Abstract
With the large volume of clinical and epidemiological data being collected, increasingly linked to extensive genotypic data, coupled with expanding high-performance computational resources, there are considerable opportunities for comprehensively exploring the networks of connections that exist between the phenome and the genome. These networks can be identified through Phenome-Wide Association Studies (PheWAS) where the association between a collection of genetic variants, or in some cases a particular clinical lab variable, and a wide and diverse range of phenotypes, diagnoses, traits, and/or outcomes are evaluated. This is a departure from the more familiar genome-wide association study (GWAS) approach, which has been used to identify single nucleotide polymorphisms (SNPs) associated with one outcome or a very limited phenotypic domain. In addition to highlighting novel connections between multiple phenotypes and elucidating more of the phenotype-genotype landscape, PheWAS can generate new hypotheses for further exploration, and can also be used to narrow the search space for research using comprehensive data collections. The complex results of PheWAS also have the potential for uncovering new mechanistic insights. We review here how the PheWAS approach has been used with data from epidemiological studies, clinical trials, and de-identified electronic health record data. We also review methodologies for the analyses underlying PheWAS, and emerging methods developed for evaluating the comprehensive results of PheWAS including genotype-phenotype networks. This review also highlights PheWAS as an important tool for identifying new biomarkers, elucidating the genetic architecture of complex traits, and uncovering pleiotropy. There are many directions and new methodologies for the future of PheWAS analyses, from the phenotypic data to the genetic data, and herein we also discuss some of these important future PheWAS developments.
Keywords: genotype-phenotype networks, pleiotropy, phenomics, International Classification of Disease Codes (ICD), Electronic Health Record (EHR)
Introduction
A dynamic network exists between the genome, gene products, signaling pathways, intermediate phenotypes, and outcome traits, and this complexity can be leveraged to develop a clearer picture of the etiology of complex traits. Through exploring the relationships between genetic variation and a wide range of phenotypic measurements at multiple levels, we can integrate these complex and comprehensive results to gain a clearer picture of the genotype-phenotype landscape. There is ample evidence of many genetic variants being associated with multiple traits, indicating the potential for pleiotropy. The NHGRI genome-wide association study (GWAS) catalog shows many single nucleotide polymorphisms (SNPs) associated with more than one phenotype and/or phenotypic domain [1]. Autoimmune diseases have shown considerable overlap in genetic regions with evidence of association [2,3], as has metabolic syndrome [4].
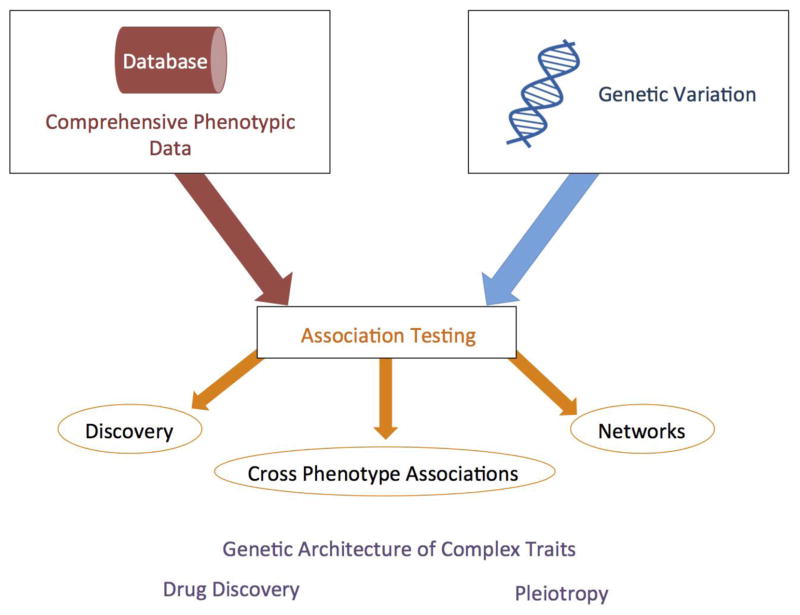
To identify dynamic networks of phenotypic and genotypic connections, Phenome-Wide Association Studies (PheWAS) can be used to evaluate the association between any number of single nucleotide polymorphisms (SNPs) and a wide range of phenotypic variables in a high-throughput manner (Figure 1). PheWAS began with investigation of the association between multiple SNPs and de-identified electronic health record (EHR) data [5], and has now been used successfully several times with EHR data [6–12]. Since then, PheWAS has been used with epidemiological study data and clinical trials data [13–15]. PheWAS can be used for comprehensively investigating the association between genetic variation and a wide array of outcome traits in any study design with a multitude of phenotypic data such as epidemiological cohorts, clinical trials and animal breeding research. PheWAS is complementary to the Genome-Wide Association Study (GWAS) approach, which investigates the association between genetic variation and one outcome/phenotype, or a limited phenotypic domain. However, GWAS cannot provide the additional information that exists when using a wide range of genotypic and phenotypic data assessed concurrently from the same dataset. If associations are found between a single SNP and multiple phenotypes, showing potential pleiotropy, there are a range of reasons for these associations that may uncover important biology. Further, PheWAS has the potential for assisting clinical and drug discovery through identifying both novel SNP phenotype associations and relationships between single variants and multiple phenotypes including identifying potential side effects.
