Abstract
Assembly of the cell surface IL-10 receptor complex is the first step in initiating IL-10 signaling pathways that regulate intestinal inflammation, viral persistence, and even tumor surveillance. The discovery of IL-10 homologs in the genomes of herpes viruses suggests IL-10 signaling pathways can be manipulated at the level of the receptor complex. This chapter will describe our current molecular understanding of IL-10 receptor assembly based on crystal structures and biochemical analyses of cellular and viral IL-10 receptor complexes.
1 Introduction
Interleukin-10 (IL-10) is the founding member of the IL-10 cytokine family, which includes IL-19, IL-20, IL-22, IL-24, and IL-26 (Fickenscher et al. 2002; Kotenko 2002; Pestka et al. 2004; Trivella et al. 2010; Zdanov 2010). With the interferons (type I, IFNα/β; type II, IFNγ; and type III, IFNλ or IL28/IL29), the IL-10 family forms the class 2 cytokine family, where membership is based on conserved cysteine positions in their receptor sequences (Bazan 1990; Ho et al. 1993; Walter 2004). IL-10 is a unique class 2 cytokine because it potently inhibits the production of pro-inflammatory cytokines such as IFNγ, tumor necrosis factor α (TNFα), IL-1β, and IL-6 in a several cell types and prevents dendritic cell (DC) maturation in part by inhibiting the expression of IL-12 (Chang et al. 2004; Moore et al. 2001). IL-10 also inhibits the expression of MHC and co-stimulatory molecules important for cell-mediated immunity (Moore et al. 2001). However, IL-10 also exhibits immunostimulatory activities that include the ability to stimulate IFNγ production in CD8+ T cells activated with anti-CD3/anti-CD28, or other cytokine cocktails (Mumm et al. 2011; Santin et al. 2000). IL-10 is also a potent growth and differentiation factor for B-cells, mast cells and thymocytes (Moore et al. 2001; Rousset et al. 1992; Thompson-Snipes et al. 1991).
IL-10 cellular responses require the specific recognition and assembly of a heterodimeric cell surface complex comprised of IL-10R1 and IL-10R2 chains (Fig. 1) (Kotenko et al. 1997). IL-10R1 is an ~80,000 kDa protein with an extracellular ligand binding domain (ECD) of 227 residues, a transmembrane helix of 21 residues, and an intracellular domain (ICD) of 322 amino acids (Liu et al. 1994). The ECD of IL-10R2 is about the same length as IL-10R1, consisting of 201 residues (Lutfalla et al. 1993). However, the ICD of IL-10R2 consists of only 83 residues. The IL-10R1 ECD forms specific high affinity interactions (KD = 50-200pM) with IL-10, while IL-10R2 is a low affinity (~mM) shared receptor that participates in receptor complexes with other class 2 cytokine family members (Donnelly et al. 2004; Tan et al. 1993). Thus, in addition to pairing with IL-10R1 to form the IL-10 signaling complex, IL-10R2 forms IL22R1/IL-10R2 (Kotenko et al. 2001; Xie et al. 2000), IL-20R1/IL-10R2 (Hor et al. 2004; Sheikh et al. 2004), and IL28R1/IL-10R2 (Kotenko et al. 2003; Sheppard et al. 2003) heterodimers that are activated by IL-22, IL-26, and IL-28a,b/IL29, respectively.
Fig. 1. Sequential Assembly of the IL-10/IL-10R1/IL-10R2 Signaling Complex.
a 1:1 IL-10/IL-10R1 complex. b 1:2 IL10/IL-10R1 complex. c 1:2:2 IL-10/IL-10R1/IL-10R2 complex. As noted in the text, an engineered monomeric cIL-10 can induce cell signaling (Josephson et al. 2000). Thus, IL-10R2 can engage the 1:1 IL-10/IL-10R1 complex. The Intracellular receptor domain and JAK/TYK kinases are shown schematically. The triangle represents the docking site/s for unknown proteins required for TNF-α inhibition (Riley et al. 1999). d Enlarged image of one cIL-10 subunit bound to IL-10R1 and IL-10R2 in the ternary signaling complex, which denotes the location of the three binding interfaces that regulate complex formation.
While the IL-10R1 and IL-10R2 ECDs bind IL-10, the ICDs are constitutively associated with JAK1 and TYK2 kinases, respectively (Finbloom and Winestock 1995; Ho et al. 1995). IL-10 receptor binding activates JAK1 and TYK2, which phosphorylates themselves and IL-10R1 ICD tyrosines Y446IL-10R1 and Y496IL10R1. IL-10R1 phosphotyrosines provide docking sites that predominantly recruit and activate, via additional phosphorylation, the transcription factor STAT3 although STAT1 and STAT5 can also be activated (Donnelly et al. 1999; Finbloom and Winestock 1995; Weber-Nordt et al. 1996; Wehinger et al. 1996). The STAT3 docking sites in IL-10R1 appear to be sufficient to induce IL-10-mediated proliferative responses (Riley et al. 1999). In contrast, inhibition of TNFα requires STAT3 and additional intracellular molecules that recognize serines located in the C-terminal 30 residues of IL-10R1 (Riley et al. 1999; Takeda et al. 1999). A docking site for one or more E3 ligases has also been found in the IL-10R1 IDC that promotes ubiquitination and destruction of the receptor (Jiang et al. 2011; Wei et al. 2006). Removal of this region of IL-10R1 increased the potency of IL-10 on cells by 10-100 fold (Ho et al. 1995). Thus, the IL-10R1 IDC engages multiple proteins / protein complexes that regulate its expression level and diverse biological activities. In contrast, the only known function of the IL-10R2 IDC is to provide a docking site for TYK2. Thus, all “IL-10” specific cellular functions appear to reside in the IL-10R1 chain, while IL-10R2 provides a generic activation signal that can be used to activate signaling pathways associated with the four different R1 chains (e.g. IL-10R1, IL-20R1, IL-22R1, and IL-28R1).
The diverse biology of IL-10 observed in cell culture studies is also observed in animal models and in humans. In particular, disruption of the IL-10 signaling pathway results in severe inflammatory disease, which was first observed in IL-10 knockout (KO) mice that spontaneously developed inflammatory bowel disease (IBD) (Kuhn et al. 1993). The IBD phenotype was dependent on T-cells and gut bacteria, suggesting IL-10 controls immune responses to commensal intestinal pathogens (Rennick and Fort 2000; Sellon et al. 1998). Consistent with the studies in animals, mutations in IL-10, IL-10R1, and IL-10R2 that disrupt IL-10 signaling have been identified in infants/children, who suffer from early onset IBD (Engelhardt et al. 2013; Glocker et al. 2009; Grundtner et al. 2009).
The possibility that IL-10 signaling could promote viral infection was established by identifying an open reading frame in the Epstein Barr virus (EBV) genome that encoded an IL-10 homolog (ebvIL-10) (Moore et al. 1990). Analysis of another persistent herpes virus, cytomegalovirus (CMV), revealed it also harbored an IL-10 homolog (cmvIL-10) in its genome (Kotenko et al. 2000; Lockridge et al. 2000). Subsequent studies have established ebvIL-10 and cmvIL-10 exhibit distinct receptor binding and biological activity profiles that may promote viral persistence (Ding et al. 2000; Ding et al. 2001; Liu et al. 1997; Raftery et al. 2004; Rousset et al. 1992; Yoon et al. 2005). In contrast to EBV and CMV, other persistent human pathogens, including HIV, upregulate cellular IL-10, which contributes to T-cell exhaustion (Blackburn and Wherry 2007; Duell et al. 2012; Wilson and Brooks 2011). Remarkably, antibody mediated disruption of IL-10 signaling has been shown to enhance T cell responses and eliminate persistent viral infection in animal models. (Brooks et al. 2006; Ejrnaes et al. 2006). Thus, strategies to efficiently block IL-10 signaling may have therapeutic potential against persistent viral infections.
While these studies are consistent with the dogma that IL-10 is an immunosuppressive cytokine, IL-10 clearly has potent immunostimulatory activities, which are observed in studies in animals and humans. Strikingly, tumor immune surveillance was shown to be weakened in IL-10 KO mice, whereas transgenic overexpression of IL-10 protected mice from carcinogenesis (Mumm et al. 2011). Furthermore, injection of PEGylated IL-10 into Her2 transgenic mice led to tumor rejection that was dependent on activated CD8+ T cells that expressed IFNγ and granzyme B. (Mumm et al. 2011). Consistent with the animal studies, the administration of lipopolysaccharide (LPS) and IL-10 to humans reduced serum TNFα levels, but also elevated IFNγ and granzyme B (Lauw et al. 2000).
These brief examples demonstrate IL-10 receptor signaling plays an essential role in inflammation, viral pathogenesis, and cancer. To understand the complex nature of IL-10 activation, this review will describe the results of structural and biochemical studies to define molecular mechanisms that control IL-10 receptor assembly and subsequent cellular responses.
