Abstract
SCUDO (Signature-based ClUstering for DiagnOstic purposes) is an online tool for the analysis of gene expression profiles for diagnostic and classification purposes. The tool is based on a new method for the clustering of profiles based on a subject-specific, as opposed to disease-specific, signature. Our approach relies on construction of a reference map of transcriptional signatures, from both healthy and affected subjects, derived from their respective mRNA or miRNA profiles. A diagnosis for a new individual can then be performed by determining the position of the individual's transcriptional signature on the map. The diagnostic power of our method has been convincingly demonstrated in an open scientific competition (SBV Improver Diagnostic Signature Challenge), scoring second place overall and first place in one of the sub-challenges.
INTRODUCTION
An important task in molecular biology is the accurate classification of biological specimens on the base of their gene expression profiles. Methods for accurate and reproducible classification are increasingly required for the potential biomedical applications of expression profiles, such as disease classification, diagnosis and monitoring. In this paper we describe SCUDO, a web server implementing an original signature identification method. Given a set of expression profiles from control and affected subjects, SCUDO verifies the existence of a signature (actually, a set of signatures, as explained below) that can be used to classify the profiles according to a phenotype of interest, such as the control/affected status. The method is composed of a sequence of simple processing steps, none of which is original; however their combination results in a robust and novel model of profile data analysis, that corresponds to a different way of exploiting similarities and differences among biological samples for classification purposes. Specifically, the originality of the method is that it introduces a new concept of subject-specific signatures, as opposed to disease-specific signatures, that also includes the benefits of rank-based classification, data dimensionality reduction and the power of network analysis in its design.
Our method was tested against a selection of the best current approaches in the course of the SBV IMPROVER Diagnostic Signature Challenge, an open competition designed to assess and verify computational approaches for classifying clinical samples based on gene expression (1). In an effort to increase robustness of the outcome with respect to chance and disease specificity, the SBV Challenge required a diagnosis for all of four diseases, namely Psoriasis, Multiple Sclerosis (MS), Chronic Obstructive Pulmonary Disease (COPD) and Lung Cancer. For each disease, the organizers (a team of researchers from IBM Research and Philip Morris International) suggested a list of publicly available mRNA expression datasets to use as possible training sets and then solicited a healthy/affected diagnosis for a previously unpublished set of profiles (test set). The submitted predictions were scored by the organizers according to a combination of three previously selected metrics: the Area Under the Precision-Recall Curve (AUPR), the Belief Confusion Metric (BCM) and the Correct Class Enrichment Metric (CCEM).
The method implemented in SCUDO ranked second overall, and first in the MS sub-challenge, out of 54 submissions. Interestingly, the MS Diagnostic appeared to be the most difficult of the four, according to the plots of the distribution of random versus observed scores prepared by the organizers. The overall first placement was achieved by the winning team through a skillful application of Linear Discriminant Analysis (LDA); the third place was obtained by a team that applied Lasso Regression (2). (See (3) and (4), respectively, for the details of the winning methods and of the scoring algorithm and score distribution plots.)
The method on which SCUDO is based represents a completely new way of addressing the expression profile classification problem. Contrary to current practices, instead of evaluating a set of profiles by means of a common yardstick (a single list of highly discriminating mRNA/miRNA species), our method first seeks to summarize the characteristics of each profile by means of a rank-based signature and then it performs a systematic, all-to-all signature comparison. The comparison consists in the measurement of a distance that quantifies the degree of similarity between any two signatures using a simplified version of the Enrichment Score (ES) (5). The result of the comparison is a distance matrix, which is then visualized in the form of a map of individual signatures. In such a map, the spontaneous emergence of groups (i.e. clusters of closely connected nodes) of transcriptionally similar subjects is typically observed. In a diagnostic application, the observed groups segregate the subjects according to their control/affected phenotype. A diagnostic classification for an unseen individual can subsequently be performed by determining the position of the individual's transcriptional signature relative to the control/affected groups. Further details of the method are described in (6).
