Abstract
Inhibition of γ-secretase cleavage of the amyloid precursor protein (APP) is a prime target for the development of therapeutics for treating Alzheimer’s disease; however, complete inhibition of this activity would also impair the processing of many other proteins, including the APP homologues, amyloid precursor-like protein (APLP) 1 and 2. To prevent unwanted side effects, therapeutically useful γ-secretase inhibitors should specifically target APP processing while sparing cleavage of other γ-substrates. Thus, since APLP1 and APLP2 are more similar to APP than any of the other known γ-secretase substrates and have important physiological roles in their own right, we reasoned that comparison of the effect of γ-secretase inhibitors on APLP processing should provide a sensitive indicator of the selectivity of putative inhibitors. To address this issue, we have optimized microsome and cell culture assays to monitor the γ-secretase proteolysis of APP and APLPs. Production of the γ-secretase-generated intracellular domain (ICD) occurs more rapidly from APLP1 than from either APLP2 or APP, suggesting that APLP1 is a better γ-substrate and that substrate recognition is not restricted to the highly conserved amino acid sequences surrounding the ε-site. As expected, the well-characterized γ-secretase modulator, fenofibrate, did not inhibit ICD release, whereas a related compound, FT-9, inhibited γ-secretase both in microsomes and in whole cells. Importantly, FT-9 displayed a preferential effect, inhibiting cleavage of APP much more effectively than cleavage of APLP1. These findings suggest that selective inhibitors can be developed and that screening of compounds against APP and APLPs should assist in this process.
The molecular pathways leading to dementia of the Alzheimer type are not well understood, but substantial data indicate that the amyloid β-protein (Aβ)1 plays a central role in Alzheimer’s disease (AD) pathogenesis (1). Aβ is produced by proteolytic processing of the amyloid precursor protein (APP) by the action of two aspartyl proteases, termed β -amyloid cleaving enzyme (BACE1 or β-secretase) (2, 3) and γ-secretase (4). In addition, APP is also cleaved by an activity termed α-secretase (5, 6). Cleavage by α-secretase and cleavage by BACE1 appear to be mutually exclusive (7), and proteolysis by either is a prerequisite for γ-cleavage (8). BACE1 acts at two sites, producing 99- or 89-amino acid C-terminal fragments (CTF) (2), and α-cleavage creates an 83-residue CTF (6). All three CTFs serve as substrates for γ-secretase, a unique protease composed of at least four transmembrane proteins [presenilin, nicastrin, anterior pharynx-defective 1 (Aph-1), and presenilin enhancer 2 (Pen-2)] that is capable of cleaving within protein domains buried deep in the hydrophobic environment of the membrane (9,10). γ-Processing of APP occurs in a stepwise fashion (11), with the first cleavage (ε-cleavage) releasing the 49–50-residue APP intracellular C-terminal domain (AICD) (12–14). The second cleavage occurs six residues C-terminal of AβVal40 and is termed the ζ-site (15,16). The final cut occurs at the γ-site and gives rise to Aβ or p3, the most common forms of which are Aβ40 and p340. The consequence of this series of γ-mediated reactions is the equimolar production of Aβ and APP ICD (17).
Therapeutic inhibition of either BACE1 or γ-secretase should prove useful for the treatment of AD, and these are areas of intense research. However, developing effective inhibitors presents a serious challenge (18–20). In the case of therapeutic targeting of γ-secretase, perhaps the biggest obstacle is the fact that γ-secretase is known to process at least 40 other substrates (21, 22), many of which mediate important physiological functions. γ-Secretase activity is essential both for proper development and during adulthood; complete ablation of γ-secretase activity by either chemical or genetic manipulation is to be avoided at all costs, since it would cause a blockade of the Notch signaling pathway which would in turn lead to numerous potentially lethal toxicities, including immunological dysfunctions and gut dyshomeostasis (23, 24). Interestingly, some nonsteroidal anti-inflammatory drugs (NSAIDs) and related compounds, collectively termed γ-secretase modulators (GSMs), have the ability to shift the cleavage specificity of γ-secretase, either increasing (25) or decreasing (26–28) the level of production of the disease-associated Aβ42 without altering cleavage of Notch (29) or ErbB-4 (27). However, how various GSMs affect the processing of other substrates has not been established and could have potential liabilities, possibly interfering with important physiological pathways (30).
Of the known γ-substrates, amyloid precursor like protein 1 (APLP1) and 2 (APLP2) share the greatest degree of structural similarity with APP and undergo proteolytic processing that results in the liberation of ICDs, p3-like, and Aβ-like peptides (13, 31–34). Thus, we reasoned that compounds capable of discriminating between APP and APLP1 or APLP2 should also be capable of discriminating between APP and other less related γ-substrates. Moreover, genetic ablation studies have revealed that APP and APLP2 have distinct physiological functions and that APLP2 has the key physiological role among the APP family of proteins (35–37). While it remains uncertain that the ICDs of APLP1 or −2 play an important transcriptional role (31, 38, 39), prudence dictates that therapeutic inhibition of γ-secretase should specifically target APP processing while sparing cleavage of APLPs. Using cell culture and microsomal assays to monitor processing of APP and APLPs, we have found that the γ-secretase cleavage of APP and APLPs appears sufficiently different to find molecules that can selectively block APP processing. Specifically, APLP1 is a better substrate for γ-secretase than either APLP2 or APP, and the γ-secretase cleavage of APLP1 is less sensitive to inhibition by an NSAID-like γ-secretase inhibitor (GSI), FT-9, than the γ-secretase cleavage of either APP or APLP2. Identification of a compound that inhibits γ-secretase processing of APP but which has little effect on APLP1, coupled with our finding that the kinetics of γ-secretase cleavage are different for each of the APP family members, suggests that it should be possible to develop therapeutic inhibitors that specifically target γ-secretase cleavage of APP while sparing processing of other substrates.
EXPERIMENTAL PROCEDURES
Reagents
Restriction enzymes were from New England Biolabs (Ipswich, MA). (2S)-2-{[(3,5-Difluorophenyl)acetyl]amino}-N-[(3S)-1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-1,4-benzodia-zepin-3-yl]propanamide (compound E) and N-[N-(3,5-difluoro-phenacetyl)-l-alanyl]-S-phenylglycine tert-butyl ester (DAPT) were gifts of M. Wolfe (Center for Neurologic Diseases), and 2-[3-(3,5-dichlorophenoxy)phenyl]propanoic acid (FT-9) was synthesized as described previously (40). Both propan-2-yl 2-[4-(4-chlorobenzoyl)phenoxy]-2-ethylpropanoate (fenofibrate) and 2-(3-fluoro-4-phenylphenyl)propanoate (R-flurbiprofen) were from Sigma-Aldrich (Sigma-Aldrich Ireland Ltd., Dublin, Ireland). All other chemicals were from Sigma-Aldrich (Sigma-Aldrich Ireland Ltd.), and tissue culture reagents were from Gibco (Invitrogen, Carlsbad, CA).
Constructs and Transfections
The coding sequences of full-length wild-type human APP695, APLP1650, and APLP2751 were PCR-amplified from existing constructs (41) using primers designed to replace the stop codon with an AgeI restriction site: APP F, gTACAAgCTTgCCATgCTgCCCggTTTggCACTgC-TCC; APP R, gATCACCggTgTTCTgCATCTgCTCAAAgAAC; APLP1 F, gTAAAAgCTTgCCATggggCCCgCCAgCCCCgCT-gCT; APLP1 R, gATCACCggTgggTCgTTCCTCCAggAAgCg; APLP2 F, gTAAggATCCgCCATggCggCCACCgggACCgCg; APLP2 R, gATCACCggTAATCTgCATCTgCTCCAgg. PCR products were cloned into the pcDNA4/myc-His vector (Invitrogen) between HindIII and AgeI sites (for APP and APLP1) or BamHI and AgeI sites (for APLP2), in frame with a sequence encoding a six-His C-terminal tag present in the plasmid, and the sequences of all constructs were verified. The constructs were transfected into Chinese hamster ovary (CHO) cells using Lipofectamine 2000 (Invitrogen), and monoclonal cell lines stably expressing the gene of interest were produced by limiting dilution into zeocin-containing medium. For each gene, at least 13 different clones were tested for expression of the His-tagged protein, and those with the closest matching expression of full-length proteins were used in subsequent experiments.
Cultured Cells
CHO cells stably overexpressing His-tagged APP (D4G), APLP1 (D7G), and APLP2 (D7F) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), 6 mM glutamine, 100 units/mL penicillin, 100 µg/mL streptomycin, 10 µg/mL l-proline, and 250 µg/mL zeocin (Invitrogen).