2. Structures of IL-10 and IL-10 Receptor Complexes
2.1. Cellular IL10 (cIL-10)
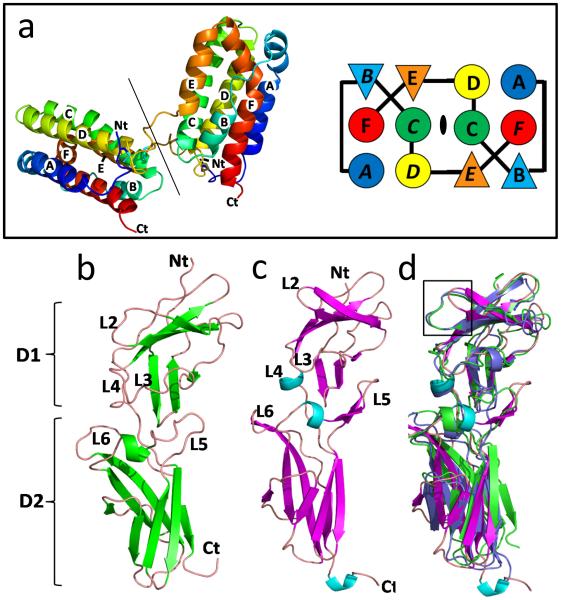
Cellular human IL-10 (cIL-10) is a 160 amino acid polypeptide that adopts a domain-swapped dimer with its subunits oriented at ~90° with respect to one another (Fig. 2a) (Walter and Nagabhushan 1995; Zdanov et al. 1995; Zdanov et al. 1996). Helices A, B, C, and D, from one peptide chain assemble with helices E and F, from the twofold related chain, to form two helical bundles containing six α-helices. The helices are connected by three tight turns (BC, CD, and EF loops) and two longer 12-residue loops (AB and DE loops) located at either ends of the bundles. Two intra-chain disulfide bonds link the N-terminus, and helix C, to the DE loop in each subunit. Thus, the disulfide bonds stabilize the assembly of helices A-D, but do not cross-link the two domain-swapped chains of the dimer.
Fig. 2. Structure of cIL-10 and IL-10 Receptors.
a Ribbon and schematic diagrams showing the structure and topology of the IL-10 domain swapped dimer. Overall structures of IL-10R1 (b), IL-10R2 (c), and a superposition of the receptor chains (d). Structural differences in the L2 receptor loops are highlighted in the box.
The subunit structure of cIL-10 is similar to crystal structures of other IL-10 family cytokines (IL-19, IL-20, IL-22, IL-28 and IL-29), and based on sequence similarity, similar to IL-24 and IL-26 (Chang et al. 2003; Gad et al. 2009; Logsdon et al. 2012; Miknis et al. 2010; Nagem et al. 2002; Xu et al. 2005). However, with the possible exception of IL-26, cIL-10 is the only IL-10 family member that adopts a domain-swapped dimeric structure. To address the necessity of cIL-10 dimer formation to induce IL-10 signaling, a monomeric cIL-10 (cIL-10M1) was engineered by inserting a 6 amino acid linker peptide into the DE loop (Josephson et al. 2000). Despite its monomeric structure, cIL-10M1 was able to induce short term cellular proliferation, albeit with ~8-18 fold lower activity than the cIL-10 dimer. However, maximal proliferative responses, equivalent to cIL-10, could be achieved with higher concentrations of cIL-10M1. These studies suggest a 1:1 cIL-10M1/IL-10R1 interaction is sufficient to engage the IL-10R2 chain and induce cIL-10 cellular responses.
2.2. cIL-10/IL-10R1 Complex
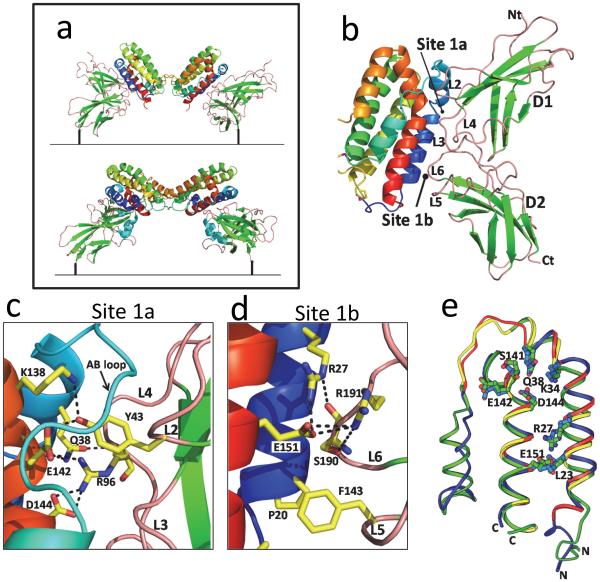
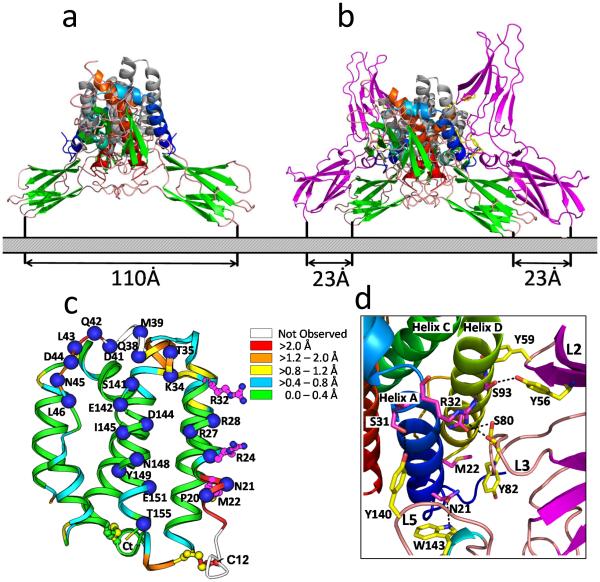
The structure of the cIL-10/IL-10R1 revealed two IL-10R1 chains bind the twofold related surfaces of IL-10, comprised of helix A, the AB loop, and helix F ( the site 1 interface, Fig. 3), to form a 1:2 cIL-10/IL-10R1 binary complex (Fig. 1b). The IL-10R1 ECD consists of two β-sandwich domains (D1 and D2) oriented at ~90° to one another (Fig. 2b). IL-10R1 contacts IL-10 through five loops (L2-L6), which are located on the convex surface of the receptor, at the junction of the D1 and D2 domains (Fig. 3b). IL-10R1 recognizes IL-10 in a “vertical” orientation, meaning the helical bundle axis of each subunit is parallel to the long axis of IL-10R1. To form these interactions, each IL-10R1 in the complex must rotate ~60° (counter-clockwise) away from a vector perpendicular to the plane of the cell membrane (Fig. 4a). In this orientation, the C-terminal ends of the two receptors are separated by ~110Å and incapable of activating a JAK mediated signaling cascade. The IL-10/IL-10R1 recognition paradigm is a unique feature of class 2 cytokines, relative to the class 1 cytokine family typified by growth hormone, IL-2 and IL6, where the helical bundle axis is recognized in a horizontal orientation, relative to the long axis of their respective receptors (de Vos et al. 1992; Josephson et al. 2001; Wang et al. 2009).
Fig. 3. Structure of the IL-10/IL-10R1 Binary Complex.
a Comparison of the binary cIL-10/IL-10R1 (top) and cmvIL-10/IL-10R1 complexes (bottom). b Enlarged region of the cIL-10/IL-10R1 complex showing the site 1 interface. c Key molecular contacts in the site 1a interface enlarged from Figure 3b. Amino acid sidechains described in the text are shown with carbon atoms and bonds colored yellow, oxygen atoms red, and nitrogen atoms blue. The amino acids are labeled using single letter amino acid codes (e.g. Glu = E). d Important contacts in the site 1b interface, enlarged from Figure 3b. Amino acid side chains described in the text are colored and labeled as described in Figure 3c. e Backbone superposition of site 1 (helices A, B and F) from cIL-10 (green) and cmvIL-10 (blue). cIL-10 and cmvIL-10 residues that donate sidechains to the site 1 interface are colored on the protein backbone yellow and red, respectively. The sidechains of amino acids conserved in the cIL-10 and cmvIL-10 site 1 interface are shown with carbon atoms and bonds colored green for cIL-10 and blue for cmvIL-10. For all sidechains shown, oxygen atoms are red and nitrogen atoms are blue.
Fig. 4. IL-10R1 Binding Induces Conformational Changes in the IL-10R2 Binding Site (Site 2).
a Orthogonal view, relative to Figure 1b, of the IL-10/IL-10R1 binary complex. b Orthogonal view, relative to Figure 1c, of the IL-10/IL-10R1/IL-10R2 signaling complex derived from a data-driven computational docking strategy (Yoon et al. 2010). c Ribbon diagram of one subunit of IL-10 colored to show the extent of conformational changes observed upon IL-10R1 binding. Distances in the legend correspond to distances between α-carbon atoms in unbound cIL-10 and IL-10R1-bound cIL-10 (Yoon et al. 2006). “Not observed” regions (white) correspond to residues that were not observed in the final electron density maps of unbound cIL-10, but were present in the structure of the cIL-10/sIL-10R1 complex. Residues that form the IL-10R1 binding site 1 are denoted by blue spheres. The sidechains of residues important for IL-10R2 binding are shown in magenta. d Enlargement of the site 2 interface (see Fig. 1d), which shows interactions between aromatic IL-10R2 residues (yellow) and IL-10 residues determined to be important for IL-10R2 binding by mutagenesis (magenta). For all sidechains shown, oxygen atoms are colored red and nitrogen atoms are blue.