Figure 1. Overview of PheWAS.
PheWAS can be used to evaluate the association between a comprehensive set of phenotypes and genetic variation. A relational database is useful for organizing and working with the phenotypic data. The phenotypic data can be collected through multiple types of studies, including epidemiological studies, de-identified electronic health records, clinical trials data, and animal breeding research. Genetic variation can be single nucleotide polymorphisms (SNPs), but any genetic variation that can be evaluated for association with phenotypic variation can be used. The association testing results can be evaluated multiple ways, and while not shown, a relational database can assist with analyses of results. Novel discoveries can be identified along with cross-phenotype associations. Networks of connections between SNPs, genes, phenotypes, can be explored. These results can provide more information about the genetic architecture of complex traits, highlight biologically important pleiotropy, and can support drug discovery.
PheWAS is not limited to associations between SNPs and phenotypes. PheWAS is now being extended to explore the relationship between other genetic variation, such as the relationship between copy-number variation and a wide range of measurements, as well as the relationship between mitochondrial variation and outcome [16]. PheWAS can be used with common frequency SNPs, but also can be applied to low-frequency variation as more and more tools for using low frequency variants are being introduced. The PheWAS approach has also been extended to explore the association between single laboratory variables and a wide range of phenotypes [17]. Further, environmental information can be used in a similar way to PheWAS to evaluate a wide range of environmental exposures for further study in Environment Wide Association Studies (EWAS) [18,19] and Dietary-wide Association Studies (DWAS) [20], and these results in turn can be used in PheWAS to explore the relationship between environmental exposures and genetic variation with a wide range of phenotypic outcomes.
Within this review we describe the characteristics and methodology used for PheWAS studies to date. We include discussion of methods that have been effectively used to set up various types of PheWAS studies, as well as to evaluate the potential thousands of results that can arise from PheWAS studies. We also highlight some of the challenges, limitations, and future directions for phenome-wide association studies.
PheWAS: Context
PheWAS, as a methodology, builds on several disciplines. The use of biological screens has been an important workhorse in science for a long time, and aspects of PheWAS share commonality with various kinds of screens for hypothesis generation. For example, inducing mutations in bacterial strains and then screening those strains for novel traits of interest has been an important way to identify genes and genetic pathways for further research [21]. In PheWAS, the individuals have a variety of natural genetic variation, and researchers can identify in a high-throughput way any indications of modified traits as a result of that genetic variation. Expression quantitative trait loci (eQTL) experiments that evaluate the association between SNPs and comprehensive gene expression also share common themes with PheWAS, when each gene expression variable is considered as a phenotype [22].
The focus on a much wider array of phenotypes, termed phenomics, is also an important part of the history and rationale behind the PheWAS approach [23–26]. Much of GWAS has focused solely on disease case-control status, or one or a small set of very highly related phenotypic variables, such as lipid levels. The field of phenomics has championed the idea of considering more phenotypic information, defining the human phenome as a systematically and comprehensive measured set of phenotypes, including qualitative and quantitative traits that capture clinical, biochemical, and imaging traits. This requires measuring phenotypes such as disease outcome, i.e. case control status, but also considering an array of other phenotypic variables, including intermediate phenotypes such as clinical lab variables. Identifying correlations between comprehensively collected phenotypes provides important information about the landscape of health and disease, showing conditions that do and do not co-occur. These data can be leveraged to understand the dynamic networks underlying health and disease. Another rationale for phenomics is to better partition groups of individuals into more homogeneous categories for study by also analyzing other traits they do or do not have. This information can provide increased precision for genetic study. The genetic etiology of disease outcome may arise from multiple different pathways, reflected in different sets of phenotypes between individuals with the same clinically defined outcome. Finally, phenomics can identify common pathways across multiple diseases, by identifying individuals with shared outcomes, thus developing a “disease network” of shared or correlated traits [27–29]. Phenomics has contributed to PheWAS through underscoring the importance of broad collections of phenotypes, and using that information in a coordinated and unified fashion to develop understanding of the relationships between phenotypic variables and disease.