ICD in Vitro Generation (ICDivg) Assay
Cells were grown to confluence in 10 cm dishes (Sarsted, Niimbrecht, Germany), such that one dish would provide enough microsomal membranes for one experimental condition. Confluent cells were washed twice with 5 mL of ice-cold phosphate-buffered saline (PBS), scraped into 600 µL of hypotonic lysis buffer [10 mM MOPS (pH 7) containing 10 mM KC1 5 mM EDTA, 1 mM EGTA, 120 µg/mL Pefabloc, and 2 mM 1,10-phenanthroline] and homogenized on ice with 30 passes of a Dounce homogenizer at 6000 rpm. The resultant suspension was centrifuged at 1000g and 4 °C for 15 min to remove nuclei and cell debris, and microsomes were isolated from the postnuclear supernatant by centrifugation at 16000g and 4 °C for 40 min. The microsomal pellet was resuspended in 40 µL of assay buffer [150 mM sodium citrate (pH 6.8) containing 5 mM EDTA, 1 mM EGTA, 2 mM 1,10-phenanthroline, and 250 µg/mL human recombinant insulin (41–43)] containing test compound in dimethyl sulfoxide (DMSO) or vehicle alone. In all cases, the concentration of DMSO was kept constant at 1 % (v/v). Samples were incubated in a water bath at 37 °C for 0–160 min, after which they were placed at −20 °C to stop the reaction. Subsequently, samples were thawed on ice and centrifuged at 150000g and 4 °C for 75 min in an Optima centrifuge using a TLA55 rotor (Beckman Coulter, Fullerton, CA). The upper 35 µL of the supernate was collected, snap-frozen in liquid nitrogen, and reduced to dryness in an Eppendorf concentrator (Eppendorf, Hamburg, Germany) at room temperature for 1 h. Each concentrate was then reconstituted in 14 µL of 1× tris-tricine sample buffer [62.5 mM Tris-HCl (pH 6.8), 10% glycerol, and 2% SDS] and boiled at 100 °C for 10 min. To maximize the detection of ICDs produced at early time points and to minimize pipetting errors, the entire sample was loaded in a single well of a precast Novex 10 to 20% polyacrylamide tris-tricine gel, and samples were electrophoresed and transferred as described below.
Compound Treatments and Whole Cell Lysates
Cells were seeded in six-well plates at a density of 600000 cells/well (Greiner Bio-One, Frickenhausen, Germany), grown to confluence, and then cultured overnight (~14 h) in the presence of test compound or vehicle. The following day, cells were washed twice with 2 mL of ice-cold PBS per well and lysed using 100 µL of lysis buffer [50 mM Tris base (pH 7.6), 150 mM NaCl, 2 mM EDTA, and 2% NP-40, containing 5 µg/mL leupeptin, 5 µg/ mL aprotinin, 2 µg/mL pepstatin, 120 µg/mL Pefabloc, and 2mM 1,10-phenanthroline] per well. Protein concentrations were measured using a BCA protein assay kit (Pierce, Rockford, IL), and equal amounts of proteins were used for immunoblotting.
Western Blot Analysis
ICDivg samples were electrophoresed on precast No vex 10 to 20% polyacrylamide tris-tricine gels (Invitrogen), while cell lysates were analyzed on precast Nu-PAGE 4 to 12% polyacrylamide bis-tris gels using MES running buffer (Invitrogen) or 10% polyacrylamide tris-glycine gels (44). Proteins were transferred onto nitrocellulose (0.2 µm pore size, Sigma-Aldrich Ireland Ltd.) at a constant rate of 400 mA and 4 °C for 2 h. Membranes were then blocked for 1 h at room temperature with 5% skim milk (Fluka, Sigma-Aldrich Ireland Ltd.) in Tris-buffered saline containing 0.05% Tween 20 (TBST), washed twice for 10 min with TBST to remove traces of blocker, and incubated overnight with primary antibodies diluted in TBST containing 5% skim milk or 3% bovine serum albumin. After incubation with primary antibodies, blots were washed four times for 15 min with TBST, incubated with appropriate HRP-linked secondary antibodies (Amersham, GE Healthcare, Chalfont St. Giles, U.K.) for 1 h at room temperature, washed as described above, and visualized using an ECL kit (Pierce) and Hyperfilm MP (Amersham, GE Healthcare).
Antibodies
Novel rabbit polyclonal antibodies W1NT, W1CT, and W2CT were raised to synthetic peptides encoding regions of the APLP1 ectodomain (EPDPQRSRRCLRDPQR) and C-terminal domain (NPTYRFLEERP) and the APLP2 C-terminal domain (NPTYKYLEQMQI), each of which was conjugated to keyhole limpet hemocyanin via an N-terminal cysteine. The specificity of these antibodies was confirmed using cells overexpressing APLP1 or APLP2 and brain homogenates from mice null for APLP1 or APLP2 (not shown). Monoclonal antibody 22C11 (Chemicon, Millipore, Billerica, MA), which recognizes the N-terminus of APP, and the polyclonal antiserum C8, which recognizes the C-terminus of APP (a gift of D. Selkoe, Center for Neurologic Diseases), and D2-II (hereby termed 2NT), which recognizes the ectodomain of APLP2 (Calbiochem, EMD Chemicals Inc., Gibbstown, NJ), have been described previously (45–47). Monoclonal antibody TetraHis was from Qiagen (Crawley, U.K.). Anti-nicastrin (NCT 164) and anti-presenilin 1 CTF (MAB 5232) antibodies were a kind gift of P. Fraering (École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland).
Data Analysis
Band intensities were quantified using Scion Image (Scion Corp., Frederick, MD), and data were analyzed using two-way ANOVA (GraphPad Prism, GraphPad Software, La Jolla, CA).
RESULTS
Selection and Characterization of Cells Stably Expressing APP, APLP1, and APLP2
Since the primary sequences of CTFs, the actual substrates of γ-secretase, have not yet been unambiguously described for APLP1 and APLP2, we have prepared cell lines stably expressing full-length human versions of the respective proteins, so that CTFs could be produced by endogenous α- and β-secretase activities. To be able to use a single antibody for the simultaneous detection of each protein, constructs expressing a C-terminal six-His tag were employed, thus obviating differences arising due to the use of three different antibodies with different avidities and affinities. A total of 13 APP-His, 18 APLPl-His, and 33 APLP2-His clones were isolated and analyzed. Lysates of each of the 64 clones were tested for expression of exogenous protein, and the three clones showing the closest matching levels of full-length protein (clones D4G, D7G, and D7F) were used in subsequent experiments (Figure 1 of the Supporting Information).
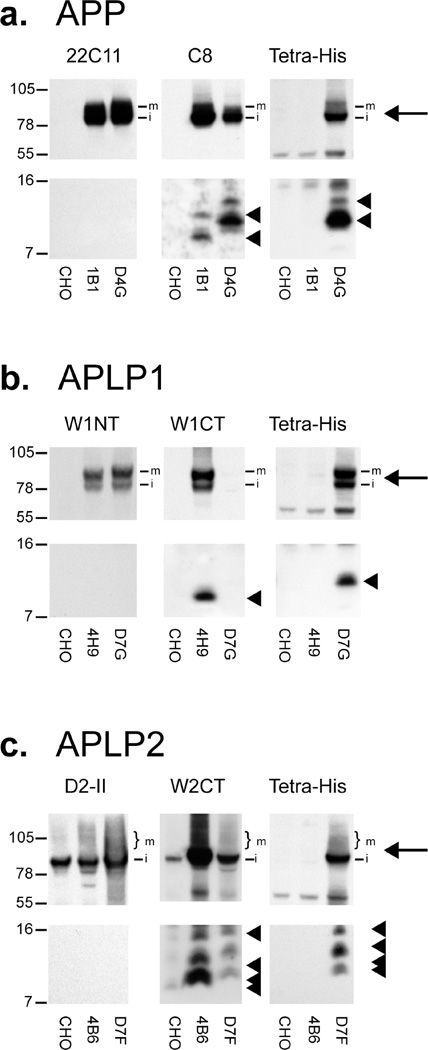
In cells expressing untagged APP, bands migrating at ~ 100 and ~93 kDa, which correspond to the mature and immature forms of APP, respectively, were detected using 22C11 and C8, but not TetraHis (Figure 1a, top panels). As expected, cells expressing His-tagged APP produced bands of ~ 100 and ~93 kDa that were detected by all three antibodies. Interestingly, His-tagged APP is less well detected by C8 than untagged APP, probably resulting from partial disruption of the C8 epitope because of its proximity to the six-His tag. However, detection of APP with 22C11, which recognizes an epitope in the N-terminal portion of the protein and thus is unaltered by the six-His tag, revealed that APP expression is similar between the two cell lines (Figure 1a). Both untagged and tagged APP generated two major CTF bands (Figure 1a, bottom panels). The untagged CTFs had calculated molecular masses of ~11.5 and ~9.1 kDa, while the corresponding six-His-tagged CTFs had slightly higher calculated molecular masses of ~ 13.2 and ~10.9 kDa, respectively (Figure 1a, bottom panels). This difference is most probably due to a combination of additional mass (0.93 kDa) and positive charge.
Figure 1.
C-Terminal six-His-tagged APP, APLP1, and APLP2 are expressed and processed in a manner highly similar to that of untagged proteins. Lysates from selected clones of cell lines expressing APP-His, APLP1-His, and APLP2-His (clones D4G, D7G, and D7F, respectively) were compared with lysates of cell lines expressing untagged versions of the same proteins (clones 1B1,4H9, and 4B6 for APP, APLP1, and APLP2, respectively) and with a lysate of naive CHO cells. Lysates were analyzed with a panel of antibodies recognizing either an epitope lying in the N-terminal ectodomain (22C11 for the APP N-terminus, W1NT for the APLP1 N-terminus, and D2-II for the APLP2 N-terminus), an epitope located at the very C-terminus (C8 for the APP C-terminus, W1CT for the APLP1 C-terminus, and W2CT for the APLP2 C-terminus), or an antibody specific for the six-His C-terminal tag (Tetra-His). (a–c) Full-length proteins (top panels) are indicated by arrows (m, mature form; i, immature form), while CTFs (bottom panels) are indicated by arrowheads.