2.3. Viral IL-10/IL-10R1 Complexes
The viral IL-10 homologs ebvIL-10 and cmvIL-10 share 83% and 27% sequence identity with cIL-10, respectively (Hsu et al. 1990; Kotenko et al. 2000; Lockridge et al. 2000). As suggested by their high sequence identity, the overall ebvIL-10/IL-10R1 structure is almost identical to the cIL-10/IL-10R1 complex (Yoon et al. 2005). In contrast, the cmvIL-10/IL-10R1 complex adopts a significantly different structure from cIL-10/IL-10R1 (Jones et al. 2002). The largest differences between cIL-10/IL-10R1 and cmvIL-10/IL-10R1 complexes occur in the subunit orientations of cmvIL-10 and cIL-10, whereas the structures of IL-10R1 and the site 1 binding interfaces are very similar (Fig. 3a). Like cIL-10 and ebvIL-10, cmvIL-10 folds as a domain swapped dimer. However, the two peptide chains in cmvIL-10 are cross-linked by an inter-chain disulfide bond that is not found in cIL-10 or ebvIL-10 (Jones et al. 2002; Yoon et al. 2005; Zdanov et al. 1997). As a result of structural changes within the dimer interface, the subunits of cmvIL-10 are oriented at an angle of 130°, compared to 90° for cIL-10. The change in subunit orientation causes the IL-10R1 chains, in the cmvIL-10/IL-10R1 complex, to rotate counter clockwise an additional ~25° relative to their position in the cIL-10/IL-10R1 complex (Fig. 3a). Despite the change in orientations of the IL-10R1s in the cmvIL-10/IL-10R1 complex, the C-terminal ends of the IL-10R1 chains are separated by 105Å, which is essentially identical to their separation in the cIL-10/IL-10R1 complex (110Å). Conservation of the twofold relationship and similar spacing of the receptor C-termini suggests these features are important for optimal IL-10 signal transduction.
The distinct orientation of the IL-10R1s in the cmvIL-10/IL-10R1 complex (Fig. 3a, e.g. almost parallel with the cell membrane) could alter the gene expression profile of cmvIL-10 relative to cIL-10. In support of this hypothesis, structural analyses of erythropoietin (EPO), and EPO peptide mimetics, bound to EPO receptor (EPOR), revealed agonist or antagonist activity was correlated, not with dimerization of the receptor chains, but the orientation of EPOR in the different complexes (Livnah et al. 1998). Despite these observations, no significant differences in biological activity have been found between cmvIL-10 and cIL-10. Interestingly, cIL-10 and ebvIL-10, which share identical complex structures, have markedly different biological activities (Ding et al. 2001; Liu et al. 1997; Rousset et al. 1992; Yoon et al. 2012). In the case of ebvIL-10, the functional differences between ebvIL-10 and cIL-10 are largely due to ebvIL-10’s reduced binding affinity for IL-10R1 and not large structural changes in the receptor complex (Ding et al. 2001; Liu et al. 1997; Yoon et al. 2005).
2.4. cIL-10/IL-10R1 Site 1 Interface
Residues in the cIL-10/IL-10R1 site 1 interface are mostly polar (~70%) and cluster into two structurally distinct interaction surfaces, site Ia and site Ib (Josephson et al. 2001). Site Ia is centered on the bend in helix F and includes the AB loop while site Ib is located near the N-terminus of helix A and the C-terminus of helix F (Fig. 3b). Receptor binding loops L2-L4 from the IL-10R1 D1 domain interact exclusively with site Ia, while loops L5 and L6 from the D2 domain interact with site Ib.
Site Ia accounts for approximately 67% of the total buried surface area (~1000Å2) in the site I interface (Fig. 3c). IL-10R1 residues Tyr-43, Arg-76, and Arg-96 make extensive interactions in the interface. The phenolic group of Tyr-43 buries the most surface area of any IL-10R1 residue into a shallow cavity created by helix F and the AB loop. The base of the cavity is formed by cIL-10 residues Glu-142 and Lys-138 that each form hydrogen bonds to the OH of Tyr-43. Adjacent to Tyr-43, the N of Gly-44IL-10R1 forms a hydrogen bond with the carbonyl oxygen of AB loop residue Asp-44IL-10. An extensive hydrogen bond/salt-bridge network is also formed between Arg-96IL10R1 and Asp-144IL-10, Gln-38IL-10, and the carbonyl oxygen of Ser-141IL-10.
Site Ib is centered on the ion pair formed between Arg-27IL-10 and Glu-151IL-10 located on helices A and F, respectively (Fig. 3d). Glu-151IL-10 forms a series of hydrogen bonds with Ser-190IL-10R1 and Arg-191IL-10R1 located on the L6 loop. Additional interactions are formed between the carbonyl oxygens of Ser-190IL-10R1 and Arg-191IL-10R1 with the Nε atoms of Arg-27IL-10 and Arg-24IL-10, respectively. The interface also contains a small hydrophobic surface comprised of Phe-143IL-10R1 that packs into a cleft formed by Pro-20IL-10 and Ile-158IL-10. Additional hydrophobic surface area is donated to the interface through packing of the aliphatic portions of Arg-191IL-10R1 and Arg-27IL-10(Josephson et al. 2001).
2.5. IL-10R2 Chain
Although IL-10R2 exhibits low affinity for cIL-10, its overall architecture is similar to IL-10R1 (Figs. 2c, 2d) (Yoon et al. 2010). Like IL-10R1, IL-10R2 consists of two β-sandwich domains (D1 and D2) that are oriented at ~90° to one another. However, the IL-10R2 L2 and L5 loops adopt significantly different conformations than observed in IL-10R1. In particular, the L2IL-10R2 adopts a β-hairpin structure and L5IL-10R2 forms a “thumb-like” structure that extends away from the rest of the molecule. The conformations of the L2IL-10R2 hairpin and L5IL-10R2 thumb give rise to two clefts (L2/L3 and L3/L5 clefts) that are not observed in IL-10R1, or the ECDs of IL-22R1, IL-20R1 or IL-28R1 (Bleicher et al. 2008; Jones et al. 2008; Logsdon et al. 2012; Miknis et al. 2010; Yoon et al. 2010). In addition to unique structures, the L2, L3, and L5 loops are lined with aromatic residues (Tyr-56, Tyr-59, Tyr-82, Tyr-140, and Trp-143) that allow IL-10R2 to recognize IL-10, as well as IL-22, IL-26, IL-28a,b and IL-29 (Fig 4d) (Yoon et al. 2010). In support of these findings, structure-function studies, performed on the class 1 shared receptors gp130 and IL2γc, demonstrated aromatic residues Phe-169gp130 and Tyr-103γc form critical contacts at the center of the IL-6/gp130, IL-2/γc and IL-4/γc interfaces (Wang et al. 2009). Comparisons between IL-10R2, gp130, and IL2γc demonstrate Phe-169gp130, Tyr-103γc and Tyr-82IL10R2 are structurally conserved, which suggests the promiscuous binding cytokine receptors share a common origin (Yoon et al. 2010).
2.6. Model of the cIL-10/IL-10R1/IL-10R2 Signaling Complex
A structure-based model of the cIL-10/IL-10R1/IL-10R2 signaling complex has been generated using a mutagenesis / computational docking strategy (Fig. 1d)(Yoon et al. 2010). In the final model, the cIL-10 dimer assembles a symmetric complex containing two IL-10R1 chains and two IL-10R2 chains (Figs. 1c, 4b). In the ternary complex, IL-10R2 forms two contacts with the cIL-10/IL-10R1 binary complex. A site 2 interface between cIL-10 and IL-10R2 and a site 3 interface between the D2 domains of IL-10R1 and IL-10R2 (Fig. 1d). At this time, no experimental data directly confirms the formation of the site 3 interface. This is because mutational data used for the docking experiment characterized the cIL-10/IL-10R2 site 2 interface but not residues in the putative site 3 contact (Yoon et al. 2010). Interestingly, modeling IL-10R2 onto the recently determined IL-20/IL-20R1/IL-20R2 ternary complex structure, another IL-10 family cytokine, suggests the site 3 interface may not form (Logsdon et al. 2012). Small angle x-ray scattering experiments confirmed IL-10R2 adopts a rigid structure with a fixed D1/D2 domain orientation (Logsdon et al. 2012). Thus, IL-10R2 flexibility cannot be used to explain problems with the docking model. This suggests IL-20R2 and IL-10R2 recognition strategies may be very different. Interestingly, the crystal structure of another class 2 cytokine ternary complex, IFNα2/IFNAR1/IFNAR2, revealed the membrane proximal domain of the IFNAR1 chain, equivalent to the IL-10R2 D2 domain, is disordered (Thomas et al. 2011). Together, the data suggests the IL-10R1/IL-10R2 site 3 interface either does not form or is extremely weak and/or transient in nature.
In contrast to site 3, the structural model of site 2 is consistent with mutational analysis of cIL-10 and IL-10R2, which provides insight into how IL-10R2 is recruited into the binary cIL-10/IL-10R1 complex (Yoon et al. 2010; Yoon et al. 2006). The main features of the site 2 interface are three IL-10R2 binding loops (L2, L3, and L5, Fig. 4d) that recognize cIL-10 helices A and D in a “vertical” manner, as described for the cIL-10/IL-10R1 interaction (Fig. 4b). Met-22cIL-10, located between helices A and D, forms the center of the interface and packs against L3IL-10R2 residue Tyr-82IL-10R2 (Fig. 4d). On either side of L3IL-10R2, L2IL-10R2 and L5IL-10R2 form contacts with helices D and A, respectively. As predicted from analysis of the IL-10R2 structure, IL-10R2 aromatic residues, that when mutated to alanine disrupt cIL-10 binding, form several key contacts in the interface (Yoon et al. 2010; Yoon et al. 2006). In particular, L3 residue Ser-80IL10R2 forms contacts with Arg-32cIL-10, L2 residue Tyr-56IL10R2 forms contacts with Ser-93cIL-10 and L5 residue Tyr-140IL-10R2 contacts Ser-31cIL-10 (Fig. 4d).