Pleiotropy is another underlying principle that can be explored through PheWAS. There are multiple definitions for pleiotropy [30]; however, pleiotropy broadly speaking refers to the effect of genetic variation on more than one trait. It is challenging to identify if genetic variation that affects one phenotype also independently affects another phenotype, as there are many biological mechanisms that could underlie the association between genetic variation and multiple phenotypes [31]. Identifying cross-phenotype (CP) associations still provides important insights, and can uncover true pleiotropy for some associations. CP associations provide clues and putative genetic contexts for associations across multiple traits, helping to focus research. For example, in a CP association, is there something about the genetic variant affecting a molecular process at the DNA level, or is the genetic variant affecting a genetic pathway, or is a physiological network being perturbed that is resulting in a co-occurrence of traits related to a single genetic variant? We can we learn about the biology of the interrelationships between genetic architecture and outcome if we explore PheWAS results on a SNP-phenotype, pathway, and network level. Further, if a CP association shows decreased risk for one phenotype, and increased risk with another phenotype, important insights can be gained. For example, if a drug is developed for a specific gene where a SNP is present that increases risk of one phenotype, are there CP associations identified through PheWAS that show decreased risk for other phenotypes? Would this change the priority of this gene in drug development, would treatment result in unintended side effects or consequences due to the potentially pleiotropic behavior of this loci? Or would insights from PheWAS result in a clinician having knowledge to prescribe a second drug to counter effect potential problems? As previously mentioned, there is considerable evidence of many CP associations existing within human genetic architecture, when comparing the results of multiple GWAS and thus far with PheWAS, and identifying CP associations is an important starting point for discovery, data exploration, and hypothesis generation. Further, results are often justified as more plausible for one phenotype if a variant is known to associate with another phenotype.
Electronic Health Record Based PheWAS
The first PheWAS were performed with de-identified EHR data linked to genotypic data. PheWAS are still described alternately as a phenome-scan, linking genetic data to comprehensive phenotypic data for association testing [32]. Expanding on the idea of a phenome-scan, before the term PheWAS was coined, a clinical phenome-scan was discussed [33]. The idea was to use comprehensive electronic medal records to generate a clinical phenome-scan for each subject, to ask the question “which gene is associated with a given disease”, instead of “which disease is associated with a given gene?” The clinical phenome-scan became a reality with the introduction of de-identified EHR data linked to DNA samples, such as through BioVU, the Vanderbilt DNA databank [34]. Clinical Electronic Health Records (EHR) contain a wide array of information about each patient, from billing codes, to free text entered by the clinician about a patients health, to clinical lab measurement values collected across multiple visits, and can include potential imaging data [35,36]. Thus, EHR provide an incredible resource of phenotypic data.
The first EHR PheWAS was performed using International Classification of Disease Codes (ICD) codes in BioVU, showing the feasibility of using ICD codes in a high-throughput way for evaluating genotype-phenotype associations [5]. ICD are used for billing purposes, and provide a way to document disease, symptoms, causes of injury and diseases, and procedures for patients. Using ICD-9 codes (the 9th revision of the ICD codes), this proof-of-principle study by Denny et al. in 2010, used five SNPs and defined 776 case/control phenotypes based on the presence/absence of each ICD-9 code for each individual in the study. If an individual had an ICD-9 code they were considered a “case”, and other individuals were considered a “control” if they did not have that specific ICD-9 code or any other ICD-9 codes that had been considered exclusionary for that specific ICD-9 code. The five SNPs were chosen for their previously reported disease associations, such as atrial fibrillation and coronary artery diseases. Four of seven expected SNP-disease associations replicated previously reported associations in the literature, and the authors also identified 19 previously unknown associations with p < 0.01.
Since the original ICD-9 code based PheWAS there have been several other PheWAS using de-identified electronic medical record data. Thus far, all of these studies have used a moderate number of SNPs chosen for specific reasons, and the majority of these studies have focused on data from European Americans. For example, Denny et al. first identified SNPs associated with hypothyroidism case/control status via GWAS, then used significantly associated SNPs from the GWAS to perform a PheWAS, using ICD-9 codes [6]. This study showed the efficacy of defining an algorithm for identifying cases and controls for hypothyroidism that used medication information, ICD-9 codes, and clinical lab data from the EHR of 5 different clinics in the Electronic Medical Records and Genomics (eMERGE) network. Then the study showed the utility of exploring comprehensive ICD-9 based PheWAS associations with four SNPs identified from the preliminary GWAS to identify novel and related phenotypic associations in addition to hypothyroidism. In a similar study, GWAS was used to identify variants influencing circulating platelet numbers and mean platelet volume, and then the PheWAS approach used significantly associated SNPs from the GWAS with ICD-9 code based case/control status identifying potentially pleiotropic associations with myocardial infarction, autoimmune, and hematalogic diagnoses [9]. Hebbring et al. used the PheWAS approach with data from the Marshfield Clinic’s Personalized Medicine Research Project (PMRP) de-identified biorepository [7]. In this study, authors investigated the association between a single SNP within the human leukocyte antigen gene HLA-DRB1, and both ICD-9 and relevant “V” codes to define case/control status. V codes are another type of billing code possible within EHR data representing a group of supplementary factors influencing health status and contact with health services, such as V86 “estrogen receptor status”. The authors found this SNP replicated a known relationship with multiple sclerosis, but also identified alcohol-induced cirrhosis of the liver as well as erythematous conditions. Another PheWAS was performed using 3,144 SNPs from the NHGRI GWAS catalog [1] and 1,358 EMR-derived phenotypes. This study replicated known associations, while identifying potentially novel associations suggestive of pleiotropy [10].
