRNA) speciation in Alzheimer’s disease (AD) cerebrospinal fluid (CSF) and extracellular fluid (ECF) Int J Biochem Mol Biol. 2012;3:365–373. [PMC free article] [PubMed] [Google Scholar]
- 22.Lukiw WJ, Dua P, Pogue AI, Eicken C, Hill JM. Upregulation of micro RNA-146a (miRNA-146a), a marker for inflammatory neurodegeneration, in sporadic Creutzfeldt-Jakob disease (sCJD) and Gerstmann-Straussler-Scheinker (GSS) syndrome. J Toxicol Environ Health A. 2011;74:1460–1468. doi: 10.1080/15287394.2011.618973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 24.Cui JG, Zhao Y, Lukiw WJ. Isolation of high spectral quality RNA using run-on gene transcription; application to gene expression profiling of human brain. Cell Mol Neurobiol. 2005;25:789–794. doi: 10.1007/s10571-005-4035-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui JG, Li YY, Zhao Y, Bhattacharjee S, Lukiw WJ. Differential regulation of interleukin-1 receptor-associated kinase-1 (IRAK-1) and IRAK-2 by microRNA-146a and NF-kappaB in stressed human astroglial cells and in Alzheimer disease. J Biol Chem. 2010;285:38951–38960. doi: 10.1074/jbc.M110.178848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krol J, Krzyzosiak WJ. Structure analysis of microRNA precursors. Methods Mol Biol. 2006;342:19–32. doi: 10.1385/1-59745-123-1:19. [DOI] [PubMed] [Google Scholar]
- 27.Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 28.Saetrom P, Snøve O, Nedland M, Grünfeld TB, Lin Y, et al. Conserved microRNA characteristics in mammals. Oligonucleotides. 2006;16:115–144. doi: 10.1089/oli.2006.16.115. [DOI] [PubMed] [Google Scholar]
- 29.Shi B, Gao W, Wang J. Sequence fingerprints of microRNA conservation. PLoS One. 2012;7:e48256. doi: 10.1371/journal.pone.0048256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang DY, Kumar S, Hedges SB. Divergence time estimates for the early history of animal phyla and the origin of plants, animals and fungi. Proc Biol Sci. 1999;266:163–171. doi: 10.1098/rspb.1999.0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hedges SB. The origin and evolution of model organisms. Nat Rev Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- 32.Lukiw WJ. Circular RNA (circRNA) in Alzheimer’s disease (AD) Front Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexandrov PN, Pogue A, Bhattacharjee S, Lukiw WJ. Retinal amyloid peptides and complement factor H in transgenic models of Alzheimer’s disease. Neuroreport. 2011;22:623–627. doi: 10.1097/WNR.0b013e3283497334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 35.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams MJ, Lefkowitz EJ, King AM, Carstens EB. Recently agreed changes to the statutes of the International Committee on Taxonomy of Viruses. Arch Virol. 2014;159:175–180. doi: 10.1007/s00705-013-1748-x. [DOI] [PubMed] [Google Scholar]
- 37.Hammann C, Steger G. Viroid-specific small RNA in plant disease. RNA Biol. 2012;9:809–819. doi: 10.4161/rna.19810. [DOI] [PubMed] [Google Scholar]
- 38.Diener TO. Subviral pathogens of plants: viroids and viroidlike satellite RNAs. FASEB J. 1991;5:2808–2813. doi: 10.1096/fasebj.5.13.1717335. [DOI] [PubMed] [Google Scholar]
- 39.Diener TO. Discovering viroids--a personal perspective. Nat Rev Microbiol. 2003;1:75–80. doi: 10.1038/nrmicro736. [DOI] [PubMed] [Google Scholar]
- 40.Ding B. The biology of viroid-host interactions. Annu Rev Phytopathol. 2009;47:105–131. doi: 10.1146/annurev-phyto-080508-081927. [DOI] [PubMed] [Google Scholar]