3. IL-10R1 and IL-10R2 Binding Properties and Impact on Signaling
3.1. Binding and Sensor Properties of IL-10R1 and IL-10R2
The affinity of cIL-10 for cell surface IL-10R1 ranges from 50-200pM when measured on different cell lines (Ding et al. 2001; Liu et al. 1997; Tan et al. 1993). On a biacore chip surface, the KD of the cIL-10/IL-10R1 interaction was determined to be 240pM (Yoon et al. 2012). Thus, cell surface binding and in vitro surface plasmon resonance (SPR) analyses are in good agreement. Although IL-10R2 is essential for the biological activity of cIL-10 (Kotenko et al. 1997), cIL-10 cell binding affinity does not change whether the IL-10R2 chain is expressed on cells or not (Ding et al. 2001). Subsequent SPR studies estimated the cIL-10/IL10R2 binding affinity to be approximately 3mM (Logsdon et al. 2002). Additional in vitro binding studies measured an ~13-fold increase in the affinity of soluble IL-10R2 (KD = 234μM) for the cIL-10/IL-10R2 binary complex, relative to cIL-10 alone (Yoon et al. 2005). Even with the ~13-fold affinity increase, IL-10R2 interaction is approximately four orders of magnitude weaker than the cIL-10/IL-10R1 site 1 interaction.
As a result of the disparate IL-10R1 and IL-10R2 affinities, each receptor chain has a distinct function in activating IL-10 cellular responses. Specifically, IL-10R1 functions as the IL-10 binding chain which controls cell specificity and cellular targeting of IL-10 to immune cells that selectively express the IL-10R1 chain (Nagalakshmi et al. 2004; Wolk et al. 2002). The second function of the IL-10R1 chain is to regulate receptor occupancy time, which is controlled by the kinetics of the IL-10/IL-10R1 interaction. In contrast to IL-10R1, IL-10R2 functions as a sensor chain, which efficiently “senses” IL-10 bound to IL-10R1 (.e.g. the IL-10/IL-10R1 complex). Thus, the role of the IL-10R2 chain is to activate signaling based on the kinetics of the IL-10/IL-10R1 interaction. Because of its singular function, IL-10R2 can be used in similar signaling strategies of the other IL-10R2 binding cytokines, IL-22, IL-26, IL-28a/b and IL-29 (Donnelly et al. 2004; Jones et al. 2008; Yoon et al. 2010). This allows specific signaling responses from 6 cytokines using 5 receptors. Thus, the promiscuous binding IL-10R2, with its singular function, reduces the number of unique receptor chains required for IL-10 family cytokine signaling.
Not surprisingly, changes in site 1 (IL-10/IL-10R1) or site 2 (IL-10/IL-10R2) interfaces result in different biological outcomes (Ding et al. 2001; Raftery et al. 2004; Yoon et al. 2012; Yoon et al. 2006). Disruptions in IL-10 site 1, increases the effective concentration of the ligand required to induce one half of measured maximal biological responses (EC50). Despite increased EC50 values, site 1 mutants can still induce biological responses equivalent to cIL-10, at high protein concentrations (Yoon et al. 2012). In contrast, mutations in IL-10 site 2 cannot induce the same response levels observed for cIL-10, despite the addition of extremely high protein concentrations (Yoon et al. 2006). However, consistent with the low affinity of the IL10R2 chain, most mutations in IL-10 site 2 have little impact on IL-10 signaling, whereas most mutations made in the high affinity site 1 cause a measurable difference in biological activity. Overall, these findings are consistent with the roles of IL-10R1 and IL-10R2, as binding and sensor chains, respectively.
3.2. Reduced IL-10/IL-10R1 Affinity Prevents Signaling on Cells with Low IL-10R1 levels
The impact of reduced IL-10/IL-10R1 binding affinity on IL-10 cellular responses has been predominantly studied using ebvIL-10, which exhibits ~1000-fold lower affinity for IL-10R1 than cIL-10 (Ding et al. 2001; Liu et al. 1997; Yoon et al. 2012). Due to its weak affinity, ebvIL-10 was unable to signal on thymocytes, which express very low levels of IL-10R1, whereas cIL-10 signaled normally (Ding et al. 2001). Thus, ebvIL-10/IL-10R1 binding affinity is insufficient to assemble enough ebvIL-10/IL-10R1/IL-10R2 complexes to reach the threshold required for signaling. While EBV has presumably engineered ebvIL-10 to not be responsive on cells with low IL-10R1 levels, regulation of cell surface IL-10R1 levels appears to be a normal mechanism to regulate IL-10 signaling. Specifically, human neutrophils do not respond to cIL-10 unless they are activated by danger signals, such as LPS, which upregulates IL-10R1 expression allowing cIL-10 signaling (Crepaldi et al. 2001; Tamassia et al. 2008). In contrast to neutrophils, activated dendritic cells (DC) express lower levels of IL-10R1 than immature DC (Corinti et al. 2001; Kalinski et al. 1998). In contrast to IL-10R1, IL-10R2 has been shown to be constitutively expressed on essentially all cells (Nagalakshmi et al. 2004; Wolk et al. 2002).
3.3. Reduced IL-10/IL-10R1 Affinity Stimulates Signaling on Cells with High IL-10R1 levels
Although ebvIL-10 cannot signal on cells expressing low IL-10R1 levels, ebvIL-10 signaling is enhanced, relative to cIL-10, on cells (e.g. human B-cells) that express high IL-10R1 levels (Liu et al. 1997; Rousset et al. 1992; Yoon et al. 2012). To address this counter intuitive observation, a series of ebvIL-10/cIL-10 chimeras were expressed, purified, and tested for receptor binding and biological activity (Yoon et al. 2012). A unique feature of this study was the ebvIL-10/cIL-10 chimeras were produced as monomers and dimers to further address the importance of the IL-10 dimer in activating IL10 cellular responses. Yoon et al. found monomeric ebvIL-10/cIL-10 chimeras stimulated short-term proliferative responses that were directly proportional to IL-10R1 affinity (Yoon et al. 2012). Thus, IL-10 monomers exhibiting weak affinity for IL-10R1 exhibited weak cellular responses and monomers with high affinity for IL-10R1 exhibited enhanced biological activity. In contrast to experiments performed with the monomers, the biological activity of ebvIL-10/cIL-10 dimers was inversely proportional to IL-10R1 affinity. For example, dimers with high affinity for IL-10R1 (e.g. cIL-10) exhibited lower biological activity than dimers with reduced affinity for IL-10R1 (e.g. ebvIL-10). Thus, the ebvIL-10 dimer was essential for its enhanced signaling properties on cells expressing high levels of IL-10R1.
Receptor binding kinetics revealed ebvIL-10 exhibits very transient interactions (~10 seconds) with IL-10R1, before the complex falls apart (Yoon et al. 2012). In contrast, cIL-10/IL-10R1 interactions are very stable ( ~50 minutes). As a result, ebvIL-10 is very inefficient in forming the 1:2 ebvIL-10/IL-10R1 complex (see Fig. 1b). This suggests ebvIL-10 may activate cellular responses primarily by formation of 1:1 ebvIL-10/IL-10R1 complexes that are recognized by IL-10R2 as shown by Josephson et al. (Josephson et al. 2000) (e.g. Fig. 1a). How transient receptor binding enhances biological activity remains to be determined. One hypothesis is transient ebvIL-10/IL-10R1 interactions prevent the engagement of the IL-10R1 ubiquitination machinery, which prevents IL-10R1 internalization and signal termination (Jiang et al. 2011; Wei et al. 2006). A second hypothesis suggests that cells that express high levels of IL-10R1 capture the high affinity cIL-10 in non-functional cIL-10/IL-10R1 complexes that reduce its biological activity. This idea originated from biochemical studies that demonstrated the soluble cIL-10/IL-10R1 complex contains two cIL-10 dimers and four sIL-10R1 molecules (Tan et al., 1995). The solution stoichiometry of the complex is also observed in the cIL-10/IL-10R1 crystals, where two 1:2 cIL-10/IL-10R1 complexes are positioned adjacent to one another such that they block IL-10R2 binding (Josephson et al. 2001).