All of these aforementioned studies either applied some form of multiple-hypothesis testing correction, or provided information about some of the most significant potentially novel results, in addition to replicating known associations. However, with the multiple hypothesis testing burden incurred in PheWAS, another approach being used to help discern results that are less likely to be by chance alone, is through seeking replication of PheWAS results across more than one dataset. Using the replication approach across multiple studies, approximately ~100,000 SNPs relevant to autoimmunity and the immune system have been investigated for association with ICD-9 codes in European Americans from two sites: BioVU and Geisinger. The authors again replicated known associations, but also identified a series of potential associations for immune and autoimmune relevant SNPs [11].
Recently two studies have been conducted by picking SNPs that are much more likely to have functional consequences on the protein encoded by a gene. Using public repositories of evidence for SNP functionality can provide a way to focus on SNPs more likely to impact phenotype. One PheWAS within the Marshfield PMRP used 105 presumed functional stop-gain and stop-loss variants, and identified a nonsense variant in ARMS2 associated with age-related macular degeneration [12]. In another PheWAS from the eMERGE network, Verma et al.[37], used multiple sources of information to identify 25 SNPs known or highly likely to be stop-gain inducing variants. This stop-gain PheWAS was undertaken in part to investigate using functional information about SNPs as the pre-filter for SNPs for PheWAS, as well as identifying clinically relevant associations to guide further development of phenotypic algorithms for validation. Unlike the aforementioned studies, this eMERGE based functional stop-gain variant study used both the full dataset using principle components to adjust for global ancestry of individuals, as well as analyses stratified by observer reported European American and African American ancestry. Within this study the comprehensive phenotypic algorithms developed by the eMERGE network were used in addition to ICD-9 based phenotypes. The authors identified a total of 84 associations replicating across the two datasets evaluated a p < 0.01, with the same 3 digit ICD-9 code and same direction of effect; 16 SNPs also showed evidence of pleiotropy [Verma et al. in preparation].
Epidemiological Study Based PheWAS
The PheWAS approach can also be used within large population based epidemiological studies linked to genotypic data [38]. For example, the first PheWAS using epidemiological study data was through the Population Architecture Using Genomics and Epidemiology (PAGE) network [13]. The phenotypic and genotypic data for this study came from five different sites within PAGE. Unlike the majority of ICD-9 code based PheWAS focused on European Americans, this study had data across four major racial/ethnic groups: European Americans, African Americans, Hispanics/Mexican-Americans, and Asian/Pacific Islanders. In a departure from case/control PheWAS based solely on ICD-9 codes, there were 4,706 comprehensive measurements for multiple traits, laboratory measures, and intermediate biomarkers. Thus, some of the phenotypic data was dichotomous in nature, while other phenotypes were quantitative in nature such as lipid measurements. A total of 83 SNPs, previously known to associate with specific traits, were used if genotyped at two or more PAGE study sites.
In contrast to ICD-9 based PheWAS the PAGE study faced multiple unique challenges. First was the use of the quantitative phenotypic data in a high-throughput way. Depending on the regression method chosen for associations, there can be assumptions of normality for the variables. The authors used standard regression (linear and logistic) and thus ran all associations with quantitative variables on both un-transformed and natural-log-transformed variables. Some of the variables were categorical with more than two states, so authors chose to create dichotomous variables in high-throughput fashion by turning any of these variables into dichotomous variables. The data was collected across race/ethnicity and thus was stratified by race/ethnicity before all associations were calculated. Another challenge was how to seek replication for results across studies, when phenotypes might be related (e.g. diabetes status) but were not determined identically across sites. It was not possible to harmonize all phenotypes, thus phenotype “binning” was used to assign phenotypes across studies into categories. In this way total of 111 PheWAS results were identified that had significant associations for two or more PAGE study sites with consistent direction of effect and a significance threshold of p<0.01 for the same racial/ethnic group, SNP, and phenotype-class.
In a similar fashion, a PheWAS was performed utilizing the diverse genotypic and phenotypic data existing across multiple populations in the National Health and Nutrition Examination Surveys (NHANES), conducted by the Centers for Disease Control and Prevention (CDC), and accessed by the Epidemiological Architecture for Genes Linked to Environment (EAGLE) study [14]. Comprehensive tests of association in Genetic NHANES used 80 SNPs and 1,008 phenotypes (grouped into 196 phenotype classes), stratified by race-ethnicity for non-Hispanic whites, non-Hispanic blacks, and Mexican Americans were calculated. Genetic NHANES contains two datasets: NHANES III collected between 1991–1994 and Continuous NHANES collected between 1999–2002. The authors identified 69 PheWAS associations that replicated across the two datasets for the same SNP, phenotype-class, direction of effect, and race-ethnicity at p < 0.01, allele frequency > 0.01, and sample size > 200. This study was the first to investigate PheWAS results in the context of networks, linking SNPs to genes, and then linking genes to pathways such as Kyoto Encyclopedia of Genes (KEGG) [39], and visualizing these networks using Cytoscape [40].