- 41.Rocheleau L, Pelchat M. The Subviral RNA Database: a toolbox for viroids, the hepatitis delta virus and satellite RNAs research. BMC Microbiol. 2006;6:24. doi: 10.1186/1471-2180-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Navarro B, Gisel A, Rodio ME, Delgado S, Flores R, et al. Viroids: how to infect a host and cause disease without encoding proteins. Biochimie. 2012;94:1474–1480. doi: 10.1016/j.biochi.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 43.Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer’s disease temporal lobe neocortex. Neurosci Lett. 2009;459:100–104. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- 44.Ritchie W, Legendre M, Gautheret D. RNA stem-loops: to be or not to be cleaved by RNAse III. RNA. 2007;13:457–462. doi: 10.1261/rna.366507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, et al. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 46.Li YY, Cui JG, Dua P, Pogue AI, Bhattacharjee S, et al. Differential expression of miRNA-146a-regulated inflammatory genes in human primary neural, astroglial and microglial cells. Neurosci Lett. 2011;499:109–113. doi: 10.1016/j.neulet.2011.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li YY, Cui JG, Hill JM, Bhattacharjee S, Zhao Y, et al. Increased expression of miRNA-146a in Alzheimer’s disease transgenic mouse models. Neurosci Lett. 2011;487:94–98. doi: 10.1016/j.neulet.2010.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cui JG, Hill JM, Zhao Y, Lukiw WJ. Expression of inflammatory genes in the primary visual cortex of late-stage Alzheimer’s disease. Neuroreport. 2007;18:115–119. doi: 10.1097/WNR.0b013e32801198bc. [DOI] [PubMed] [Google Scholar]
- 49.Lukiw WJ, Andreeva TV, Grigorenko AP, Rogaev EI. Studying micro RNA Function and Dysfunction in Alzheimer’s Disease. Front Genet. 2013;3:327. doi: 10.3389/fgene.2012.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reijerkerk A, Lopez-Ramirez MA, van Het Hof B, Drexhage JA, Kamphuis WW, et al. MicroRNAs regulate human brain endothelial cell-barrier function in inflammation: implications for multiple sclerosis. J Neurosci. 2013;33:6857–6863. doi: 10.1523/JNEUROSCI.3965-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gallego JA, Gordon ML, Claycomb K, Bhatt M, Lencz T, et al. In vivo microRNA detection and quantitation in cerebrospinal fluid. J Mol Neurosci. 2012;47:243–248. doi: 10.1007/s12031-012-9731-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Machida A, Ohkubo T, Yokota T. Circulating microRNAs in the cerebrospinal fluid of patients with brain diseases. Methods Mol Biol. 2013;1024:203–209. doi: 10.1007/978-1-62703-453-1_16. [DOI] [PubMed] [Google Scholar]
- 53.Madathil SK, Nelson PT, Saatman KE, Wilfred BR. MicroRNAs in CNS injury: potential roles and therapeutic implications. Bioessays. 2011;33:21–26. doi: 10.1002/bies.201000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Müller M, Kuiperij HB, Claassen JA, Küsters B, Verbeek MM. MicroRNAs in Alzheimer’s disease: differential expression in hippocampus and cell-free cerebrospinal fluid. Neurobiol Aging. 2014;35:152–158. doi: 10.1016/j.neurobiolaging.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 55.Aronica E, Fluiter K, Iyer A, Zurolo E, Vreijling J, et al. Expression pattern of miR-146a, an inflammation-associated microRNA, in experimental and human temporal lobe epilepsy. Eur J Neurosci. 2010;31:1100–1107. doi: 10.1111/j.1460-9568.2010.07122.x. [DOI] [PubMed] [Google Scholar]
- 56.Saba R, Gushue S, Huzarewich RL, Manguiat K, Medina S, et al. MicroRNA 146a (miR-146a) is over-expressed during prion disease and modulates the innate immune response and the microglial activation state. PLoS One. 2012;7:e30832. doi: 10.1371/journal.pone.0030832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Erickson MA, Banks WA. Blood-brain barrier dysfunction as a cause and consequence of Alzheimer’s disease. J Cereb Blood Flow Metab. 2013;33:1500–1513. doi: 10.1038/jcbfm.2013.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sagare AP, Bell RD, Zlokovic BV. Neurovascular defects and faulty amyloid-Î2 vascular clearance in Alzheimer’s disease. J Alzheimers Dis. 2013;33(Suppl 1):S87–S100. doi: 10.3233/JAD-2012-129037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao Y, Bhattacharjee S, Jones BM, Dua P, Alexandrov PN, et al. Regulation of TREM2 expression by an NF-κB-sensitive miRNA-34a. Neuroreport. 