4. Structural Mechanisms Regulating IL-10/IL-10R1/IL-10R2 Assembly
4.1. Conserved IL-10 Site 1 Residues
To identify essential molecular features of IL-10 required for IL-10 receptor binding, the diverse sequences and structures of the cellular and viral IL-10s were compared, since all share the ability to engage IL-10R1 and IL-10R2 and initiate IL-10 signaling responses (Slobedman et al. 2009). This analysis revealed the subunit structures of ebvIL-10 and cmvIL-10 exhibit root mean square deviations (r.m.s.d.) of 0.5Å and 1.9Å, respectively, with cIL-10 (Yoon et al. 2005). The structural comparison identified eight residues (Leu-23, Arg-27, Lys-34, Gln-38, Ser-141, Asp-142, Asp-144, Glu-151, see Fig. 3d) whose sequence and structure were conserved in the site 1 interface (Jones et al. 2002). Not surprisingly, these residues participate in the extensive hydrogen binding networks observed in site 1a (Fig. 3c) and site 1b (Fig. 3d). However, they also form inter-peptide salt bridges between helices A and F in the domain-swapped dimer (Fig. 3e) (Jones et al. 2002; Zdanov et al. 1995). Thus, these conserved residues promote the folding and stability of IL-10, as well as forming critical interactions with IL-10R1.
4.2. IL-10/IL-10R1 Specificity: A Two-Point Recognition Model
While conserved residues in the IL-10 site 1 interface are important for IL-10R1 binding, they do not fully explain why IL-10 is specific for IL-10R1 and not other class 2 cytokine receptors, such as IL-22R1. To identify key molecular features that control IL-10/IL-10R1 specificity, structural comparisons of cellular and viral IL-10/IL-10R1 complexes (Jones et al. 2002; Josephson et al. 2001; Yoon et al. 2005), and IL-22/IL-22R1 complexes (Bleicher et al. 2008; Jones et al. 2008) were performed. This analysis identified the site 1a L2/AB loop recognition motif, which consists of Tyr-43IL-10R1 and Gly-44IL-10R1, as a critical specificity determinant (Fig. 3c). This is because the precise geometry of the extensive site 1a hydrogen bonds, positions the IL-10 subunit for additional IL-10R1 interactions in site 1b (Fig. 3d), which cannot be accurately reproduced in noncognate complexes (e.g., IL-22/sIL-10R1 or IL-10/sIL-22R1) without steric clashes between other regions of the molecules. Thus, the requirement for specific contacts in two spatially distinct regions of the interface, site1a and site 1b (Fig. 3b), provides critical constraints that ensure specificity between IL-10/IL-10R1 and other IL-10 family complexes, such as IL-22/IL-22R1.
4.3. IL-10 Conformational Changes Regulate IL-10/IL-10R1 Affinity and IL-10R2 Recruitment
A static picture of IL-10/IL-10R1 binding does not explain ebvIL-10 binding affinity or recruitment of IL-10R2 into the IL-10/IL-10R1 complex. Both cIL-10 and ebvIL-10 share the 8 conserved residues within the IL-10R1 binding epitope. However, ebvIL-10 exhibits ~1000-fold lower affinity for IL-10R1 than cIL-10 (Ding et al. 2001; Liu et al. 1997). Comparison of the amount of surface area buried in the cIL-10R1/IL-10R1 and ebvIL-10/IL-10R1 site 1 interfaces are very similar (Yoon et al. 2005). Thus, buried surface area does not explain the differences in IL-10R1 affinity between cIL-10 and ebvIL-10. However, closer examination revealed the ebvIL-10 AB loop is partially disordered in the ebvIL10/IL-10R1 complex (Yoon et al. 2005). In contrast, the cIL-10 AB loop is completely ordered in the cIL-10/IL-10R1 complex (Josephson et al. 2001; Yoon et al. 2005). Furthermore, the cIL-10 and ebvIL-10 AB loops are completely disordered in structures determined without the IL-10R1 chain (Fig. 4c) (Walter and Nagabhushan 1995; Zdanov et al. 1997). Taken together, this data suggests IL-10R1 binding promotes the ordering of the cIL-10 and ebvIL-10 AB loops. However, in the case of ebvIL-10, the transition to the ordered state is only partially completed. As a result of the increased mobility of the ebvIL-10 AB loop, the hydrogen bonds in site 1a are less precise (e.g. increased hydrogen bond lengths and poor geometry) than observed in the cIL-10/IL-10R1 complex, which as described in the two-point recognition model, disrupts interactions in site 1b and overall ebv/IL-10R1 affinity (Yoon et al. 2005).
To substantiate this model, a series of ebvIL-10/cIL-10 chimeras were tested for IL-10R1 binding affinity (Yoon et al. 2012). This study revealed changing just two residues in the ebvIL-10 AB loop (V43ebvIL-10 and A87ebvIL-10) to cIL-10 residues (L43cIL-10 and I87cIL-10) almost completely restored “cIL-10-like” IL-10R1 affinity (Yoon et al. 2012). Remarkably, residues L43cIL-10 and I87cIL-10 point into the IL-10 hydrophobic core and make no direct contacts with IL-10R1 (Yoon et al. 2005). Thus, ebvIL-10 contains packing defects in its hydrophobic core, which prevents the AB loop from effectively “locking” into an orientation that precisely positions ebvIL-10 site 1a and site1b IL-10R1 contacts for high affinity binding.
Conformational changes in cIL-10 that occur upon IL-10R1 binding were determined by comparing crystal structures of unbound cIL-10 and IL-10R1-bound cIL-10 (Fig. 4c) (Yoon et al. 2006). In addition to “ordering” the AB loop, IL-10R1 binding also induces conformational changes (exceeding 2Å) in the N-terminus of helix A and in the CD loop, which corresponds to the IL-10R2 site 2 binding site (Figs. 1d, 4d) (Yoon et al. 2006). Specific IL-10R2 binding residues (Fig. 4d) that undergo large conformational changes include Asn-21cIL-10, Met-22cIL-10, and Arg-32cIL-10, which are adjacent to IL-10R1 site 1a and site 1b. These conformational changes provide a structural mechanism to explain the ~13-fold increase in IL-10R2 affinity for the cIL-10/IL-10R1 complex compared to cIL-10 alone (Logsdon et al. 2002). The conformational coupling between cIL-10 site 1 and site 2 revealed from crystal structure analysis is further validated by mutations in cIL-10 site 2, which increase the affinity of the site 1 cIL-10/IL-10R1 interaction (Yoon et al. 2006).
5. Conclusions
Using crystal structures, receptor binding, protein engineering and functional studies, a unifying mechanism of IL-10 receptor assembly is beginning to appear. These data provide a molecular framework for understanding the diverse biological functions of IL-10 under normal and pathologic conditions. To date, the structural data has been used to design non-functional versions of rhesus cytomegalovirus IL-10 (RhcmvIL-10) for use in vaccination strategies against RhCMV, the nonhuman primate model of CMV (Crough and Khanna 2009; Yue and Barry 2008). Immunization of RhCMV-infected rhesus macaques with these non-function RhcmvIL-10 mutants stimulated antibodies that neutralize wildtype RhcmvIL-10 biological activity, but do not cross react with rhesus cellular IL-10 (Logsdon et al. 2011). Given the critical role of IL-10 in persistent viral infections, vaccine induced neutralization of cmvIL-10 biological activity could be an exciting new strategy to prevent CMV infection or re-infection.
Structural elucidation of the IL-10 signaling complex has also played an important role in understanding IL-10, IL-10R1, and IL-10R2 mutations, which are found in patients suffering from early onset inflammatory bowel disease (Eberhardt et al. 2012; Grundtner et al. 2009). Further studies will refine the ability to predict the biological properties of newly discovered mutations and might ultimately lead to short term therapies that restore IL-10 signaling to these patients while hematopoietic stem cell therapy is being considered.
While excellent progress has been made elucidating molecular mechanisms of IL-10 receptor binding and formation of the extracellular signaling complex, the accompanying intracellular steps required for activation remain largely unknown (Fig. 1). This problem is not specific to the IL-10 receptor complex, but is a general problem in the cytokine structural biology field. It should be noted that crystal structures of JAK1 and TYK2 kinase (JH1) domains, and a JAK pseudo kinase (JH2) domain, have been determined (Bandaranayake et al. 2012; Chrencik et al. 2010; Williams et al. 2009). However, this represents only ~50% of the full length JAK and TYK proteins (Haan et al. 2010). Furthermore, how these proteins assemble with the IL-10R1 and IL-10R2 ICDs, or how the JAKs alter their conformations upon IL-10 binding to the ECDs remains unknown. These are important fundamental questions that could lead to new strategies to harness IL-10 biological activities for therapies to treat inflammatory disease, viral infections, and even cancer.
Acknowledgement
This review is supported in part by the National Institutes of Health Grants AI047300, AI0473-S1, AI049342, and AI097629.