The pattern for tagged and untagged full-length APLP1 and APLP2 was similar to that seen with APP (Figure 1b,c, top panels). Cells expressing APLP1 and APLP1-His produced bands of ~90 and ~83 kDa, each of which was detected by W1NT; the tagged protein was detected by TetraHis but not W1CT, and the untagged protein was detected by W1CT but not TetraHis (Figure 1b, top panels). Again, tagged APLP1 cannot be detected with W1CT probably because of disruption of the W1CT epitope, which lies at the very C-terminus of APLP1, close to the six-His sequence. However, APLP1 detection with W1NT indicated that the levels of tagged and untagged full-length APLP1 were similar (Figure 1b). In cells expressing APLP2, a prominent band migrating at ~93.2 kDa was detected with 2NT, and as expected, the His-tagged protein was also detected by W2CT and TetraHis whereas the untagged protein was not detected by TetraHis (Figure 1c, top panels). For both tagged and untagged APLP2, a specific smear likely indicative of various post-translational modifications (47) was detected. As was the case for APP-His and APLPl-His, APLP2-His was also less well detected than its untagged counterpart when a C-terminal antibody was used, and use of an antibody directed to the N-terminal ectodomain provided evidence that the levels of full-length APLP2 and full-length APLP2-His were similar. The electrophoretic migration of full-length APP family proteins on tris-tricine and tris-glycine polyacrylamide gels differed significantly with molecular masses estimated from tris-glycine gels (~108 and ~100 kDa for APP, ~92 and ~82 kDa for APLP1, and ~99 kDa for APLP2) higher than those determined using tris-tricine gels (Figure 1 of the Supporting Information) and more in keeping with those reported previously (41).
Both untagged and tagged APLP1 generated a single CTF (Figure 1b, bottom panels), migrating at ~9.5 and ~11.2 kDa, respectively. The untagged CTF could be detected only by using the W1CT antibody, while the tagged CTF was detected only by using the TetraHis antibody. Processing of untagged and tagged APLP2 was also similar with four major CTF bands detected in each (Figure 1c, bottom panels). Untagged APLP2 CTFs were detected migrating at ~14.5, ~11.9, ~10.3, and ~9.6 kDa, while tagged APLP2 CTFs migrated slightly slower, producing molecular mass estimates of ~15.5, ~13.0, ~11.1, and ~ 10.6 kDa.
Production of APLP1 ICD Is Faster Than That of APLP2 ICD and APP ICD
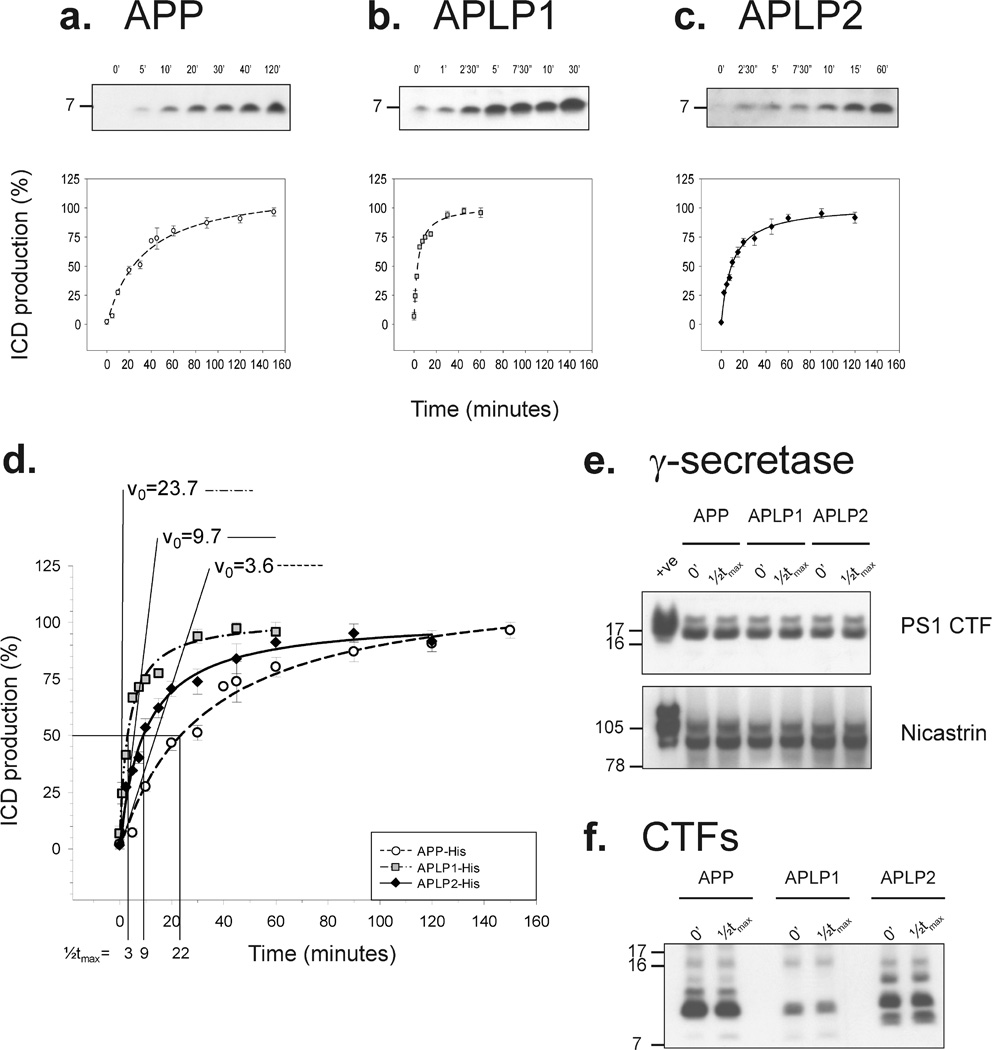
Microsomes from each cell line were incubated at 37 °C for 0–160 min, and the soluble phase was analyzed by Western blotting (Figure 2a – c). For each cell, line a single band migrating at ~7 kDa was detected, the production of which was time-dependent, with the initial reaction velocities (v0) different for each of the three proteins (Figure 2d). ICD production was faster from APLP1 (v0 = 23.7% ICD/min) than from either APLP2 (v0 = 9.7% ICD/min) or APP (v0 = 3.6% ICD/ min). Consequently, the plateau level was reached faster for APLP1 ICD production than for APLP2 or APP production. The time required to achieve half of the maximum amount of ICD produced, ½tmax, was 3 min for APLP1, 9 min for APLP2, or 22 min for APP (Figure 2d). The observed differences in the rates of ICD production were confirmed in additional experiments using other APP-His-, APLP1-His-, and APLP2-His-expressing clones (not shown). Moreover, these differences were not due to variance in the levels of the γ-secretase complex in the different cell lines, since the levels of markers of active γ-secretase [PS1 CTF and mature nicastrin (48,49)] were similar for all three cell lines (Figure 2e), and these remained constant during the course of the assay (Figure 2e). Further, the plateau in ICD production seen for all three proteins is not a result of substrate exhaustion, since the levels of CTFs remained high throughout the reaction (Figure 2f).
Figure 2.
Characterization of the ICDivg assay. Microsomes derived from APP-His-, APLPl-His-, and APLP2-His-expressing cells were incubated for different amounts of time and ICD levels measured by immunoblotting. Representative Western blots and calculated curves describing the time dependency of ICD production for each cell line are shown (a—c). Values are given as the percentage of ICD production, where 100% is set to respective mean plateau levels. Each experimental point is the mean of at least six independent experiments with duplicate samples in each experiment. APLP1 ICDs (dashed–dotted line with gray squares) are produced faster than both APLP2 ICDs (solid line with black diamonds) and APP ICDs (dashed line with open circles) (d). The initial velocity (v0) for the γ-secretase activity of the three substrates and the time required to achieve half of the maximal amount of ICD produced (½tmax) are indicated (d). Lysates of microsomes at time zero and after an incubation for the respective ½tmax were analyzed for the γ-secretase components PS1 CTF and nicastrin (e), and for the levels of the respective CTFs using TetraHis antibody (f). Neither the levels of γ-secretase nor CTFs changed significantly during the course of the reaction.
To exclude differences resulting due to the addition of the six-His tag, we also conducted pilot studies using cell lines expressing untagged human wild-type APP695 and untagged human APLP1650. These experiments revealed results highly similar to those obtained for the tagged proteins (for untagged APP, ½tmax = 20.5 min and v0 = 3.7 ICD/min; for untagged APLP1,½tmax = 3.7 min and v0 = 20.2 ICD/min) (compare Figure 2a,b,d with Figure 4a of the Supporting Information).