Clinical Trial based PheWAS
Clinical trials collect a wide range of clinical laboratory measurements, and also often include questionnaire/survey based variables. With coupled genotypic information, there is an opportunity for PheWAS to identify novel pharmacogenomic associations. The first PheWAS using clinical trials data has now been performed; this was also the first PheWAS to use genome-wide genotypic data. This study used data from the AIDS Clinical Trials Group (ACTG) human, using immunodeficiency virus (HIV) clinical trials datasets. As proof-of-concept, the authors focused on 27 laboratory tests from antiretroviral therapy-naïve individuals. To test for replication, data from four trials were divided into two equally proportioned datasets for discovery/replication. Final analyses involved 2,547 individuals and 5,954,294 genotyped and imputed polymorphisms. A total of 11,156 (0.18%) single nucleotide polymorphisms (SNPs) had associations of p-value < 0.01 in both datasets with same direction of association. Twenty SNPs replicated associations with identical or related phenotypes reported in the NHGRI GWAS Catalog [1], including several previously reported only in HIV-negative cohorts, as well as potentially novel associations [15].
Moving Beyond SNPs
The PheWAS approach is not limited to evaluating the association between nuclear DNA based SNPs and a range of phenotypic data. For instance there was one study using PheWAS in African Americans, linked to de-identified EHR data and mitochondrial SNPs[16]. Further, clinical laboratory variables can be used in high-throughput association with an array of diagnoses to identify important clinically relevant biomarkers. For example, one study investigated the association between autoantibodies as well as risk alleles for autoimmune disease and clinical diagnoses in rheumatoid arthritis cases and controls[8]. By exploring other clinical diagnoses related to biomarkers and SNPs, more insight can be developed about the bigger picture of the relationship between measureable clinical laboratory variables, genetic architecture, and the interrelationships between diseases and diagnoses. Similarly, a study was done to investigate the association between thiopurine S-methyltransferase (TPMT) activity in patients on thiopurine drugs and a series of ICD-9/ICD-10 codes [17].
Challenges and Promise for Complex Comprehensive Phenotypic Data
Phenotypic Data
One of the most important aspects of PheWAS, and one of the greatest challenges, is obtaining and using comprehensive data in the most high-throughput way possible. In EHR data, the majority of PheWAS has been performed using ICD-9 codes; the dichotomous nature of such variables make high-throughput translation of these data to association testing relatively straight forward. Much more challenging is taking continuous variables and using these in PheWAS. Hurdles faced with continuous variables include assessing phenotypic variables in a high-throughput manner for unusual or suspect data points such as extreme outliers, as well as agreement in measurement units. For example, there may be phenotypic measurements that are not possible for a specific clinical lab variable that could cause problems with further analyses, or variability in measurement units for a specific variable when multiple sites are collecting data. If the regression approach used for the particular PheWAS relies on an assumption of normality of quantitative phenotypic data, high throughput evaluation of normality and/or transformation of those variables may be necessary. As mentioned, in a PheWAS from the PAGE network[13], because linear regression was used, all associations for quantitative variables were run with the data untransformed and natural log transformed as natural log transformation is one of the most common ways to adjust phenotypic for normality. In the case of the AIDS clinical trials PheWAS, where linear regression was also used, a more limited number of clinical measurements were used such that the phenotypes could be carefully evaluated for normality, transformed as necessary, outliers could identified, and other issues such as different measurement units for the same clinical laboratory variable could be explicitly addressed [15]. In the future, other regression approaches such as robust regression could be used to reduce the impact of assumptions of normality on association testing in PheWAS. Seeking replication across more than one dataset for a large number of phenotypic variables without carefully harmonizing phenotypes across datasets can also be a challenge for PheWAS. Some phenotypes are easier to compare across datasets than others, such as lipid level measurements when compared to a possible variety of metrics of diabetes status. Key for PheWAS has been the use of relational databases to assist with the process of organizing phenotypic data, using queries to pull summary information about phenotypic data, migrating data into a form supporting high-throughput visualization of summary information of phenotypic data, as well as shifting these data via queries into formats appropriate for association testing.
For both epidemiological and EHR/clinical data, development of additional methods for automatic and high-throughput phenotype extraction and evaluation will be critical moving forward. For example, there are clinical measurements collected longitudinally for patients, and if these phenotypes were “mined” in a high-throughput way, they would add an incredible wealth of data to PheWAS analyses. Continuous variables can provide more power for association testing, when compared to dichotomous variables, and may highlight important associations yet been discovered within EHR data. These data are promising, but also challenging, as these variables often change with age, health status, and drug use, making interpretation difficult.
While PheWAS can be run with a variety of different types of data sets, improving phenotype harmony across studies can assist with PheWAS association testing. For example, PhenX (https://www.phenxtoolkit.org/) is a toolkit providing standard measures for phenotypic traits and environmental exposures [41]. This kit, if used across studies, can provide critical guidance to researchers in collection and standardization of high quality phenotypic data for use in PheWAS. This would also make comparing PheWAS results across studies much easier. Automatic term identification may also become an important tool for identifying phenotypes across datasets that have similar, but different, identifiers for further phenotype binning or harmonization [42].