References
- Bandaranayake RM, Ungureanu D, Shan Y, Shaw DE, Silvennoinen O, Hubbard SR. Crystal structures of the JAK2 pseudokinase domain and the pathogenic mutant V617F. Nat Struct Mol Biol. 2012;19:754–9. doi: 10.1038/nsmb.2348. doi: 10.1038/nsmb.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan JF. Shared architecture of hormone binidng in type I and type II interferon receptors. Cell. 1990;61:753–754. doi: 10.1016/0092-8674(90)90182-e. [DOI] [PubMed] [Google Scholar]
- Blackburn SD, Wherry EJ. IL-10, T cell exhaustion and viral persistence. Trends Microbiol. 2007;15:143–6. doi: 10.1016/j.tim.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Bleicher L, de Moura PR, Watanabe L, Colau D, Dumoutier L, Renauld JC, Polikarpov I. Crystal structure of the IL-22/IL-22R1 complex and its implications for the IL-22 signaling mechanism. FEBS Lett. 2008;582:2985–92. doi: 10.1016/j.febslet.2008.07.046. doi: 10.1016/j.febslet.2008.07.046. [DOI] [PubMed] [Google Scholar]
- Brooks DG, Trifilo MJ, Edelmann KH, Teyton L, McGavern DB, Oldstone MB. Interleukin-10 determines viral clearance or persistence in vivo. Nat Med. 2006;12:1301–9. doi: 10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Magracheva E, Kozlov S, Fong S, Tobin G, Kotenko S, Wlodawer A, Zdanov A. Crystal structure of interleukin-19 defines a new subfamily of helical cytokines. J Biol Chem. 2003;278:3308–13. doi: 10.1074/jbc.M208602200. [DOI] [PubMed] [Google Scholar]
- Chang WL, Baumgarth N, Yu D, Barry PA. Human cytomegalovirus-encoded interleukin-10 homolog inhibits maturation of dendritic cells and alters their functionality. J Virol. 2004;78:8720–31. doi: 10.1128/JVI.78.16.8720-8731.2004. doi: 10.1128/JVI.78.16.8720-8731.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrencik JE, Patny A, Leung IK, Korniski B, Emmons TL, Hall T, Weinberg RA, Gormley JA, Williams JM, Day JE, Hirsch JL, Kiefer JR, Leone JW, Fischer HD, Sommers CD, Huang HC, Jacobsen EJ, Tenbrink RE, Tomasselli AG, Benson TE. Structural and thermodynamic characterization of the TYK2 and JAK3 kinase domains in complex with CP-690550 and CMP-6. J Mol Biol. 2010;400:413–33. doi: 10.1016/j.jmb.2010.05.020. doi: 10.1016/j.jmb.2010.05.020. [DOI] [PubMed] [Google Scholar]
- Corinti S, Albanesi C, la Sala A, Pastore S, Girolomoni G. Regulatory activity of autocrine IL-10 on dendritic cell functions. J Immunol. 2001;166:4312–8. doi: 10.4049/jimmunol.166.7.4312. [DOI] [PubMed] [Google Scholar]
- Crepaldi L, Gasperini S, Lapinet JA, Calzetti F, Pinardi C, Liu Y, Zurawski S, de Waal Malefyt R, Moore KW, Cassatella MA. Up-regulation of IL-10R1 expression is required to render human neutrophils fully responsive to IL-10. J Immunol. 2001;167:2312–22. doi: 10.4049/jimmunol.167.4.2312. [DOI] [PubMed] [Google Scholar]
- Crough T, Khanna R. Immunobiology of human cytomegalovirus: from bench to bedside. Clin Microbiol Rev. 2009;22:76–98. doi: 10.1128/CMR.00034-08. Table of Contents. doi: 10.1128/CMR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos AM, Ultsch M, Kossiakoff AA. Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science. 1992;255:306–12. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- Ding Y, Qin L, Kotenko SV, Pestka S, Bromberg JS. A single amino acid determines the immunostimulatory activity of interleukin 10. J Exp Med. 2000;191:213–24. doi: 10.1084/jem.191.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Qin L, Zamarin D, Kotenko SV, Pestka S, Moore KW, Bromberg JS. Differential IL-10R1 expression plays a critical role in IL-10-mediated immune regulation. J Immunol. 2001;167:6884–92. doi: 10.4049/jimmunol.167.12.6884. [DOI] [PubMed] [Google Scholar]
- Donnelly RP, Dickensheets H, Finbloom DS. The interleukin-10 signal transduction pathway and regulation of gene expression in mononuclear phagocytes. J Interferon Cytokine Res. 1999;19:563–73. doi: 10.1089/107999099313695. [DOI] [PubMed] [Google Scholar]
- Donnelly RP, Sheikh F, Kotenko SV, Dickensheets H. The expanded family of class II cytokines that share the IL-10 receptor-2 (IL-10R2) chain. J Leukoc Biol. 2004 doi: 10.1189/jlb.0204117. [DOI] [PubMed] [Google Scholar]
- Duell BL, Tan CK, Carey AJ, Wu F, Cripps AW, Ulett GC. Recent insights into microbial triggers of interleukin-10 production in the host and the impact on infectious disease pathogenesis. FEMS Immunol Med Microbiol. 2012;64:295–313. doi: 10.1111/j.1574-695X.2012.00931.x. doi: 10.1111/j.1574-695X.2012.00931.x. [DOI] [PubMed] [Google Scholar]
- Eberhardt MK, Chang WL, Logsdon NJ, Yue Y, Walter MR, Barry PA. Host immune responses to a viral immune modulating protein: immunogenicity of viral interleukin-10 in rhesus cytomegalovirus-infected rhesus macaques. PLoS One. 2012;7:e37931. doi: 10.1371/journal.pone.0037931. doi: 10.1371/journal.pone.0037931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejrnaes M, Filippi CM, Martinic MM, Ling EM, Togher LM, Crotty S, von Herrath MG. Resolution of a chronic viral infection after interleukin-10 receptor blockade. J Exp Med. 2006;203:2461–72. doi: 10.1084/jem.20061462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt KR, Shah N, Faizura-Yeop I, Kocacik Uygun DF, Frede N, Muise AM, Shteyer E, Filiz S, Chee R, Elawad M, Hartmann B, Arkwright PD, Dvorak C, Klein C, Puck JM, Grimbacher B, Glocker EO. Clinical outcome in IL-10- and IL-10 receptor-deficient patients with or without hematopoietic stem cell transplantation. J Allergy Clin Immunol. 2013;131:825–30. doi: 10.1016/j.jaci.2012.09.025. doi: 10.1016/j.jaci.2012.09.025. [DOI] [PubMed] [Google Scholar]
- Fickenscher H, Hor S, Kupers H, Knappe A, Wittmann S, Sticht H. The interleukin-10 family of cytokines. Trends Immunol. 2002;23:89–96. doi: 10.1016/s1471-4906(01)02149-4. [DOI] [PubMed] [Google Scholar]
- Finbloom DS, Winestock KD. IL-10 induces the tyrosine phosphorylation of tyk2 and Jak1 and the differential assembly of STAT1 alpha and STAT3 complexes in human T cells and monocytes. J Immunol. 1995;155:1079–90. [PubMed] [Google Scholar]
- Gad HH, Dellgren C, Hamming OJ, Vends S, Paludan SR, Hartmann R. Interferon-lambda is functionally an interferon but structurally related to the interleukin-10 family. J Biol Chem. 2009;284:20869–75. doi: 10.1074/jbc.M109.002923. doi: 10.1074/jbc.M109.002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schaffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D, Hatscher N, Pfeifer D, Sykora KW, Sauer M, Kreipe H, Lacher M, Nustede R, Woellner C, Baumann U, Salzer U, Koletzko S, Shah N, Segal AW, Sauerbrey A, Buderus S, Snapper SB, Grimbacher B, Klein C. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–45. doi: 10.1056/NEJMoa0907206. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundtner P, Gruber S, Murray SS, Vermeire S, Rutgeerts P, Decker T, Lakatos PL, Gasche C. The IL-10R1 S138G loss-of-function allele and ulcerative colitis. Genes Immun. 2009;10:84–92. doi: 10.1038/gene.2008.72. doi: 10.1038/gene.2008.72. [DOI] [PubMed] [Google Scholar]
- Haan C, Behrmann I, Haan S. Perspectives for the use of structural information and chemical genetics to develop inhibitors of Janus kinases. J Cell Mol Med. 2010;14:504–27. doi: 10.1111/j.1582-4934.2010.01018.x. doi: 10.1111/j.1582-4934.2010.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho AS, Liu Y, Khan TA, Hsu DH, Bazan JF, Moore KW. A receptor for interleukin 10 is related to interferon receptors. Proc Natl Acad Sci U S A. 1993;90:11267–71. doi: 10.1073/pnas.90.23.11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho AS, Wei SH, Mui AL, Miyajima A, Moore KW. Functional regions of the mouse interleukin-10 receptor cytoplasmic domain. Mol Cell Biol. 1995;15:5043–53. doi: 10.1128/mcb.15.9.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hor S, Pirzer H, Dumoutier L, Bauer F, Wittmann S, Sticht H, Renauld JC, de Waal Malefyt R, Fickenscher H. The T-cell lymphokine interleukin-26 targets epithelial cells through the interleukin-20 receptor 1 and interleukin-10 receptor 2 chains. J Biol Chem. 2004;279:33343–51. doi: 10.1074/jbc.M405000200. doi: 10.1074/jbc.M405000200. [DOI] [PubMed] [Google Scholar]
- Hsu DH, de Waal Malefyt R, Fiorentino DF, Dang MN, Vieira P, de Vries J, Spits H, Mosmann TR, Moore KW. Expression of interleukin-10 activity by Epstein-Barr virus protein BCRF1. Science. 1990;250:830–2. doi: 10.1126/science.2173142. [DOI] [PubMed] [Google Scholar]