The signature map is the output of SCUDO. The map is in the form of a graph where the nodes correspond to signatures/subjects and the length of a connecting edge encodes the level of similarity between the connected nodes (short edge = high similarity; no edge = negligible similarity). SCUDO accepts two separate datasets as input, respectively a testing and a training dataset, but it can also be used with a single (training) dataset. If a testing set is specified, SCUDO runs the same rank-based classification algorithm on it, but it skips the feature selection step, using instead the list of features (genes or miRNAs) already selected for the training dataset. The optimization of the signature length is left to the user and is performed by trial and error; in practice the user needs to try and select the n1 and n2 values that provide the clearest separation between the affected and the control subjects (n1 = number of up-regulated genes to consider, n2 = number of down-regulated genes, n1+ n2 being the total signature length). Crucially, we have shown that our method is robust to variations of its input parameter values, therefore the selection process is not critical and satisfactory values can be found with only a few iterations (6). The expected result is the emergence of a partitioning of the set of samples in separate, clearly visible groups on the basis of signature similarity. While for the training set the sample classification is known in advance and thus the correctness of the partitioning is immediately verified by the appearance of homogeneously colored groups, for the test set in general the sample classification is the desired final output and needs to be deduced from the graph. The classification problem for the test set is then reduced to the simpler task of identifying the control/affected phenotype associated to each of the emerging groups in the test graph, in what we call the labeling step. This relatively simple labeling step can be carried out by means of empirical methods that rely on some form of previous knowledge. One form of previous knowledge consists of a small number of samples for which the classification is known; by adding these samples to the test set, one can deduce the labeling of the groups based on the membership of the known samples. Another form of previous knowledge is represented by a list of literature-derived disease genes; by computing the average expression values of these genes across all the members of a group, one can deduce the labeling of the groups by comparing averages and taking a majority vote. We used both these methods for the labeling of samples in the SBV Improver competition.
Our rank-based signature method is quite general and we have tested it successfully with a large assortment of data types: mRNA and miRNA profiles for various diseases/treatments/experiments, obtained from biopsies, blood and urine and even some non-biological multidimensional datasets (i.e. engineering data). With SCUDO we are making available an online implementation of our new method to encourage experimentation and discovery of novel applications.
THE SCUDO INTERFACE: THE INPUT
SCUDO accepts a testing and a training dataset as input (Figure 1), but it can also be used with a single (training) dataset, in case the user is not interested in the testing step. If a testing set is specified, SCUDO produces a map of the profiles using the same list of features (genes or miRNAs) that were used in producing the training profiles map. In a sense, the testing set is used to validate the quality of the feature list, obtained examining the training set, as a starting point in producing discriminating signatures for a different set of profiles.
Figure 1.
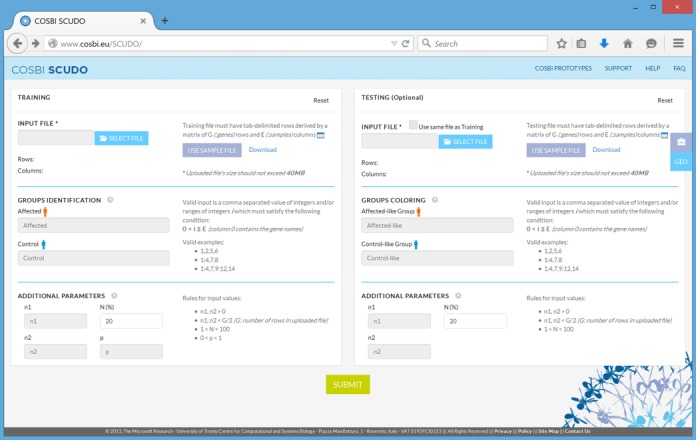
Snapshot of the SCUDO start page. The input data fields for the training set are on the left, the ones for the testing set on the right.
A microarray expression dataset to be analyzed needs to have been previously normalized; the choice of the algorithm is not critical (i.e. either MAS5 or RMA can be used for Affymetrix platforms; either calibrated or uncalibrated data can be used for RT-PCR profiles), the data does not need to be log transformed. The data must be in a tab-separated text format, with one gene or probeset per row, and one sample (i.e. a patient profile) per column; the testing and training datasets must have the same number of rows and the same gene/probeset on corresponding rows. The first column of the file is reserved for a gene/probeset identifier while the first row of the file is used for sample identifiers. Both set of identifiers must be unique character strings without spaces; the leftmost field of the first row, corresponding to the probeset ID column, needs to be filled with a placeholder character string. Assuming that the user is using both a training and a testing dataset, the typical analysis workflow is as follows: upload the two data files, specify the algorithm parameters, launch the analysis, examine the two resulting maps (one for the training profiles, the other for the testing profiles). After inspecting the maps, the user can repeat the same process with different parameter values, for example with a different signature size. The input parameters to be specified on the start page are:
column ranges corresponding to the two classes of profiles (i.e. control, affected) in the training dataset
n1 = number of genes from top ranked genes to include in signature
n2 = number of genes from bottom ranked genes to include in signature
pval = P-value to be used for the initial feature selection
N = percentage of all of the signature-to-signature distances that will be used for drawing the map of subjects.
The user has the option to use the same file for the training and testing dataset, presumably selecting different ranges of columns as the training and the testing sets. The column ranges specified by the user for the testing set are also used to assign different colors to the two groups of samples. Only the specified columns for both sets are used for the analysis.