More methods can be used with PheWAS that combine information from multiple phenotypes before association testing. As these methods emerge, more of these may prove useful in leveraging the rich phenotypic data of PheWAS, while also reducing the number of statistical tests and type-I error rate. Principle components for example, are one approach for transforming a group of phenotypic variables into a smaller number of variables. Matthew Stephens has introduced a method for working with multiple related phenotypes, using Bayesian methods [43]. Graph-guided fused lasso (GFlasso) has also been introduced as a method for working with a large number of correlated phenotypes [44] . The CAPE method has been developed to analyze pleiotropy in addition to gene-gene epistatic interactions [45,46]. Other approaches beyond PheWAS include a study assessing the results of multiple studies, taking correlation between a range of metabolic traits and inflammatory markers into account [4].
Networks
While single SNP-phenotype and CP associations can be investigated in PheWAS, an additional powerful feature of PheWAS is the ability to explore the networks of connections that exist in the results. PheWAS uncovers links between SNPs, gene regions, and phenotypes that can define, these connections. Because genes are in pathways, SNP, gene, and phenotypic results can also be connected to pathway information. These networks can be visualized with software such as Cytoscape[40] and Gephi[47]. Using networks, shifts the understanding of genetic architecture from individual results to a higher level of complex interrelationships between traits and genetic variation [48,49].
Multiple Hypothesis Testing
PheWAS can incur a substantial multiple hypothesis testing burden, and thus increased type-1 error, due to the number of association tests. The Bonferroni correction is the p-value adjustment primarily used with GWAS to correct for multiple testing where a p-value (usually 0.05 or 0.01) is divided by the number of tests of association to determine an adjusted p-value cutoff. Correcting for multiple hypothesis testing using a Bonferroni correction within GWAS is problematic for multiple reasons. These include the assumption of independence of tests even though in reality there are correlated SNPs in GWAS, adequately balancing both the risk of false positive findings and false negative findings, and the importance of disease or outcome context and how that will impact association testing [50,51]. For PheWAS, both phenotypes and genotypes can be correlated. Power from one result to another can vary in part due to variations in sample size for the specific phenotype, as well as the effect size of different genetic variants. In addition, phenotype-class binning of results can result in different numbers of sub-phenotypes in each bin for potential replication. Some PheWAS studies have sought replication to partially control for type-1 error, however choosing the right p-value cutoff for comparing results across studies remains a challenge.
An important tool for PheWAS may be permutation testing. With permutation testing the connection between genotypic data and phenotypic data can be shuffled, resulting in the correlations between SNPs being maintained and correlations between phenotypes for each individual being maintained. The link between the genotypic data and phenotypic data is the only information shuffled. The p-values across the PheWAS within the null data can be evaluated and compared with the p-values of the non-permuted data, providing empirical support of the significance of results.
Environmental Data
Environmental data will be important to incorporate into PheWAS. Already the PheWAS approach has been applied using environmental variables, EWAS[18,19] and dietary variables (DWAS) [20]. The EWAS and DWAS approach can be used to identify important environmental variables for further study, and then PheWAS can be used that includes relevant environmental exposures as interaction terms.
Conclusions
PheWAS are a purposely high-throughput approach, for exploration and hypothesis generation. With PheWAS the complex interrelationships between genetic architecture and multiple outcomes can be explored. PheWAS can be used with a variety of data types. Within this review we have covered the methods of PheWAS to date, as well as important considerations and future directions. The PheWAS approach is quickly becoming an important tool for using more of rich data collections, providing a way to highlight important directions for new discovery.
Acknowledgments
MD Ritchie has received research grants from the NIH and Geisinger Health Systems.