Direct Inhibition of γ-Secretase Affects the Production of ICDs from APP-His, APLPl-His, and APLP2-His CTFs Similarly
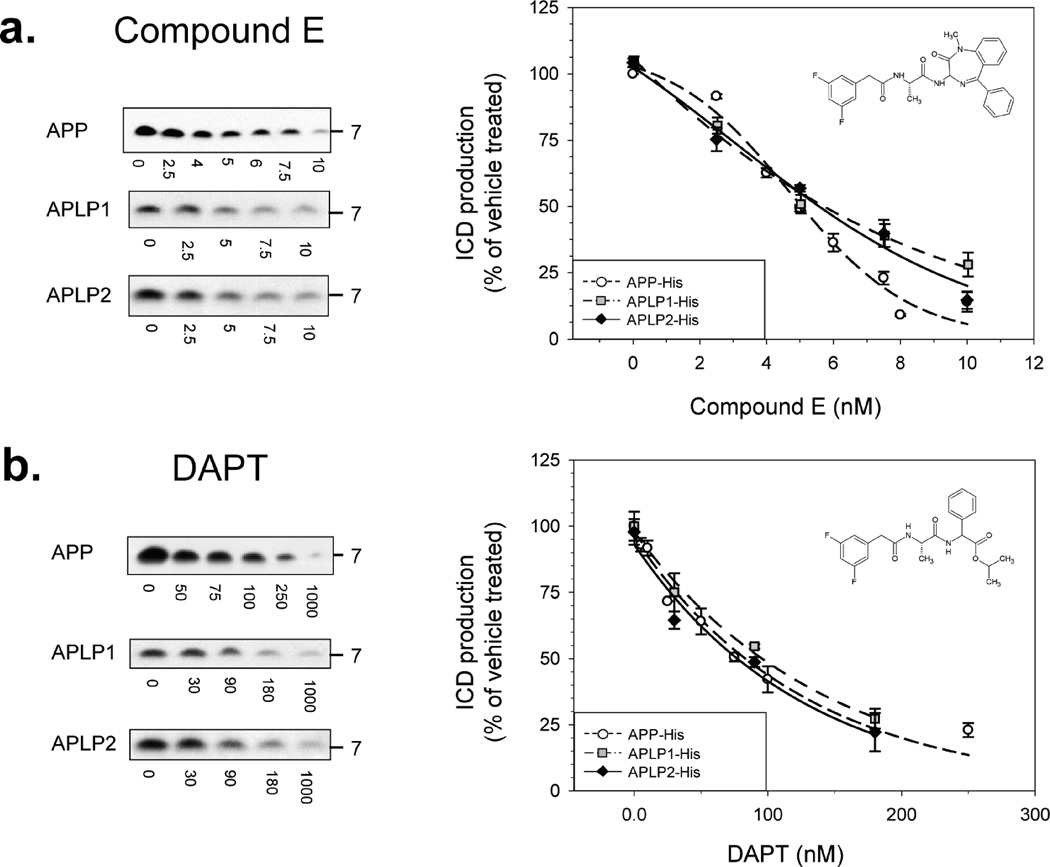
Having optimized the conditions of our assay, we next examined the effect of GSIs and GSMs on the production of APP, APLP1, and APLP2 ICDs. First, we tested two structurally and pharmacologically distinct GSIs: compound E, a potent, non-transition state, non-competitive peptidomimetic inhibitor (50, 51), and DAPT, a peptidomimetic active site-directed inhibitor (52, 53). Compound E caused a dose-dependent inhibition of ICD production, starting at low nanomolar concentrations, with the extent of inhibition similar for all the substrates considered (Figure 3a). The calculated concentrations needed to achieve 50% inhibition (IC50) of γ-secretase processing of APP, APLP1, and APLP2 were 5.0,5.3, and 5.3 nM, respectively. Moreover, given that compound E is a highly selective inhibitor, these results are further evidence that the ~7 kDa immunoreactive bands detected are genuine γ-secretase-generated ICDs. As with compound E, DAPT showed a dose-dependent inhibition of ICD production with the extent of inhibition almost identical for APP, APLP1, and APLP2. The calculated IC50 values for inhibition of γ-secretase cleavage of APP, APLP1, and APLP2 were 83.1, 95.7, and 82.7 nM, respectively (Figure 3b).
Figure 3.
Dose-dependent inhibition of γ-secretase cleavage of APP, APLP1, and APLP2. γ-Secretase processing of APP (dashed line with open circles), APLP1 (dashed–dotted line with gray squares), and APLP2 (solid line with black diamonds) are similarly inhibited by compound E (a) and DAPT (b). Microsomes of each cell line were incubated for the appropriate ½tmax with different concentrations of inhibitor, and ICD levels were measured. For each compound, the molecular structure, representative Western blots, and calculated inhibition curves are shown. Each experimental point is expressed as the mean of at least four independent experiments with duplicate samples at each dose ± the standard error.
Fenofibrate Does Not Inhibit γ-Secretase Cleavage of APP-His, APLP1-His, and APLP2-His
NSAIDs have been reported to act as γ-secretase modulators both in vitro and in vivo (25, 26, 29). In particular, some NSAIDs have been shown to increase the level of Aβ42 while others were reported to decrease Aβ42 levels; in both cases, the total Aβ was virtually unchanged (26,54). We decided to test the effect of two NSAIDs: one that is known to decrease the level of Aβ42 production and the other that is known to increase the level of Aβ42 production.
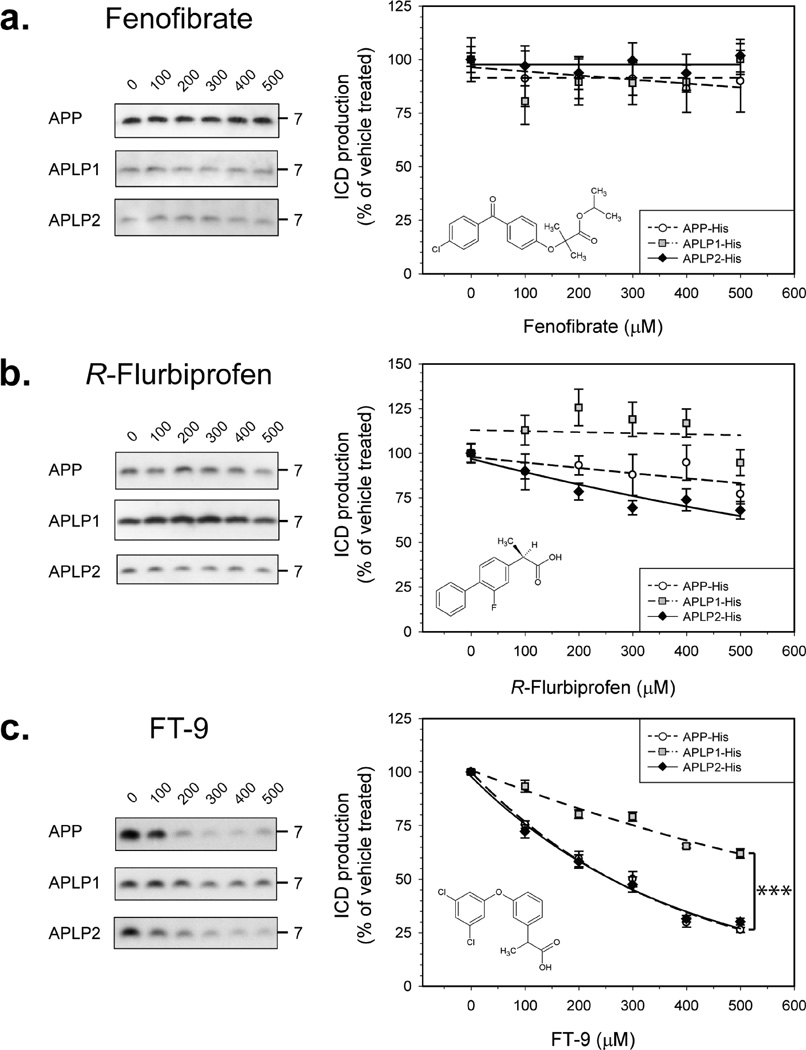
Fenofibrate is a GSM that increases the level of production of Aβ42 and correspondingly reduces the level of production of Aβ38, while leaving the levels of Aβ40 almost unchanged (25). Even at very high concentrations (500 µM), fenofibrate did not inhibit γ-secretase, and ICD production from APP, APLP1, and APLP2 was unaffected by this compound (Figure 4a). R-Flurbiprofen exerts γ-secretase modulatory effects opposite from those of fenofibrate, reducing Aβ42 levels while causing a corresponding increase in Aβ38 levels (54). Concentrations of R-flurbiprofen of ≥ 200 µM caused a significant decrease in the level of γ-cleavage of APLP2, and at very high concentrations, R-flurbiprofen weakly inhibited γ-secretase processing of APP but γ-secretase cleavage of APLP1 was unaffected by R-flurbiprofen (Figure 4b).
Figure 4.
Aβ42-lowering GSMs differentially inhibit γ-secretase cleavage of some substrates. Microsomes of each cell line were incubated for the appropriate ½tmax, with different concentrations of test compound, and ICD levels were measured. For each compound, the molecular structure, representative Western blots, and calculated inhibition curves are shown. Each experimental point is expressed as the mean of at least four independent experiments with duplicate samples at each dose ± the standard error. All compounds were tested in the range between 100 and 500 µM; higher concentrations were not possible because of solubility issues. Fenofibrate did not inhibit ICD production in the concentration range examined (a). R-Flurbiprofen showed a weak inhibitory effect on APP and APLP2 but did not alter APLP1 ICD production (b). FT-9 acts as a γ-secretase inhibitor, causing similar dose-dependent inhibition of the γ-secretase cleavage of APP and APLP2, but has a less potent effect on γ-secretase processing of APLP1 (c). Key: dashed line with open circles for APP, dashed–dotted line with gray squares for APLP1, and solid line with black diamonds for APLP2.
A Compound Structurally Related to Fenofibrate, FT-9, Is a Partially Selective γ-Secretase Inhibitor in both Microsomes and Whole Cells
Encouraged by the small but significant differential effects of R-flurbiprofen, we investigated the effects of another Aβ42-reducing GSM, FT-9 (40). At concentrations of ≥ 100 µM, FT-9 behaves as a GSI, decreasing ICD production of all three ICDs (Figure 4c). Strikingly, the inhibitory effect was much stronger for APP and APLP2 than for APLP1. At the highest concentration tested (500 µM), FT-9 caused a 70–75% reduction of both APP and APLP2 ICDs, while production of APLP1 ICDs was only reduced by ~38% [P < 0.0001 (Figure 4c)]. The IC50 values against APP ICD and APLP2 ICD were ~260 µM, whereas the estimated IC50 against APLP1 ICD was almost 3 times greater. To exclude possible artifacts arising from the use of a single clonal line, we tested the effects of FT-9 on other APP-, APLP1-, and APLP2-expressing cells, and in these experiments, we again observed the same effect: FT-9 caused a more robust inhibition of APP and APLP2 cleavage than APLP1 cleavage. Similarly, when we examined the effect of FT-9 on microsomes from cells expressing untagged APP or untagged APLP1, we again found that FT-9 more potently inhibited γ-secretase processing of APP (Figure 4b of the Supporting Information).