In the current version of SCUDO, the optimization of input parameters such as the signature length and the P-value of the feature selection step is left to the user and is performed by trial and error. In a typical session, the user needs to try and select the n1 and n2 values that provide the clearest separation between the affected and the control subjects in the output maps, starting with the suggested value of P-value (0.01 for mRNA, 0.1 for miRNA profiles). Based on our experience with a large number of representative datasets, good starting values for n1 and n2 are 250/250 for mRNA, 25/25 for miRNA and these values can be repeatedly doubled or halved until good separation is achieved; if unsuccessful, a more stringent P-value can be tried. The P-value is not critical because it does not represent a threshold for inclusion of crucial features in a signature; rather, the main purpose of the initial feature selection step is to prevent pollution of signature composition by non-informative features (i.e. to remove the probesets/genes that are stuck high or low). The value of N is finally adjusted to improve clarity of the map by either reducing the clutter (smaller N) or increasing the number of connecting edges (larger N) as needed.
THE SCUDO INTERFACE: THE OUTPUT
The output of the tool is a map for each of the input dataset (see Figure 2). The nodes in the map correspond to subjects and the length of a connecting edge encodes the level of similarity between the connected nodes (short edge = high similarity; no edge = negligible similarity). The map is in the form of a network drawn using Cytoscape Web, which allows manipulation and setting of display attributes. The desired outcome of the analysis is a map of the testing set in which the profiles are clearly divided in two groups composed of nodes of the same color; such result would give high confidence in the fact that using the selected feature list the user will be able to satisfactorily classify a new set of profiles using signatures composed of the n1 + n2 most differentially expressed genes of each profile.
Figure 2.
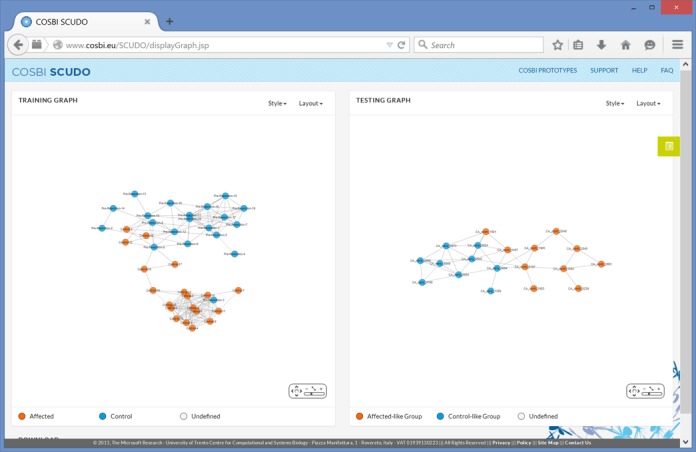
Snapshot of the SCUDO result page; the map for the training data is on the left, the one for the testing data on the right.
Once the user is satisfied with the result, he/she can download the maps as graphical files, or as network files (edge list) ready to be imported in Cytoscape (7) or similar tools. A Cytoscape plugin such as clusterMaker (8) can be useful in identifying clusters in those maps in which the boundaries between groups are not immediately apparent (as in the right map of Figure 2); see reference (6) for an example of use of such plugin. The user can also download a file containing the subject specific signatures (one column per subject) plus two ‘consensus signatures’ (one for each of the two groups indicated by the user). The consensus signature for each group is computed as follows: the rank of each gene in a profile is computed separately for each subject by sorting expression values from the most expressed to the least expressed, then the columns of gene ranks corresponding only to the subjects included in the group are summed row-wise and the resulting rank sums are sorted to provide a new set of ‘consensus ranks’; the signature for the group is then composed by the first n1 and the last n2 gene IDs ordered according to the new consensus rank list. The consensus signature is reminiscent of a disease gene list although derived using our unconventional approach and can be used to study the underlying molecular mechanism of a disease (for an instance of such an application see (9)).
SAMPLE DATA
A sample set of expression profiles and corresponding suggested parameter values are available in the initial page. The sample datasets provide an interesting example of the versatility of SCUDO. The two datasets comprise profiles of circulating miRNA in patient with early breast cancer and matching controls published by two different research groups (GEO accession numbers GSE22981 and GSE41526, including 40 and 70 profiles respectively). GSE41256 was used as the training set and GSE22981 as the testing set, because the larger size of the former improves the performance of the feature selection step. The two datasets were used as downloaded from GEO (GSE Series Matrix format) except for the required changes in the formatting; also, in the first set we had to remove six rows corresponding to miRNA species that were missing from the other set (both set use the same platform, Illumina Human v2 MicroRNA Expression Beadchip). The recommended signatures sizes are 20/20 for both sets; the recommended P-value of 0.001, empirically found to work best, is uncharacteristically small for SCUDO, probably reflecting the existence of high noise levels or confounding signals in the two datasets.