Footnotes
Conflict of Interest
SA Pendergrass declares no conflicts of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, have been highlighted as:
•• Of major importance
• Of importance
- 1.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. PNAS. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cotsapas C, Voight BF, Rossin E, Lage K, Neale BM, Wallace C, et al. Pervasive Sharing of Genetic Effects in Autoimmune Disease. PLoS Genet. 2011;7:e1002254. doi: 10.1371/journal.pgen.1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knight JC. Genomic modulators of the immune response. Trends Genet. 2013;29:74–83. doi: 10.1016/j.tig.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kraja AT, Chasman DI, North KE, Reiner AP, Yanek LR, Kilpeläinen TO, et al. Pleiotropic genes for metabolic syndrome and inflammation. Mol Genet Metab. 2014 doi: 10.1016/j.ymgme.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5••.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–10. doi: 10.1093/bioinformatics/btq126. First PheWAS, used electronic health record data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denny JC, Crawford DC, Ritchie MD, Bielinski SJ, Basford MA, Bradford Y, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet. 2011;89:529–42. doi: 10.1016/j.ajhg.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hebbring SJ, Schrodi SJ, Ye Z, Zhou Z, Page D, Brilliant MH. A PheWAS approach in studying HLA-DRB1*1501. Genes Immun. 2013;14:187–91. doi: 10.1038/gene.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao KP, Kurreeman F, Li G, Duclos G, Murphy S, Guzman R, et al. Associations of autoantibodies, autoimmune risk alleles, and clinical diagnoses from the electronic medical records in rheumatoid arthritis cases and non-rheumatoid arthritis controls. Arthritis Rheum. 2013;65:571–81. doi: 10.1002/art.37801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shameer K, Denny JC, Ding K, Jouni H, Crosslin DR, de Andrade M, et al. A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum Genet. 2014;133:95–109. doi: 10.1007/s00439-013-1355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31:1102–10. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verma A, Kuivaniemi H, Tromp G, Carey DJ, Gerhard GS, Crowe JE, et al. Exploring the relationship between immune system related genetic variants and complex traits and disease through a Phenome-Wide Association Study (PheWAS) In Preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye Z, Mayer J, Ivacic L, Zhou Z, He M, Schrodi SJ, et al. Phenome-wide association studies (PheWASs) for functional variants. Eur J Hum Genet. 2014 doi: 10.1038/ejhg.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13••.Pendergrass SA, Brown-Gentry K, Dudek S, Frase A, Torstenson ES, Goodloe R, et al. Phenome-wide association study (PheWAS) for detection of pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network. PLoS Genet. 2013;9:e1003087. doi: 10.1371/journal.pgen.1003087. First PheWAS using epidemiological study data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall MA, Verma A, Brown-Gentry K, Goodloe RJ, Boston J, Wilson S, et al. Detection of pleiotropy through a phenome-wide association study (PheWAS) of epidemiologic data as part of the Environmental Architecture for Genes Linked to Environment (EAGLE) Study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15••.Moore CB, Verma A, Pendergrass SA, Verma SS, Johnson DH, Daar ES, et al. Phenome-wide Associations Study (PheWAS) Relating Pre-treatment Laboratory Parameters with Human Genetic Variants in AIDS Clinical Trials Group Protocols. Under Revision. First PheWAS using clinical trials data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Mitchell SL, Hall JB, Goodloe RJ, Boston J, Farber-Eger E, Pendergrass SA, et al. Investigating the relationship between mitochondrial genetic variation and cardiovascular-related traits to develop a framework for mitochondrial phenome-wide association studies. BioData Mining. 2014;7:6. doi: 10.1186/1756-0381-7-6. First PheWAS using mitochondrial genetic variation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neuraz A, Chouchana L, Malamut G, Le Beller C, Roche D, Beaune P, et al. Phenome-wide association studies on a quantitative trait: application to TPMT enzyme activity and thiopurine therapy in pharmacogenomics. PLoS Comput Biol. 2013;9:e1003405. doi: 10.1371/journal.pcbi.1003405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18•.Patel CJ, Bhattacharya J, Butte AJ. An Environment-Wide Association Study (EWAS) on type 2 diabetes mellitus. PLoS ONE. 2010;5:e10746. doi: 10.1371/journal.pone.0010746. First Environment-Wide Association Study (EWAS) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall MA, Dudek SM, Goodloe R, Crawford DC, Pendergrass SA, Peissig P, et al. Environment-wide association study (EWAS) for type 2 diabetes in the Marshfield Personalized Medicine Research Project Biobank. Pac Symp Biocomput. 2014:200–11. In Press PLoS Genetics. [PMC free article] [PubMed] [Google Scholar]
- 20•.Davis MA, Gilbert-Diamond D, Karagas MR, Li Z, Moore JH, Williams SM, et al. A Dietary-Wide Association Study (DWAS) of Environmental Metal Exposure in US Children and Adults. PLoS ONE. 2014;9:e104768. doi: 10.1371/journal.pone.0104768. First Dietary-Wide Association Study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shuman HA, Silhavy TJ. The art and design of genetic screens: Escherichia coli. Nat Rev Genet. 2003;4:419–31. doi: 10.1038/nrg1087. [DOI] [PubMed] [Google Scholar]
- 22.Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–94. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oti M, Huynen MA, Brunner HG. Phenome connections. Trends Genet. 2008;24:103–6. doi: 10.1016/j.tig.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 24.Bilder RM, Sabb FW, Cannon TD, London ED, Jentsch JD, Parker DS, et al. Phenomics: the systematic study of phenotypes on a genome-wide scale. Neuroscience. 2009;164:30–42. doi: 10.1016/j.neuroscience.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25•.Lanktree MB, Hassell RG, Lahiry P, Hegele RA. Phenomics: expanding the role of clinical evaluation in genomic studies. J Investig Med. 2010;58:700–6. doi: 10.231/JIM.0b013e3181d844f7. Review/idea paper about leveraging phenomics for improved understanding of disease. [DOI] [PubMed] [Google Scholar]