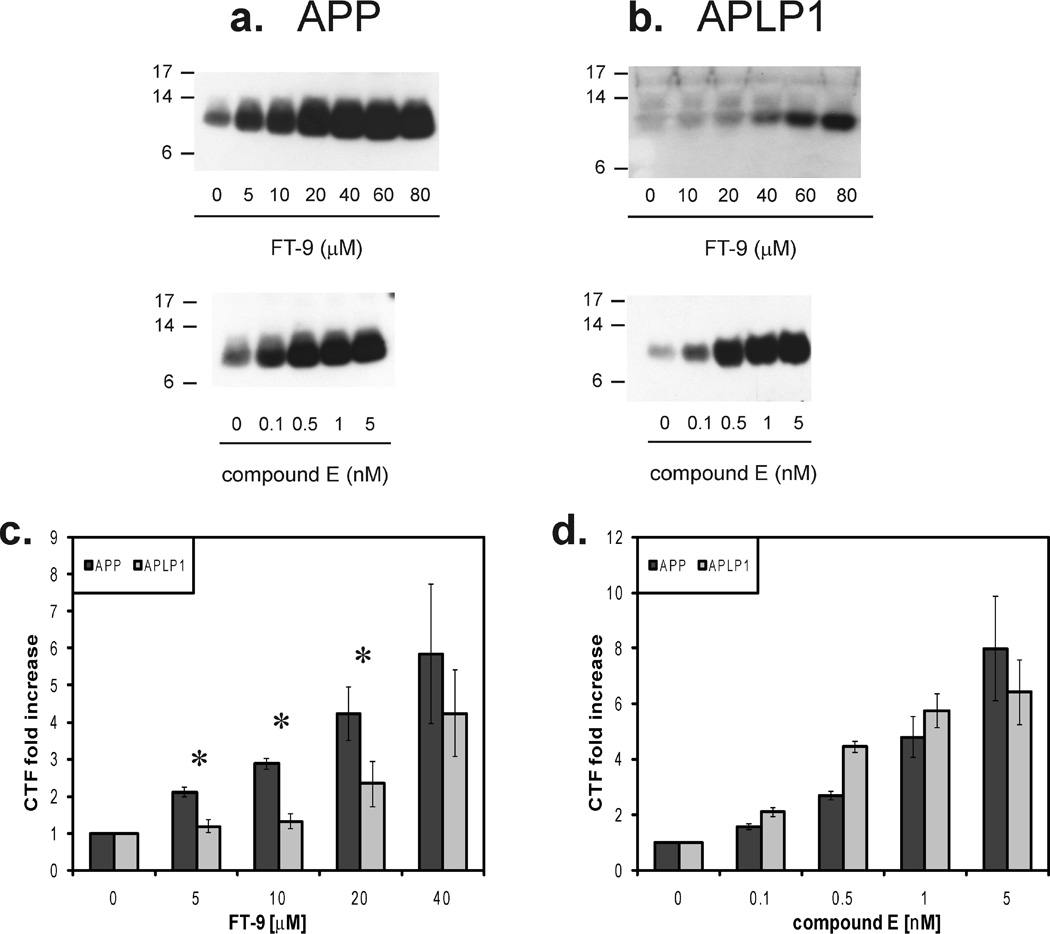
Following this, we tested the effect of FT-9 on intact cells, comparing it to the effect of the nonselective GSI, compound E (Figure 3a). Cells expressing APP-His and APLP1-His were grown to confluence and treated overnight (14 h) with different concentrations of FT-9 or compound E. Consistent with results seen using the ICDivg assay, both FT-9 (Figure 5a,b, top panels) and compound E (Figure 5a,b, bottom panels) induced a dose-dependent buildup of CTFs of both proteins. Interestingly, FT-9 caused accumulation of APP CTFs at concentrations as low as 5 µM, while accumulation of APLP1 CTFs was only significant at ≥ 20 µM (Figure 5a,b, top panels, and Figure 5c). Accordingly, the concentration of FT-9 required to cause a 2-fold increase in APP CTFs, 3.4 ± 0.4 µM, was significantly lower than that for APLP1 (25.7 ± 8.9 µM) (P = 0.001). On the other hand, compound E similarly inhibited processing of both substrates at each dose tested, and the concentration needed for a 2-fold increase in the magnitude of the CTF band signal was not significantly different, being 0.27 ± 0.01 nM for APP versus 0.24 ± 0.22 nM for APLP1 (Figure 5a,b, bottom panels, and Figure 5d). As with the microsome assay, the different effect of FT-9 was also confirmed in other clones expressing APP-His and APLP1-His (Figure 2 of the Supporting Information).
Figure 5.
FT-9 preferentially inhibits γ-processing of APP in intact cells. Cells were treated overnight with subtoxic concentrations of FT-9 or compound E, and lysates were analyzed by Western blotting with TetraHis antibody (a and b). A dose-dependent accumulation of CTFs can be seen in the two cell lines for both compounds (a and b). Quantitation of the CTFs levels demonstrates that FT-9 preferentially inhibits γ-secretase processing of APP (c), whereas compound E has no preferential effect (d). Results are presented as the means of at least three measurements ± the standard error, with the fold increase in CTFs levels determined by comparison of the CTFs detected in the vehicle control. An asterisk indicates a significant difference in the fold of increased APP and APLP1 CTFs (P ≥ 0.05).
DISCUSSION
Numerous epidemiologic studies have shown that chronic treatment with NSAIDs is associated with a decreased risk of developing AD (55,56) and that some NSAIDs can modulate and/ or inhibit γ-secretase cleavage of APP (25,26, 29). Consequently, this has led to speculation that NSAIDs may modify development of AD by decreasing the level of production of Aβ42 (30). However, initial clinical trials of the GSM, R-flurbiprofen, have proved to be disappointing (57). Although the reason for the failure of R-flurbiprofen is probably linked to the poor brain penetrance of this compound (57), optimization of NSAID-like GSMs/GSIs is essential prior to further testing in humans.
Since genetic or chemical ablation of γ-secretase in mice results in thymocyte and gastrointestinal toxicities (23, 24) associated with defective cell differentiation, most studies on the potential side effects of GSIs and GSMs have focused on Notch, yet it is possible that certain compounds may cause secondary toxicity by altering the processing of other γ-substrates. Given that the number of γ-substrates identified now stands in excess of 40 (21,22,58,59) and that it is likely that still more substrates will be discovered, it is impractical to screen compounds for their effects on all γ-substrates. Thus, a pragmatic approach would be to perform an initial specificity screen comparing modulation of γ-processing of APP versus those substrates most similar to APP. The three members of the APP family of proteins (APP, APLP1, and APLP2) are processed by the same set of proteases (32, 33, 60–63), share a high degree of structural similarity, and exhibit comparable regional and cellular distributions in the brain (64). Moreover, the highly similar transmembrane domains and protein sequences surrounding the ε-cleavage site (Figure 3 of the Supporting Information), allied with the finding that all three proteins have similar brain interactomes (65) and are capable of forming heterodimers (66), indicate that analysis of APLPs should provide a useful first-round screen to determine the specificity of GSIs/GSMs for APP.
Knockout studies indicate that both APLP1 and APLP2 have important physiological functions (36, 37, 67), and several reports have suggested that γ-secretase-produced APLP1 ICD and APLP2 ICD are involved in transcriptional regulation (31, 68). In addition, a series of studies have demonstrated that APLP1 and APLP2 can compensate for the loss of APP (36,69). Thus, it would be clearly beneficial to maintain normal APLP expression in the face of pharmacological manipulation of APP to ameliorate any negative effects resulting from changes in APP processing.
γ-Secretase cleavage of APP involves successive proteolysis at three major sites designated ε, ζ, and γ (2,4,15). In this study, we investigated the effect of known GSIs and GSMs on the initial γ-secretase cleavage that occurs at the ε-site and results in the liberation of APP ICD. Thus, our focus was on the ε-site which shows 100% similarity among APP, APLP1, and APLP2 (Figure 3 of the Supporting Information) and not on the GxxxG motif believed to regulate homo- and heterodimerization of APP (66, 70, 71) and binding of GSMs (40). Using a microsomal ICDivg assay, we found that the kinetics of γ-secretase cleavage are different for each APP family member, with APLP1 ICDs produced significantly faster than either APLP2 ICD or APP ICD. These differences in ICD production rates could not be explained by the variance in the γ-secretase levels in the different cell lines used and therefore appear to reflect innate differences in substrate specificity. In this regard, it is interesting to note that the sequences surrounding the ε-cleavage site are highly similar for all three APP family members (Figure 3 of the Supporting Information), thus suggesting that other substrate recognition parameters [e.g., differences in cellular localization (66)] must also influence cleavage at the ε-site. Our ability to discern differences in the preference of γ-secretase for substrate was only evident from time course analysis, whereas in experiments employing a fixed 2 h incubation period, it is not possible to see such effects (40, 41). Therefore, in studies of GSIs and GSMs, we used incubation times when ICD production was still linear, i.e.,½tmax.
As expected, the active site-directed GSIs, compound E and DAPT, strongly inhibited γ-secretase processing of all three proteins, yielding virtually identical dose-dependent inhibition profiles. These results are in keeping with prior reports that these GSIs similarly inhibited γ-processing of APP, Erb4, and Notch (72). Indeed, given that these GSIs are known to directly target the active site of γ-secretase, one would expect near-identical inhibition of all γ-substrates.