2013;24:318–323. doi: 10.1097/WNR.0b013e32835fb6b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jucker M, Walker LC. Pathogenic protein seeding in Alzheimer disease and other neurodegenerative disorders. Ann Neurol. 2011;70:532–540. doi: 10.1002/ana.22615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee SJ, Lim HS, Masliah E, Lee HJ. Protein aggregate spreading in neurodegenerative diseases: problems and perspectives. Neurosci Res. 2011;70:339–348. doi: 10.1016/j.neures.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Colangelo V, Schurr J, Ball MJ, Pelaez RP, Bazan NG, et al. Gene expression profiling of 12633 genes in Alzheimer hippocampal CA1: transcription and neurotrophic factor down-regulation and up-regulation of apoptotic and pro-inflammatory signaling. J Neurosci Res. 2002;70:462–473. doi: 10.1002/jnr.10351. [DOI] [PubMed] [Google Scholar]
- 63.Lukiw WJ, Bazan NG. Strong nuclear factor-kappaB-DNA binding parallels cyclooxygenase-2 gene transcription in aging and in sporadic Alzheimer’s disease superior temporal lobe neocortex. J Neurosci Res. 1998;53:583–592. doi: 10.1002/(SICI)1097-4547(19980901)53:5<583::AID-JNR8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 64.Lukiw WJ. Gene expression profiling in fetal, aged, and Alzheimer hippocampus: a continuum of stress-related signaling. Neurochem Res. 2004;29:1287–1297. doi: 10.1023/b:nere.0000023615.89699.63. [DOI] [PubMed] [Google Scholar]
- 65.Perron MP, Provost P. Protein components of the microRNA pathway and human diseases. Methods Mol Biol. 2009;487:369–385. doi: 10.1007/978-1-60327-547-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Busch S, Wu L, Feng Y, Gretz N, Hoffmann S, et al. Alzheimer’s disease and retinal neurodegeneration share a consistent stress response of the neurovascular unit. Cell Physiol Biochem. 2012;30:1436–1443. doi: 10.1159/000343331. [DOI] [PubMed] [Google Scholar]
- 67.Lukiw WJ, LeBlanc HJ, Carver LA, McLachlan DR, Bazan NG. Runon gene transcription in human neocortical nuclei. Inhibition by nanomolar aluminum and implications for neurodegenerative disease. J Mol Neurosci. 1998;11:67–78. doi: 10.1385/JMN:11:1:67. [DOI] [PubMed] [Google Scholar]
- 68.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lukiw WJ, Handley P, Wong L, Crapper McLachlan DR. BC200 RNA in normal human neocortex, non-Alzheimer dementia (NAD), and senile dementia of the Alzheimer type (AD) Neurochem Res. 1992;17:591–597. doi: 10.1007/BF00968788. [DOI] [PubMed] [Google Scholar]
- 71.Zhao Y, Cui JG, Lukiw WJ. Natural secretory products of human neural and microvessel endothelial cells: Implications in pathogenic “spreading” and Alzheimer’s disease. Mol Neurobiol. 2006;34:181–192. doi: 10.1385/MN:34:3:181. [DOI] [PubMed] [Google Scholar]
- 72.Triboulet R, Gregory RI. Autoregulatory mechanisms controlling the Microprocessor. Adv Exp Med Biol. 2010;700:56–66. [PubMed] [Google Scholar]
- 73.Zhang L, Hou D, Chen X, Li D, Zhu L, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22:107–126. doi: 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ginsberg SD, Alldred MJ, Che S. Gene expression levels assessed by CA1 pyramidal neuron and regional hippocampal dissections in Alzheimer’s disease. Neurobiol Dis. 2012;45:99–107. doi: 10.1016/j.nbd.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Loring JF, Wen X, Lee JM, Seilhamer J, Somogyi R. A gene expression profile of Alzheimer’s disease. DNA Cell Biol. 2001;20:683–695. doi: 10.1089/10445490152717541. [DOI] [PubMed] [Google Scholar]
- 76.Lukiw WJ, Gordon WC, Rogaev EI, Thompson H, Bazan NG. Presenilin-2 (PS2) expression up-regulation in a model of retinopathy of prematurity and pathoangiogenesis. Neuroreport. 2001;12:53–57. doi: 10.1097/00001756-200101220-00019. [DOI] [PubMed] [Google Scholar]
- 77.Bhattacharjee S, Lukiw WJ. Alzheimer’s disease and the microbiome. Frontiers of Cellular Neuroscience. 2013;7:153. doi: 10.3389/fncel.2013.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]