- 26•.Houle D, Govindaraju DR, Omholt S. Phenomics: the next challenge. Nature Reviews Genetics. 2010;11:855–66. doi: 10.1038/nrg2897. Detailed review of considerations for phenomics. [DOI] [PubMed] [Google Scholar]
- 27.Goh K-I, Cusick ME, Valle D, Childs B, Vidal M, Barabasi A-L. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–90. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rzhetsky A, Wajngurt D, Park N, Zheng T. Probing genetic overlap among complex human phenotypes. Proc Natl Acad Sci U S A. 2007;104:11694–9. doi: 10.1073/pnas.0704820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Barabási A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12:56–68. doi: 10.1038/nrg2918. Review of exploring networks and interactions for understanding disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paaby AB, Rockman MV. The many faces of pleiotropy. Trends Genet. 2013;29:66–73. doi: 10.1016/j.tig.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet. 2013;14:483–95. doi: 10.1038/nrg3461. Excellent review of the importance of pleiotropy and cross-phenotype associations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Jones R, Pembrey M, Golding J, Herrick D. The search for genenotype/phenotype associations and the phenome scan. Paediatr Perinat Epidemiol. 2005;19:264–75. doi: 10.1111/j.1365-3016.2005.00664.x. Idea paper introducing fundamental ideas behind PheWAS focused on networks in the context of dense phenotypic and genotypic information. [DOI] [PubMed] [Google Scholar]
- 33••.Ghebranious N, McCarty CA, Wilke RA. Clinical phenome scanning. Personalized Medicine. 2007;4:175–82. doi: 10.2217/17410541.4.2.175. Idea paper introducing fundamental ideas behind PheWAS, particularly with comprehensive electronic health record data. [DOI] [PubMed] [Google Scholar]
- 34.Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84:362–9. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio TA, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15:761–71. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crawford DC, Crosslin DR, Tromp G, Kullo IJ, Kuivaniemi H, Hayes MG, et al. eMERGEing progress in genomics-the first seven years. Front Genet. 2014;5:184. doi: 10.3389/fgene.2014.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verma A, Verma SS, Pendergrass SA, Crawford DC, Crosslin DR, Kuivaniemi H, et al. Phenome-Wide Association Study (PheWAS) Identifies Clinical Associations and Pleiotropy for Functional Variants. In Preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.Pendergrass SA, Brown-Gentry K, Dudek SM, Torstenson ES, Ambite JL, Avery CL, et al. The use of phenome-wide association studies (PheWAS) for exploration of novel genotype-phenotype relationships and pleiotropy discovery. Genet Epidemiol. 2011;35:410–22. doi: 10.1002/gepi.20589. Idea paper describing PheWAS in epidemiological study based data sets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCarty CA, Huggins W, Aiello AE, Bilder RM, Hariri A, Jernigan TL, et al. PhenX RISING: real world implementation and sharing of PhenX measures. BMC Med Genomics. 2014;7:16. doi: 10.1186/1755-8794-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsu C-N, Kuo C-J, Cai C, Pendergrass SA, Ritchie MD, Ambite JL. Learning phenotype mapping for integrating large genetic data. Proceedings of BioNLP 2011 Workshop; Portland, Oregon: Association for Computational Linguistics; 2011. pp. 19–27. [Google Scholar]
- 43.Stephens M. A Unified Framework for Association Analysis with Multiple Related Phenotypes. PLoS ONE. 2013;8:e65245. doi: 10.1371/journal.pone.0065245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim S, Xing EP. Statistical Estimation of Correlated Genome Associations to a Quantitative Trait Network. PLoS Genet. 2009;5:e1000587. doi: 10.1371/journal.pgen.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tyler AL, Crawford DC, Pendergrass SA. Detecting and characterizing pleiotropy: new methods for uncovering the connection between the complexity of genomic architecture and multiple phenotypes- session introduction. Pac Symp Biocomput. 2014;19:183–7. doi: 10.1142/9789814583220_0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tyler AL, Lu W, Hendrick JJ, Philip VM, Carter GW. CAPE: an R package for combined analysis of pleiotropy and epistasis. PLoS Comput Biol. 2013;9:e1003270. doi: 10.1371/journal.pcbi.1003270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.BASTIAN M, HEYMANN S, JACOMY M. Gephi: An Open Source Software for Exploring and Manipulating Networks. International AAAI Conference on Weblogs and Social Media [Internet]; 2009. Available from: http://www.aaai.org/ocs/index.php/ICWSM/09/paper/view/154. [Google Scholar]
- 48.Darabos C, White MJ, Graham BE, Leung DN, Williams SM, Moore JH. The multiscale backbone of the human phenotype network based on biological pathways. BioData Min. 2014;7:1. doi: 10.1186/1756-0381-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Darabos C, Harmon SH, Moore JH. Using the bipartite human phenotype network to reveal pleiotropy and epistasis beyond the gene. Pac Symp Biocomput. 2014:188–99. [PMC free article] [PubMed] [Google Scholar]
- 50•.Williams SM, Haines JL. Correcting Away the Hidden Heritability. Annals of Human Genetics. 2011;75:348–50. doi: 10.1111/j.1469-1809.2011.00640.x. Important considerations for GWAS multiple testing also important for PheWAS. [DOI] [PubMed] [Google Scholar]
- 51.Sobota R, Shriner D, Kodaman N, Goodloe RJ, Zheng W, Gao T, et al. Addressing Population-Specific Multiple Testing Burdens in Genetic Association Studies. Annals of Human Genetics. doi: 10.1111/ahg.12095. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]