In contrast, both fenofibrate and R-flurbiprofen have been shown to modulate γ-secretase cleavage of APP by binding to a region within residues 28–36 of the Aβ domain and directly altering cleavage at a site that is just four to six residues from the putative GSM binding site (40). Since the ICDivg assay employed in this study measures cleavage at the ε-site, some 13–15 residues C-terminal to the γ-site, it was not surprising that these compounds had little or no effect on ICD release. Indeed, even at 500 µM fenofibrate did not alter ICD release. However, an earlier report had found that fenofibrate could inhibit APP ICD release (25); however, that study used microsomes from cells that expressed APP bearing the Swedish and London familial AD mutations, and other studies have indicated that familial AD mutations in the transmembrane domain of APP alter γ-secretase cleavage specificity and how GSMs interact with APP (29, 70) and that such effects are not relevant to wild-type APP. The results for R-flurbiprofen were in contrast to those obtained with fenofibrate, with a small but significant inhibition of both APP and APLP2 processing at ≥400 µM R-flurbiprofen.
This modest differential effect encouraged us to test another Aβ42-lowering compound, FT-9 (40). At concentrations of ≥ 100 µM, FT-9 caused a dose-dependent inhibition of APP and APLP2 ICD release. APLP1 ICD production was also inhibited, but the extent of inhibition was much lower than for either APP or APLP2 ICDs. Thus, under the conditions tested, FT-9 behaves as a GSI that can differentially inhibit APP and APLP1 γ-secretase cleavage. To confirm the veracity of these findings in a more physiological setting, we used cultured cells that express APP or APLP1 and conducted dose-dependent experiments using subtoxic concentrations of FT-9, with compound E as a non-selective GSI control. As expected for GSIs, compound E and FT-9 caused a dose-dependent accumulation of APP and APLP1 CTFs. However, while the buildup of CTFs caused by compound E was similar for APP and APLP1 at each dose tested, FT-9 displayed a more differential inhibitory effect. Specifically, as little as 5 µM FT9 caused a significant increase in APP CTFs, whereas concentrations of ≥20 µM were required to induce significant accumulation of APLP1 CTF.
Thus, as in the ICDivg assay, FT-9 preferentially inhibited γ-secretase cleavage of APP over APLP1 in living cells, establishing the precedent that a GSI can preferentially target APP processing while sparing other γ-substrates. Although this effect did not extend to APLP2, the γ-substrate most similar to APP, these findings offer hope that selective, therapeutically useful GSIs can be developed. Consequently, we suggest that screening for selective inhibitors should begin with a first-round comparison of effects on APP and APLPs, and those identified to be specific tested for their effect on other γ-secretase substrates whose loss is known to cause overt toxicity.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Michael Wolfe for the gift of compound E and DAPT and Dr. Pamela Osenkowski for useful discussions and technical help. We also thank Drs. Dennis Selkoe and Patrick Fraering for generously providing antibodies C8, and NCT 164 and MAB 5232, respectively.
Footnotes
This work was supported by a grant from the Foundation for Neurologic Diseases (D.M.W.) and by National Institutes of Health Grants AG29886 and AG20206 (T.E.G.).
Abbreviations: AD, Alzheimer’s disease; Aβ, amyloid β-protein: APP, amyloid precursor protein; APLP1, amyloid precursor-like protein 1; APLP2, amyloid precursor-like protein 2; CHO, Chinese hamster ovary; CTF, C-terminal fragment; DMSO, dimethyl sulfoxide; GSI, γ-secretase inhibitor; GSM, γ-secretase modulator; ICD, intracellular domain; ICDivg, ICD in vitro generation assay; PS, presenilin; NSAID, nonsteroidal anti-inflammatory drug.
SUPPORTING INFORMATION AVAILABLE
Supplemental figures. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Hardy J, Allsop D. Amyloid deposition as the central event in the aetiology of Alzheimer’s disease. Trends Pharmacol. Sci. 1991;12:383–388. doi: 10.1016/0165-6147(91)90609-v. [DOI] [PubMed] [Google Scholar]
- 2.Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC, Collins F, Treanor J, Rogers G, Citron M. β-Secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 3.Sinha S, Anderson JP, Barbour R, Basi GS, Caccavello R, Davis D, Doan M, Dovey HF, Frigon N, Hong J, Jacobson-Croak K, Jewett N, Keim P, Knops J, Lieberburg I, Power M, Tan H, Tatsuno G, Tung J, Schenk D, Seubert P, Suomensaari SM, Wang S, Walker D, Zhao J, McConlogue L, John V. Purification and cloning of amyloid precursor protein β-secretase from human brain. Nature. 1999;402:537–540. doi: 10.1038/990114. [DOI] [PubMed] [Google Scholar]
- 4.Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and γ-secretase activity. Nature. 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 5.Sisodia SS, Koo EH, Beyreuther K, Unterbeck A, Price DL. Evidence that β-amyloid protein in Alzheimer’s disease is not derived by normal processing. Science. 1990;248:492–495. doi: 10.1126/science.1691865. [DOI] [PubMed] [Google Scholar]
- 6.Sisodia SS. β-Amyloid precursor protein cleavage by a membrane-bound protease. Proc. Natl. Acad. Sci. U.S.A. 1992;89:6075–6079. doi: 10.1073/pnas.89.13.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haass C, Hung AY, Schlossmacher MG, Teplow DB, Selkoe DJ. β-Amyloid peptide and a 3-kDa fragment are derived by distinct cellular mechanisms. J. Biol. Chem. 1993;268:3021–3024. [PubMed] [Google Scholar]
- 8.Wolfe MS, Kopan R. Intramembrane proteolysis: Theme and variations. Science. 2004;305:1119–1123. doi: 10.1126/science.1096187. [DOI] [PubMed] [Google Scholar]
- 9.Haass C. Take five: BACE and the γ-secretase quartet conduct Alzheimer’s amyloid β-peptide generation. EMBO J. 2004;23:483–488. doi: 10.1038/sj.emboj.7600061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Strooper B. Aph-1, Pen-2, and Nicastrin with Presenilin generate an active γ-Secretase complex. Neuron. 2003;38:9–12. doi: 10.1016/s0896-6273(03)00205-8. [DOI] [PubMed] [Google Scholar]
- 11.Steiner H, Fluhrer R, Haass C. Intramembrane proteolysis by γ-secretase. J. Biol. Chem. 2008;283:29627–29631. doi: 10.1074/jbc.R800010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weidemann A, Eggert S, Reinhard FB, Vogel M, Paliga K, Baier G, Masters CL, Beyreuther K, Evin G. A novel ε-cleavage within the transmembrane domain of the Alzheimer amyloid precursor protein demonstrates homology with Notch processing. Biochemistry. 2002;41:2825–2835. doi: 10.1021/bi015794o. [DOI] [PubMed] [Google Scholar]
- 13.Gu Y, Misonou H, Sato T, Dohmae N, Takio K, Ihara Y. Distinct intramembrane cleavage of the β-amyloid precursor protein family resembling γ-secretase-like cleavage of Notch. J Biol. Chem. 2001;276:35235–35238. doi: 10.1074/jbc.C100357200. [DOI] [PubMed] [Google Scholar]
- 14.Sastre M, Steiner H, Fuchs K, Capell A, Multhaup G, Condron MM, Teplow DB, Haass C. Presenilin-dependent γ-secretase processing of β-amyloid precursor protein at a site corresponding to the S3 cleavage of Notch. EMBO Rep. 2001;2:835–841. doi: 10.1093/embo-reports/kve180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao G, Mao G, Tan J, Dong Y, Cui MZ, Kim SH, Xu X. Identification of a new presenilin-dependent ζ-cleavage site within the transmembrane domain of amyloid precursor protein. J. Biol. Chem. 2004;279:50647–50650. doi: 10.1074/jbc.C400473200. [DOI] [PubMed] [Google Scholar]
- 16.Zhao G, Cui MZ, Mao G, Dong Y, Tan J, Sun L, Xu X. γ-Cleavage is dependent on ζ-cleavage during the proteolytic processing of amyloid precursor protein within its transmembrane domain. J. Biol. Chem. 2005;280:37689–37697. doi: 10.1074/jbc.M507993200. [DOI] [PubMed] [Google Scholar]