In the two resulting maps (Figure 2) it can be observed that (i) in the training set map, SCUDO is able to group together control and affected patients with good accuracy and (ii) in the testing set, SCUDO is able to separate control and affected subjects reasonably well as long as the analysis is restricted to the subset of Caucasian American individuals. These results are of particular interest in view of the fact that the authors of the first set were not able to identify a reproducible diagnostic signature (10) and that the authors of the second set reported differences in terms of differentially expressed circulating miRNAs between Caucasian American and African American subjects (11).
DISCUSSION
SCUDO implements a new expression profile classification based on the degree of similarity between profile-specific signatures. As a consequence of being rank-based, SCUDO is quite robust to differences in lab protocols, data processing and batch effects, because it relies only on the relative ordering of the gene expression values within each profile, and not on the values themselves (for more details on this point see (6)). This robustness can be appreciated in the analysis of the provided sample data files, which are from the same Affymetrix platform but have been produced by different research groups and have undergone different normalizations. Additionally, we have shown that our method is robust to wide variations of its parameter values therefore the choice of the values is not overly critical. Our rank-based signature method is quite general and we have tested it successfully with a large assortment of data types: mRNA and miRNA profiles for various diseases/treatments/experiments, obtained from biopsies, blood and urine, and even some non-biological multidimensional datasets (i.e. engineering data). Using publicly available data, we have obtained promising results on the stratification of patients with respect to treatment response (12) and early diagnosis of cancer based on blood miRNA (13); we have preliminary results for sample classification in toxicity studies. The reasons for this generality are that being completely data driven, our method is agnostic about the details of the mechanisms producing the transcriptional response. By making available an online implementation of our new method we want to encourage experimentation with new data types and discovery of novel applications. SCUDO has been used as part of a class project by the students of a graduate course held at the University of Trento during the Spring 2014 semester, whose feedback we gratefully acknowledge.
AVAILABILITY
http://www.cosbi.eu/scudo—this website is free and open to all users.
FUNDING
Funding for open access charge: The Microsoft Research—University of Trento Centre for Computational and Systems Biology (COSBI).
Conflict of interest statement. None declared.
REFERENCES
- 1.Rhrissorrakrai K., Rice J.J., Boue S., Talikka M., Bilal E., Martin F., Meyer P., Norel R., Xiang Y., Stolovitzky G., et al. sbv IMPROVER diagnostic signature challenge—design and results. Syst. Biomed. 2013;1:196–207. [Google Scholar]
- 2.Tibshirani R. Regression shrinkage and selection via the lasso. J. Royal. Statist. Soc B. 1996;58:267–288. [Google Scholar]
- 3.Tarca A.L., Lauria M., Unger M., Bilal E., Boue S., Kumar Dey K., Hoeng J., Koeppl H., Martin F., Meyer P., et al. IMPROVER DSC Collaborators. Strengths and limitations of microarray-based phenotype prediction: lessons learned from the IMPROVER Diagnostic Signature Challenge. Bioinformatics. 2013;29:2892–2899. doi: 10.1093/bioinformatics/btt492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Norel R., Bilal E., Conrad-Chemineau N., Bonneau R., de la Fuente A., Jurisica I., Marbach D., Meyer P., Rice J.J., Tuller T., et al. sbv IMPROVER diagnostic signature challenge—scoring strategies. Syst. Biomed. 2013;1:208–216. [Google Scholar]
- 5.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 6.Lauria M. Rank-based transcriptional signatures: a novel approach to diagnostic biomarker definition and analysis. Syst. Biomed. 2013;1:35–46. [Google Scholar]
- 7.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morris J.H., Apeltsin L., Newman A.M., Baumbach J., Wittkop T., Su G., Bader G.D., Ferrin T.E. clusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC bioinformatics. 2011;12:436. doi: 10.1186/1471-2105-12-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caberlotto L., Lauria M., Nguyen T.P., Scotti M. The central role of AMP-kinase and energy homeostasis impairment in Alzheimer's disease: a multifactor network analysis. PLoS One. 2013;8:e78919. doi: 10.1371/journal.pone.0078919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leidner R.S., Li L., Thompson C.L. Dampening enthusiasm for circulating microRNA in breast cancer. PLoS One. 2013;8:e57841. doi: 10.1371/journal.pone.0057841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao H., Shen J., Medico L., Wang D., Ambrosone C.B., Liu S. A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS One. 2010;5:e13735. doi: 10.1371/journal.pone.0013735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caberlotto L., Lauria M. Systems biology meets -omic technologies: novel approaches to biomarker discovery and companion diagnostic development. Expert Rev. Mol. Diagn. 2015;15:255–265. doi: 10.1586/14737159.2015.975214. [DOI] [PubMed] [Google Scholar]
- 13.Lauria M. Rank-based miRNA signatures for early cancer detection. Biomed. Res. Int. 2014;2014:192646. doi: 10.1155/2014/192646. [DOI] [PMC free article] [PubMed] [Google Scholar]