- Jiang H, Lu Y, Yuan L, Liu J. Regulation of interleukin-10 receptor ubiquitination and stability by beta-TrCP-containing ubiquitin E3 ligase. PLoS One. 2011;6:e27464. doi: 10.1371/journal.pone.0027464. doi: 10.1371/journal.pone.0027464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BC, Logsdon NJ, Josephson K, Cook J, Barry PA, Walter MR. Crystal structure of human cytomegalovirus IL-10 bound to soluble human IL-10R1. Proc Natl Acad Sci U S A. 2002;99:9404–9. doi: 10.1073/pnas.152147499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BC, Logsdon NJ, Walter MR. Structure of IL-22 bound to its high-affinity IL-22R1 chain. Structure. 2008;16:1333–44. doi: 10.1016/j.str.2008.06.005. doi: 10.1016/j.str.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephson K, DiGiacomo R, Indelicato SR, Iyo AH, Nagabhushan TL, Parker MH, Walter MR. Design and analysis of an engineered human interleukin-10 monomer. J Biol Chem. 2000;275:25054. doi: 10.1074/jbc.275.18.13552. [DOI] [PubMed] [Google Scholar]
- Josephson K, Logsdon NJ, Walter MR. Crystal structure of the IL-10/IL-10R1 complex reveals a shared receptor binding site. Immunity. 2001;15:35–46. doi: 10.1016/s1074-7613(01)00169-8. [DOI] [PubMed] [Google Scholar]
- Kalinski P, Schuitemaker JH, Hilkens CM, Kapsenberg ML. Prostaglandin E2 induces the final maturation of IL-12-deficient CD1a+CD83+ dendritic cells: the levels of IL-12 are determined during the final dendritic cell maturation and are resistant to further modulation. J Immunol. 1998;161:2804–9. [PubMed] [Google Scholar]
- Kotenko SV. The family of IL-10-related cytokines and their receptors: related, but to what extent? Cytokine Growth Factor Rev. 2002;13:223–40. doi: 10.1016/s1359-6101(02)00012-6. [DOI] [PubMed] [Google Scholar]
- Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- Kotenko SV, Izotova LS, Mirochnitchenko OV, Esterova E, Dickensheets H, Donnelly RP, Pestka S. Identification of the functional interleukin-22 (IL-22) receptor complex: the IL-10R2 chain (IL-10Rbeta ) is a common chain of both the IL-10 and IL-22 (IL-10-related T cell-derived inducible factor, IL-TIF) receptor complexes. J Biol Chem. 2001;276:2725–32. doi: 10.1074/jbc.M007837200. [DOI] [PubMed] [Google Scholar]
- Kotenko SV, Krause CD, Izotova LS, Pollack BP, Wu W, Pestka S. Identification and functional characterization of a second chain of the interleukin-10 receptor complex. Embo J. 1997;16:5894–903. doi: 10.1093/emboj/16.19.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko SV, Saccani S, Izotova LS, Mirochnitchenko OV, Pestka S. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10) Proc Natl Acad Sci U S A. 2000;97:1695–700. doi: 10.1073/pnas.97.4.1695. doi: 97/4/1695 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–74. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- Lauw FN, Pajkrt D, Hack CE, Kurimoto M, van Deventer SJ, van der Poll T. Proinflammatory effects of IL-10 during human endotoxemia. J Immunol. 2000;165:2783–9. doi: 10.4049/jimmunol.165.5.2783. [DOI] [PubMed] [Google Scholar]
- Liu Y, de Waal Malefyt R, Briere F, Parham C, Bridon JM, Banchereau J, Moore KW, Xu J. The EBV IL-10 homologue is a selective agonist with impaired binding to the IL-10 receptor. J Immunol. 1997;158:604–13. [PubMed] [Google Scholar]
- Liu Y, Wei SH, Ho AS, de Waal Malefyt R, Moore KW. Expression cloning and characterization of a human IL-10 receptor. J Immunol. 1994;152:1821–9. [PubMed] [Google Scholar]
- Livnah O, Johnson DL, Stura EA, Farrell FX, Barbone FP, You Y, Liu KD, Goldsmith MA, He W, Krause CD, Pestka S, Jolliffe LK, Wilson IA. An antagonist peptide-EPO receptor complex suggests that receptor dimerization is not sufficient for activation. Nat Struct Biol. 1998;5:993–1004. doi: 10.1038/2965. [DOI] [PubMed] [Google Scholar]
- Lockridge KM, Zhou SS, Kravitz RH, Johnson JL, Sawai ET, Blewett EL, Barry PA. Primate cytomegaloviruses encode and express an IL-10-like protein. Virology. 2000;268:272–80. doi: 10.1006/viro.2000.0195. [DOI] [PubMed] [Google Scholar]
- Logsdon NJ, Deshpande A, Harris BD, Rajashankar KR, Walter MR. Structural basis for receptor sharing and activation by interleukin-20 receptor-2 (IL-20R2) binding cytokines. Proc Natl Acad Sci U S A. 2012;109:12704–9. doi: 10.1073/pnas.1117551109. doi: 10.1073/pnas.1117551109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon NJ, Eberhardt MK, Allen CE, Barry PA, Walter MR. Design and analysis of rhesus cytomegalovirus IL-10 mutants as a model for novel vaccines against human cytomegalovirus. PLoS One. 2011;6:e28127. doi: 10.1371/journal.pone.0028127. doi: 10.1371/journal.pone.0028127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon NJ, Jones BC, Josephson K, Cook J, Walter MR. Comparison of interleukin-22 and interleukin-10 soluble receptor complexes. J Interferon Cytokine Res. 2002;22:1099–112. doi: 10.1089/10799900260442520. [DOI] [PubMed] [Google Scholar]
- Lutfalla G, Gardiner K, Uze G. A new member of the cytokine receptor gene family maps on chromosome 21 at less than 35 kb from IFNAR. Genomics. 1993;16:366–73. doi: 10.1006/geno.1993.1199. doi: 10.1006/geno.1993.1199. [DOI] [PubMed] [Google Scholar]
- Miknis ZJ, Magracheva E, Li W, Zdanov A, Kotenko SV, Wlodawer A. Crystal structure of human interferon-lambda1 in complex with its high-affinity receptor interferon-lambdaR1. J Mol Biol. 2010;404:650–64. doi: 10.1016/j.jmb.2010.09.068. doi: 10.1016/j.jmb.2010.09.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- Moore KW, Vieira P, Fiorentino DF, Trounstine ML, Khan TA, Mosmann TR. Homology of cytokine synthesis inhibitory factor (IL-10) to the Epstein-Barr virus gene BCRFI. Science. 1990;248:1230–4. doi: 10.1126/science.2161559. [DOI] [PubMed] [Google Scholar]
- Mumm JB, Emmerich J, Zhang X, Chan I, Wu L, Mauze S, Blaisdell S, Basham B, Dai J, Grein J, Sheppard C, Hong K, Cutler C, Turner S, LaFace D, Kleinschek M, Judo M, Ayanoglu G, Langowski J, Gu D, Paporello B, Murphy E, Sriram V, Naravula S, Desai B, Medicherla S, Seghezzi W, McClanahan T, Cannon-Carlson S, Beebe AM, Oft M. IL-10 elicits IFNgamma-dependent tumor immune surveillance. Cancer Cell. 2011;20:781–96. doi: 10.1016/j.ccr.2011.11.003. doi: 10.1016/j.ccr.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi ML, Murphy E, McClanahan T, de Waal Malefyt R. Expression patterns of IL-10 ligand and receptor gene families provide leads for biological characterization. Int Immunopharmacol. 2004;4:577–92. doi: 10.1016/j.intimp.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Nagem RA, Colau D, Dumoutier L, Renauld JC, Ogata C, Polikarpov I. Crystal structure of recombinant human interleukin-22. Structure (Camb) 2002;10:1051–62. doi: 10.1016/s0969-2126(02)00797-9. [DOI] [PubMed] [Google Scholar]
- Pestka S, Krause CD, Sarkar D, Walter MR, Shi Y, Fisher PB. Interleukin-10 and Related Cytokines and Receptors. Annu Rev Immunol. 2004;22:929–979. doi: 10.1146/annurev.immunol.22.012703.104622. [DOI] [PubMed] [Google Scholar]
- Raftery MJ, Wieland D, Gronewald S, Kraus AA, Giese T, Schonrich G. Shaping phenotype, function, and survival of dendritic cells by cytomegalovirus-encoded IL-10. J Immunol. 2004;173:3383–91. doi: 10.4049/jimmunol.173.5.3383. [DOI] [PubMed] [Google Scholar]
- Rennick DM, Fort MM. Lessons from genetically engineered animal models. XII. IL-10-deficient (IL-10(−/−) mice and intestinal inflammation. Am J Physiol Gastrointest Liver Physiol. 2000;278:G829–33. doi: 10.1152/ajpgi.2000.278.6.G829. [DOI] [PubMed] [Google Scholar]
- Riley JK, Takeda K, Akira S, Schreiber RD. Interleukin-10 receptor signaling through the JAK-STAT pathway. Requirement for two distinct receptor-derived signals for anti-inflammatory action. J Biol Chem. 1999;274:16513–21. doi: 10.1074/jbc.274.23.16513. [DOI] [PubMed] [Google Scholar]