- 17.Kakuda N, Funamoto S, Yagishita S, Takami M, Osawa S, Dohmae N, Ihara Y. Equimolar production of amyloid β-protein and amyloid precursor protein intracellular domain from β-carboxyl-terminal fragment by γ-secretase. J. Biol. Chem. 2006;281:14776–14786. doi: 10.1074/jbc.M513453200. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh AK, Gemma S, Tang J. β-Secretase as a therapeutic target for Alzheimer’s disease. Neurotherapeutics. 2008;5:399–408. doi: 10.1016/j.nurt.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siemers ER, Dean RA, Friedrich S, Ferguson-Sells L, Gonzales C, Farlow MR, May PC. Safety, tolerability, and effects on plasma and cerebrospinal fluid amyloid-β after inhibition of γ-secretase. Clin. Neuropharmacol. 2007;30:317–325. doi: 10.1097/WNF.0b013e31805b7660. [DOI] [PubMed] [Google Scholar]
- 20.Imbimbo BP. γ-Secretase inhibitors and modulators as a therapeutic approach to Alzheimer’s disease. Curr. Top. Med. Chem. 2008;8:1. doi: 10.2174/156802608783334015. [DOI] [PubMed] [Google Scholar]
- 21.Parks AL, Curtis D. Presenilin diversifies its portfolio. Trends Genet. 2007;23:140–150. doi: 10.1016/j.tig.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 22.Hemming ML, Elias JE, Gygi SP, Selkoe DJ. Proteomic profiling of γ-secretase substrates and mapping of substrate requirements. PLoS Biol. 2008;6:e257. doi: 10.1371/journal.pbio.0060257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hadland BK, Manley NR, Su D, Longmore GD, Moore CL, Wolfe MS, Schroeter EH, Kopan R. γ-Secretase inhibitors repress thymocyte development. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7487–7491. doi: 10.1073/pnas.131202798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Searfoss GH, Jordan WH, Calligaro DO, Galbreath EJ, Schirtzinger LM, Berridge BR, Gao H, Higgins MA, May PC, Ryan TP. Adipsin, a biomarker of gastrointestinal toxicity mediated by a functional γ-secretase inhibitor. J. Biol. Chem. 2003;278:46107–46116. doi: 10.1074/jbc.M307757200. [DOI] [PubMed] [Google Scholar]
- 25.Kukar T, Murphy MP, Eriksen JL, Sagi SA, Weggen S, Smith TE, Ladd T, Khan MA, Kache R, Beard J, Dodson M, Merit S, Ozols VV, Anastasiadis PZ, Das P, Fauq A, Koo EH, Golde TE. Diverse compounds mimic Alzheimer disease-causing mutations by augmenting A/342 production. Nat. Med. 2005;11:545–550. doi: 10.1038/nm1235. [DOI] [PubMed] [Google Scholar]
- 26.Weggen S, Eriksen JL, Das P, Sagi SA, Wang R, Pietrzik CU, Findlay KA, Smith TE, Murphy MP, Bulter T, Kang DE, Marquez-Sterling N, Golde TE, Koo EH. A subset of NSAIDs lower amyloidogenic Aβ42 independently of cyclooxygenase activity. Nature. 2001;414:212–216. doi: 10.1038/35102591. [DOI] [PubMed] [Google Scholar]
- 27.Weggen S, Eriksen JL, Sagi SA, Pietrzik CU, Golde TE, Koo EH. Aβ42-lowering nonsteroidal anti-inflammatory drugs preserve intramembrane cleavage of the amyloid precursor protein (APP) and ErbB-4 receptor and signaling through the APP intracellular domain. J. Biol. Chem. 2003;278:30748–30754. doi: 10.1074/jbc.M304824200. [DOI] [PubMed] [Google Scholar]
- 28.Morihara T, Chu T, Ubeda O, Beech W, Cole GM. Selective inhibition of Aβ42 production by NSAID R-enantiomers. J. Neurochem. 2002;83:1009–1012. doi: 10.1046/j.1471-4159.2002.01195.x. [DOI] [PubMed] [Google Scholar]
- 29.Weggen S, Eriksen JL, Sagi SA, Pietrzik CU, Ozols V, Fauq A, Golde TE, Koo EH. Evidence that nonsteroidal anti-inflammatory drugs decrease amyloid β42 production by direct modulation of γ-secretase activity. J. Biol. Chem. 2003;278:31831–31837. doi: 10.1074/jbc.M303592200. [DOI] [PubMed] [Google Scholar]
- 30.Wolfe MS. Inhibition and modulation of γ-secretase for Alzheimer’s disease. Neurotherapeutics. 2008;5:391–398. doi: 10.1016/j.nurt.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheinfeld MH, Ghersi E, Laky K, Fowlkes BJ, D’Adamio L. Processing of β-amyloid precursor-like protein-1 and −2 by γ-secretase regulates transcription. J. Biol. Chem. 2002;277:44195–44201. doi: 10.1074/jbc.M208110200. [DOI] [PubMed] [Google Scholar]
- 32.Eggert S, Paliga K, Soba P, Evin G, Masters CL, Weidemann A, Beyreuther K. The proteolytic processing of the amyloid precursor protein gene family members APLP-1 and APLP-2 involves α-, β-, γ-, and ε-like cleavages: Modulation of APLP-1 processing by N-glycosylation. J. Biol. Chem. 2004;279:18146–18156. doi: 10.1074/jbc.M311601200. [DOI] [PubMed] [Google Scholar]
- 33.Minogue AM, Stubbs AK, Frigerio CS, Boland B, Fadeeva JV, Tang J, Selkoe DJ, Walsh DM. γ-Secretase processing of APLP1 leads to the production of a p3-like peptide that does not aggregate and is not toxic to neurons. Brain Res. 2009;1262:89–99. doi: 10.1016/j.brainres.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 34.Yanagida K, Okochi M, Tagami S, Nakayama T, Kodama TS, Nishitomi K, Jiang J, Mori K, Tatsumi S, Arai T, Ikeuchi T, Kasuga K, Tokuda T, Kondo M, Ikeda M, Deguchi K, Kazui H, Tanaka T, Morihara T, Hashimoto R, Kudo T, Steiner H, Haass C, Tsuchiya K, Akiyama H, Kuwano R, Takeda M. The 28-amino acid form of an APLP1-derived Aβ-like peptide is a surrogate marker for Aβ42 production in the central nervous system. EM BO Mol. Med. 2009 doi: 10.1002/emmm.200900026. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Koch CS, Zheng H, Chen H, Trumbauer M, Thinakaran G, van der Ploeg LH, Price DL, Sisodia SS. Generation of APLP2 KO mice and early postnatal lethality in APLP2/APP double KO mice. Neurobiol. Aging. 1997;18:661–669. doi: 10.1016/s0197-4580(97)00151-6. [DOI] [PubMed] [Google Scholar]
- 36.Heber S, Herms J, Gajic V, Hainfellner J, Aguzzi A, Rulicke T, von Kretzschmar H, von Koch C, Sisodia S, Tremml P, Lipp HP, Wolfer DP, Muller U. Mice with combined gene knock-outs reveal essential and partially redundant functions of amyloid precursor protein family members. J. Neurosci. 2000;20:7951–7963. doi: 10.1523/JNEUROSCI.20-21-07951.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herms J, Anliker B, Heber S, Ring S, Fuhrmann M, Kretzschmar H, Sisodia S, Muller U. Cortical dysplasia resembling human type 2 lissencephaly in mice lacking all three APP family members. EMBO J. 2004;23:4106–4115. doi: 10.1038/sj.emboj.7600390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao X, Sudhof TC. Dissection of amyloid-β precursor protein-dependent transcriptional transactivation. J. Biol. Chem. 2004;279:24601–24611. doi: 10.1074/jbc.M402248200. [DOI] [PubMed] [Google Scholar]
- 39.Hebert SS, Serneels L, Tolia A, Craessaerts K, Derks C, Filippov MA, Muller U, De Strooper B. Regulated intramembrane proteolysis of amyloid precursor protein and regulation of expression of putative target genes. EMBO Rep. 2006;7:739–745. doi: 10.1038/sj.embor.7400704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kukar TL, Ladd TB, Bann MA, Fraering PC, Narlawar R, Maharvi GM, Healy B, Chapman R, Welzel AT, Price RW, Moore B, Rangachari V, Cusack B, Eriksen J, Jansen-West K, Verbeeck C, Yager D, Eckman C, Ye W, Sagi S, Cottrell BA, Torpey J, Rosenberry TL, Fauq A, Wolfe MS, Schmidt B, Walsh DM, Koo EH, Golde TE. Substrate-targeting γ-secretase modulators. Nature. 2008;453:925–929. doi: 10.1038/nature07055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walsh DM, Fadeeva JV, LaVoie MJ, Paliga K, Eggert S, Kimberly WT, Wasco W, Selkoe DJ. γ-Secretase cleavage and binding to FE65 regulate the nuclear translocation of the intracellular C-terminal domain (ICD) of the APP family of proteins. Biochemistry. 2003;42:6664–6673. doi: 10.1021/bi027375c. [DOI] [PubMed] [Google Scholar]
- 42.McLendon C, Xin T, Ziani-Cherif C, Murphy MP, Findlay KA, Lewis PA, Pinnix I, Sambamurti K, Wang R, Fauq A, Golde TE. Cell-free assays for γ-secretase activity. FASEB J. 2000;14:2383–2386. doi: 10.1096/fj.00-0286fje. [DOI] [PubMed] [Google Scholar]
- 43.Pinnix I, Musunuru U, Tun H, Sridharan A, Golde T, Eckman C, Ziani-Cherif C, Onstead L, Sambamurti K. A novel γ-secretase assay based on detection of the putative C-terminal fragment-gamma of amyloid β protein precursor. J. Biol. Chem. 2001;276:481–487. doi: 10.1074/jbc.M005968200. [DOI] [PubMed] [Google Scholar]
- 44.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 45.Hoffmann J, Twiesselmann C, Kummer MP, Romagnoli P, Herzog V. A possible role for the Alzheimer amyloid precursor protein in the regulation of epidermal basal cell proliferation. Eur. J. Cell Biol. 2000;79:905–914. doi: 10.1078/0171-9335-00117. [DOI] [PubMed] [Google Scholar]