- Rousset F, Garcia E, Defrance T, Peronne C, Vezzio N, Hsu DH, Kastelein R, Moore KW, Banchereau J. Interleukin 10 is a potent growth and differentiation factor for activated human B lymphocytes. Proc Natl Acad Sci U S A. 1992;89:1890–3. doi: 10.1073/pnas.89.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santin AD, Hermonat PL, Ravaggi A, Bellone S, Pecorelli S, Roman JJ, Parham GP, Cannon MJ. Interleukin-10 increases Th1 cytokine production and cytotoxic potential in human papillomavirus-specific CD8(+) cytotoxic T lymphocytes. J Virol. 2000;74:4729–37. doi: 10.1128/jvi.74.10.4729-4737.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, Rennick DM, Sartor RB. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–31. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh F, Baurin VV, Lewis-Antes A, Shah NK, Smirnov SV, Anantha S, Dickensheets H, Dumoutier L, Renauld JC, Zdanov A, Donnelly RP, Kotenko SV. Cutting edge: IL-26 signals through a novel receptor complex composed of IL-20 receptor 1 and IL-10 receptor 2. J Immunol. 2004;172:2006–10. doi: 10.4049/jimmunol.172.4.2006. [DOI] [PubMed] [Google Scholar]
- Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, Ostrander C, Dong D, Shin J, Presnell S, Fox B, Haldeman B, Cooper E, Taft D, Gilbert T, Grant FJ, Tackett M, Krivan W, McKnight G, Clegg C, Foster D, Klucher KM. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63–8. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- Slobedman B, Barry PA, Spencer JV, Avdic S, Abendroth A. Virus-encoded homologs of cellular interleukin-10 and their control of host immune function. J Virol. 2009;83:9618–29. doi: 10.1128/JVI.01098-09. doi: 10.1128/JVI.01098-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Clausen BE, Kaisho T, Tsujimura T, Terada N, Forster I, Akira S. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10:39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- Tamassia N, Calzetti F, Menestrina N, Rossato M, Bazzoni F, Gottin L, Cassatella MA. Circulating neutrophils of septic patients constitutively express IL-10R1 and are promptly responsive to IL-10. Int Immunol. 2008;20:535–41. doi: 10.1093/intimm/dxn015. doi: 10.1093/intimm/dxn015. [DOI] [PubMed] [Google Scholar]
- Tan JC, Indelicato SR, Narula SK, Zavodny PJ, Chou CC. Characterization of interleukin-10 receptors on human and mouse cells. J Biol Chem. 1993;268:21053–9. [PubMed] [Google Scholar]
- Thomas C, Moraga I, Levin D, Krutzik PO, Podoplelova Y, Trejo A, Lee C, Yarden G, Vleck SE, Glenn JS, Nolan GP, Piehler J, Schreiber G, Garcia KC. Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell. 2011;146:621–32. doi: 10.1016/j.cell.2011.06.048. doi: 10.1016/j.cell.2011.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson-Snipes L, Dhar V, Bond MW, Mosmann TR, Moore KW, Rennick DM. Interleukin 10: a novel stimulatory factor for mast cells and their progenitors. J Exp Med. 1991;173:507–10. doi: 10.1084/jem.173.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivella DB, Ferreira-Junior JR, Dumoutier L, Renauld JC, Polikarpov I. Structure and function of interleukin-22 and other members of the interleukin-10 family. Cell Mol Life Sci. 2010;67:2909–35. doi: 10.1007/s00018-010-0380-0. doi: 10.1007/s00018-010-0380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MR. Structural analysis of IL-10 and Type I interferon family members and their complexes with receptor. Adv Protein Chem. 2004;68:171–223. doi: 10.1016/S0065-3233(04)68006-5. [DOI] [PubMed] [Google Scholar]
- Walter MR, Nagabhushan TL. Crystal structure of interleukin 10 reveals an interferon gamma-like fold. Biochemistry. 1995;34:12118–25. doi: 10.1021/bi00038a004. [DOI] [PubMed] [Google Scholar]
- Wang X, Lupardus P, Laporte SL, Garcia KC. Structural biology of shared cytokine receptors. Annu Rev Immunol. 2009;27:29–60. doi: 10.1146/annurev.immunol.24.021605.090616. doi: 10.1146/annurev.immunol.24.021605.090616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Nordt RM, Egen C, Wehinger J, Ludwig W, Gouilleux-Gruart V, Mertelsmann R, Finke J. Constitutive activation of STAT proteins in primary lymphoid and myeloid leukemia cells and in Epstein-Barr virus (EBV)-related lymphoma cell lines. Blood. 1996;88:809–16. [PubMed] [Google Scholar]
- Wehinger J, Gouilleux F, Groner B, Finke J, Mertelsmann R, Weber-Nordt RM. IL-10 induces DNA binding activity of three STAT proteins (Stat1, Stat3, and Stat5) and their distinct combinatorial assembly in the promoters of selected genes. FEBS Lett. 1996;394:365–70. doi: 10.1016/0014-5793(96)00990-8. [DOI] [PubMed] [Google Scholar]
- Wei SH, Ming-Lum A, Liu Y, Wallach D, Ong CJ, Chung SW, Moore KW, Mui AL. Proteasome-mediated proteolysis of the interleukin-10 receptor is important for signal downregulation. J Interferon Cytokine Res. 2006;26:281–90. doi: 10.1089/jir.2006.26.281. doi: 10.1089/jir.2006.26.281. [DOI] [PubMed] [Google Scholar]
- Williams NK, Bamert RS, Patel O, Wang C, Walden PM, Wilks AF, Fantino E, Rossjohn J, Lucet IS. Dissecting specificity in the Janus kinases: the structures of JAK-specific inhibitors complexed to the JAK1 and JAK2 protein tyrosine kinase domains. J Mol Biol. 2009;387:219–32. doi: 10.1016/j.jmb.2009.01.041. doi: 10.1016/j.jmb.2009.01.041. [DOI] [PubMed] [Google Scholar]
- Wilson EB, Brooks DG. The role of IL-10 in regulating immunity to persistent viral infections. Curr Top Microbiol Immunol. 2011;350:39–65. doi: 10.1007/82_2010_96. doi: 10.1007/82_2010_96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk K, Kunz S, Asadullah K, Sabat R. Cutting edge: immune cells as sources and targets of the IL-10 family members? J Immunol. 2002;168:5397–402. doi: 10.4049/jimmunol.168.11.5397. [DOI] [PubMed] [Google Scholar]
- Xie MH, Aggarwal S, Ho WH, Foster J, Zhang Z, Stinson J, Wood WI, Goddard AD, Gurney AL. Interleukin (IL)-22, a novel human cytokine that signals through the interferon receptor-related proteins CRF2-4 and IL-22R. J Biol Chem. 2000;275:31335–9. doi: 10.1074/jbc.M005304200. [DOI] [PubMed] [Google Scholar]
- Xu T, Logsdon NJ, Walter MR. Structure of insect-cell-derived IL-22. Acta Crystallogr D Biol Crystallogr. 2005;61:942–50. doi: 10.1107/S0907444905009601. [DOI] [PubMed] [Google Scholar]
- Yoon SI, Jones BC, Logsdon NJ, Harris BD, Deshpande A, Radaeva S, Halloran BA, Gao B, Walter MR. Structure and mechanism of receptor sharing by the IL-10R2 common chain. Structure. 2010;18:638–48. doi: 10.1016/j.str.2010.02.009. doi: 10.1016/j.str.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SI, Jones BC, Logsdon NJ, Harris BD, Kuruganti S, Walter MR. Epstein-Barr virus IL-10 engages IL-10R1 by a two-step mechanism leading to altered signaling properties. J Biol Chem. 2012;287:26586–95. doi: 10.1074/jbc.M112.376707. doi: 10.1074/jbc.M112.376707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SI, Jones BC, Logsdon NJ, Walter MR. Same structure, different function crystal structure of the Epstein-Barr virus IL-10 bound to the soluble IL-10R1 chain. Structure. 2005;13:551–64. doi: 10.1016/j.str.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Yoon SI, Logsdon NJ, Sheikh F, Donnelly RP, Walter MR. Conformational changes mediate interleukin-10 receptor 2 (IL-10R2) binding to IL-10 and assembly of the signaling complex. J Biol Chem. 2006;281:35088–96. doi: 10.1074/jbc.M606791200. [DOI] [PubMed] [Google Scholar]
- Yue Y, Barry PA. Rhesus cytomegalovirus a nonhuman primate model for the study of human cytomegalovirus. Adv Virus Res. 2008;72:207–26. doi: 10.1016/S0065-3527(08)00405-3. doi: 10.1016/S0065-3527(08)00405-3. [DOI] [PubMed] [Google Scholar]
- Zdanov A. Structural analysis of cytokines comprising the IL-10 family. Cytokine Growth Factor Rev. 2010;21:325–30. doi: 10.1016/j.cytogfr.2010.08.003. doi: 10.1016/j.cytogfr.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdanov A, Schalk-Hihi C, Gustchina A, Tsang M, Weatherbee J, Wlodawer A. Crystal structure of interleukin-10 reveals the functional dimer with an unexpected topological similarity to interferon gamma. Structure. 1995;3:591–601. doi: 10.1016/s0969-2126(01)00193-9. [DOI] [PubMed] [Google Scholar]
- Zdanov A, Schalk-Hihi C, Menon S, Moore KW, Wlodawer A. Crystal structure of Epstein-Barr virus protein BCRF1, a homolog of cellular interleukin-10. J Mol Biol. 1997;268:460–7. doi: 10.1006/jmbi.1997.0990. [DOI] [PubMed] [Google Scholar]
- Zdanov A, Schalk-Hihi C, Wlodawer A. Crystal structure of human interleukin-10 at 1.6 A resolution and a model of a complex with its soluble receptor. Protein Sci. 1996;5:1955–62. doi: 10.1002/pro.5560051001. [DOI] [PMC free article] [PubMed] [Google Scholar]