- 46.Haass C, Koo EH, Mellon A, Hung AY, Selkoe DJ. Targeting of cell-surface β-amyloid precursor protein to lysosomes: Alternative processing into amyloid-bearing fragments. Nature. 1992;357:500–503. doi: 10.1038/357500a0. [DOI] [PubMed] [Google Scholar]
- 47.Thinakaran G, Sisodia SS. Amyloid precursor-like protein 2 (APLP2) is modified by the addition of chondroitin sulfate glycosaminoglycan at a single site. J. Biol. Chem. 1994;269:22099–22104. [PubMed] [Google Scholar]
- 48.Takasugi N, Tomita T, Hayashi I, Tsuruoka M, Niimura M, Takahashi Y, Thinakaran G, Iwatsubo T. The role of presenilin cofactors in the γ-secretase complex. Nature. 2003;422:438–441. doi: 10.1038/nature01506. [DOI] [PubMed] [Google Scholar]
- 49.Edbauer D, Winkler E, Regula JT, Pesold B, Steiner H, Haass C. Reconstitution of γ-secretase activity. Nat. Cell Biol. 2003;5:486–488. doi: 10.1038/ncb960. [DOI] [PubMed] [Google Scholar]
- 50.Micchelli CA, Esler WP, Kimberly WT, Jack C, Berezovska O, Kornilova A, Hyman BT, Perrimon N, Wolfe MS. γ-Secretase/presenilin inhibitors for Alzheimer’s disease phenocopy Notch mutations in Drosophila . FASEB J. 2003;17:79–81. doi: 10.1096/fj.02-0394fje. [DOI] [PubMed] [Google Scholar]
- 51.Seiffert D, Bradley JD, Rominger CM, Rominger DH, Yang F, Meredith JE, Jr, Wang Q, Roach AH, Thompson LA, Spitz SM, Higaki JN, Prakash SR, Combs AP, Copeland RA, Arneric SP, Hartig PR, Robertson DW, Cordell B, Stern AM, Olson RE, Zaczek R. Presenilin-1 and −2 are molecular targets for γ-secretase inhibitors. J. Biol. Chem. 2000;275:34086–34091. doi: 10.1074/jbc.M005430200. [DOI] [PubMed] [Google Scholar]
- 52.Yagishita S, Morishima-Kawashima M, Tanimura Y, Ishiura S, Ihara Y. DAPT-induced intracellular accumulations of longer amyloid β-proteins: Further implications for the mechanism of intramembrane cleavage by γ-secretase. Biochemistry. 2006;45:3952–3960. doi: 10.1021/bi0521846. [DOI] [PubMed] [Google Scholar]
- 53.Dovey HF, John V, Anderson JP, Chen LZ, de Saint Andrieu P, Fang LY, Freedman SB, Folmer B, Goldbach E, Holsztynska EJ, Hu KL, Johnson-Wood KL, Kennedy SL, Kholodenko D, Knops JE, Larimer LH, Lee M, Liao Z, Lieberburg IM, Motter RN, Mutter LC, Nietz J, Quinn KP, Sacchi KL, Seubert PA, Shopp GM, Thorsett ED, Tung JS, Wu J, Yang S, Yin CT, Schenk DB, May PC, Altstiel LD, Bender MH, Boggs LN, Britton TC, Clemens JC, Czilli DL, Dieckman-McGinty DK, Droste JJ, Fuson KS, Gitter BD, Hyslop PA, Johnstone EM, Li WY, Little SP, Mabry TE, Miller FD, Audia JE. Functional γ-secretase inhibitors reduce β-amyloid peptide levels in brain. J. Neurochem. 2001;76:173–181. doi: 10.1046/j.1471-4159.2001.00012.x. [DOI] [PubMed] [Google Scholar]
- 54.Eriksen JL, Sagi SA, Smith TE, Weggen S, Das P, McLendon DC, Ozols VV, Jessing KW, Zavitz KH, Koo EH, Golde TE. NSAIDs and enantiomers of flurbiprofen target γ-secretase and lower Aβ42 in vivo. J. Clin. Invest. 2003;112:440–449. doi: 10.1172/JCI18162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGeer PL, Schulzer M, McGeer EG. Arthritis and anti-inflammatory agents as possible protective factors for Alzheimer’s disease: A review of 17 epidemiologic studies. Neurology. 1996;47:425–432. doi: 10.1212/wnl.47.2.425. [DOI] [PubMed] [Google Scholar]
- 56.in t’Veld BA, Ruitenberg A, Hofman A, Launer LJ, van Duijn CM, Stijnen T, Breteler MM, Stricker BH. Nonsteroidal antiinflammatory drugs and the risk of Alzheimer’s disease. N. Engl. J. Med. 2001;345:1515–1521. doi: 10.1056/NEJMoa010178. [DOI] [PubMed] [Google Scholar]
- 57.Imbimbo BP. Why Did Tarenflurbil Fail in Alzheimer’s Disease. J. Alzheimer’s Dis. 2009;17:757–760. doi: 10.3233/JAD-2009-1092. [DOI] [PubMed] [Google Scholar]
- 58.Lleo A. Activity of γ-secretase on substrates other than APP. Curr. Top. Med. Chem. 2008;8:9–16. doi: 10.2174/156802608783334060. [DOI] [PubMed] [Google Scholar]
- 59.Beel AJ, Sanders CR. Substrate specificity of γ-secretase and other intramembrane proteases. Cell. Mol. Life Sci. 2008;65:1311–1334. doi: 10.1007/s00018-008-7462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bayer TA, Paliga K, Weggen S, Wiestler OD, Beyreuther K, Multhaup G. Amyloid precursor-like protein 1 accumulates in neuritic plaques in Alzheimer’s disease. Acta Neuropathol. 1997;94:519–524. doi: 10.1007/s004010050745. [DOI] [PubMed] [Google Scholar]
- 61.Naruse S, Thinakaran G, Luo JJ, Kusiak JW, Tomita T, Iwatsubo T, Qian X, Ginty DD, Price DL, Borchelt DR, Wong PC, Sisodia SS. Effects of PS1 deficiency on membrane protein trafficking in neurons. Neuron. 1998;21:1213–1221. doi: 10.1016/s0896-6273(00)80637-6. [DOI] [PubMed] [Google Scholar]
- 62.Li Q, Sudhof TC. Cleavage of amyloid-β precursor protein and amyloid-β precursor-like protein by BACE 1. J. Biol. Chem. 2004;279:10542–10550. doi: 10.1074/jbc.M310001200. [DOI] [PubMed] [Google Scholar]
- 63.Pastorino L, Ikin AF, Lamprianou S, Vacaresse N, Revelli JP, Platt K, Paganetti P, Mathews PM, Harroch S, Buxbaum JD. BACE (β-secretase) modulates the processing of APLP2 in vivo. Mol. Cell. Neurosci. 2004;25:642–649. doi: 10.1016/j.mcn.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 64.Walsh DM, Minogue AM, Sala Frigerio C, Fadeeva JV, Wasco W, Selkoe DJ. The APP family of proteins: Similarities and differences. Biochem. Soc. Trans. 2007;35:416–420. doi: 10.1042/BST0350416. [DOI] [PubMed] [Google Scholar]
- 65.Bai Y, Markham K, Chen F, Weerasekera R, Watts J, Home P, Wakutani Y, Bagshaw R, Mathews PM, Fraser PE, Westaway D, St George-Hyslop P, Schmitt-Ulms G. The in vivo brain interactome of the amyloid precursor protein. Mol. Cell. Proteomics. 2008;7:15–34. doi: 10.1074/mcp.M700077-MCP200. [DOI] [PubMed] [Google Scholar]
- 66.Kaden D, Voigt P, Munter LM, Bobowski KD, Schaefer M, Multhaup G. Subcellular localization and dimerization of APLP1 are strikingly different from APP and APLP2. J. Cell Sci. 2009;122:368–377. doi: 10.1242/jcs.034058. [DOI] [PubMed] [Google Scholar]
- 67.Schrenk-Siemens K, Perez-Alcala S, Richter J, Lacroix E, Rahuel J, Korte M, Muller U, Barde YA, Bibel M. Embryonic stem cell-derived neurons as a cellular system to study gene function: Lack of amyloid precursor proteins APP and APLP2 leads to defective synaptic transmission. Stem Cells. 2008;26:2153–2163. doi: 10.1634/stemcells.2008-0010. [DOI] [PubMed] [Google Scholar]
- 68.Xu Y, Kim HS, Joo Y, Choi Y, Chang KA, Park CH, Shin KY, Kim S, Cheon YH, Baik TK, Kim JH, Suh YH. Intracellular domains of amyloid precursor-like protein 2 interact with CP2 transcription factor in the nucleus and induce glycogen synthase kinase-3β expression. Cell Death Differ. 2007;14:79–91. doi: 10.1038/sj.cdd.4401928. [DOI] [PubMed] [Google Scholar]
- 69.Ring S, Weyer SW, Kilian SB, Waldron E, Pietrzik CU, Filippov MA, Herms J, Buchholz C, Eckman CB, Korte M, Wolfer DP, Muller UC. The secreted β-amyloid precursor protein ectodomain APPs α is sufficient to rescue the anatomical, behavioral, and electrophysiological abnormalities of APP-deficient mice. J. Neurosci. 2007;27:7817–7826. doi: 10.1523/JNEUROSCI.1026-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Munter LM, Voigt P, Harmeier A, Kaden D, Gottschalk KE, Weise C, Pipkorn R, Schaefer M, Langosch D, Multhaup G. GxxxG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of Aβ42. EMBO J. 2007;26:1702–1712. doi: 10.1038/sj.emboj.7601616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaden D, Munter LM, Joshi M, Treiber C, Weise C, Bethge T, Voigt P, Schaefer M, Beyermann M, Reif B, Multhaup G. Homophilic interactions of the amyloid precursor protein (APP) ectodomain are regulated by the loop region and affect β-secretase cleavage of APP. J. Biol. Chem. 2008;283:7271–7279. doi: 10.1074/jbc.M708046200. [DOI] [PubMed] [Google Scholar]
- 72.Lee HJ, Jung KM, Huang YZ, Bennett LB, Lee JS, Mei L, Kim TW. Presenilin-dependent γ-secretase-like intramembrane cleavage of ErbB4. J. Biol. Chem. 2002;277:6318–6323. doi: 10.1074/jbc.M110371200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.