Abstract
This article highlights the defining principles, progress, and future directions in epigenetics research in relation to this special issue. Exciting studies in the fields of neuroscience, psychology, and psychiatry have provided new insights into the epigenetic factors (e.g. DNA methylation) that are responsive to environmental input and serve as biological pathways in behavioral development. Here we highlight the experimental evidence, mainly from animal models, that factors such as psychosocial stress and environmental adversity can become encoded within epigenetic factors with functional consequences for brain plasticity and behavior. We also highlight evidence that epigenetic marking of genes in one generation can have consequences for future generations (i.e. inherited), and work with humans linking epigenetics, cognitive dysfunction, and psychiatric disorder. Though epigenetics has offered more of a beginning than an answer to the centuries-old nature-nurture debate, continued research is certain to yield substantial information regarding biological determinants of CNS changes and behavior with relevance for the study of developmental psychopathology.
Keywords: DNA methylation, plasticity, memory, sensitive periods, transgenerational inheritance, stress
Introduction
Experiences, particularly those occurring during sensitive periods of development, are well-recognized for their ability to canalize neurobiological trajectories and yield significant consequences for life-long health and mental well-being. For some time now it has also been recognized that proper brain development and life-long function rely on the coordination of an extraordinarily complex set of neurodevelopmental events that involve genetic and environmental interactions. The past decade of behavioral epigenetics research has begun to shed light on mechanisms through which our experiences can interact with and become linked with our biology, providing a new framework to understand the brain’s ability to change as a result of experience (i.e. plasticity) and thus how behavior can arise.
Although epigenetic modifications were originally thought to only program patterns of gene expression during cellular development and differentiation, a growing body of research has forced us to realize that such modifications can occur in response to a range of environmental signals occurring not just in infancy but throughout the lifespan, and that these modifications have significance in regards to changes in gene regulation, neural plasticity, and behavior. To better understand the consequences of early- and later-life stress on epigenetic mechanisms in this capacity, this has required the utilization of experimental rodent models in which the timing and duration of exposure to stress could be manipulated and carefully controlled and the subsequent neurobiological outcomes assessed. Such experimental endeavors also revealed that acquired epigenetic information is capable of being passed to other generations in some cases, hence epigenetic alterations have emerged as a candidate biological pathway linking gene-environment interactions to multi-generational trajectories in behavioral development.
In this review, we will highlight the literature concerning these discoveries, paying particular attention to studies with implications for tenets central to the study of developmental psychopathology, particularly the examination of biological factors that facilitate behavioral change, mechanisms through which risk or protective factors operate to yield consequences for a phenotype, and objective measures of how we might define normal and abnormal development (Cicchetti, 1993, 2006; Sroufe & Rutter, 1984). We first discuss work linking epigenetics to learning and memory, the susceptibility to stress-related disorders, and cognitive impairment. We will then discuss rodent studies that have empirically demonstrated that epigenetic alterations occur in response to stress/trauma during and outside of sensitive periods of development to facilitate behavioral change. We also discuss studies in which the translation of these findings has been made to humans, and the idea that DNA methylation is a valuable biomarker indicative of norms or aberrations present at the molecular level. Finally, we end with suggestions of future directions we think are necessary to advance our understanding of epigenetics in plasticity.
Epigenetics overview
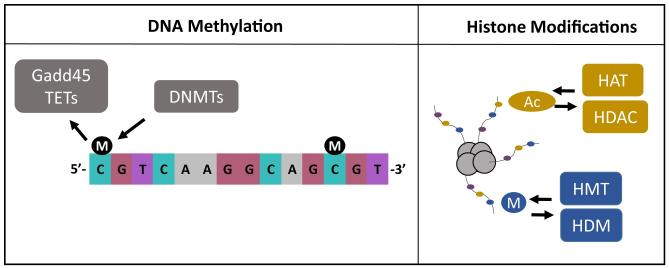
DNA methylation is an epigenetic modification that mainly occurs at cytosine residues of cytosine-guanine (CG) dinucleotides (Figure 1), though several studies have recently revealed that 5-methylcytosine (5mC) is also abundant at non-CG sites within the genome (Lister et al., 2013; Shirane et al., 2013). Once considered a static epigenetic modification responsible for programming patterns of gene expression during cellular development and differentiation, DNA methylation is now recognized for its capacity to be dynamically regulated throughout the lifespan. The predominant view in the literature is that methylation of DNA is associated with the suppression of gene transcription. The precise molecular processes through which DNA methylation can suppress gene transcription are complex, but in general methylated cytosines (cytosines are methylated via enzymes called DNA methyltransferases) can bind repressor proteins, including the methyl-binding domain protein MeCP2 and histone deacetylases (HDACs) (Moore, Le, & Fan, 2013). In line with this interpretation, most studies have been conducted under the framework that environmentally-driven increases in DNA methylation will correlate with sustained decreases in basal levels of gene expression.
Figure 1. Most commonly studied epigenetic mechanisms in plasticity and behavioral development.
The left panel is a schematic of DNA methylation occurring at CG dinucleotides, in which methyl groups (designated M) are added by DNA methyltransferase (DNMTs) enzymes. Gadd45b and TET proteins have been recently identified to actively demethylate the genome in response to environmental stimuli. Depicted in the panel on the right is epigenetic marking of histone tails, including the processes of acetylation (Ac) and methylation (M). Acetylation occurs when there is the addition of an acetyl group via an enzyme called histone acetyltransferase (HAT). Deacetylation occurs when the acetyl group is removed by enzymes called histone deacetylases (HDACs). In a similar fashion, methyl groups can be added via histone methyltransferases (HMTs) or removed by histone demethylases (HDMs).
It is important to note that while most evidence indicates that DNA methylation is associated with reduced gene activity, a handful of studies have indicated that DNA methylation can also be associated with transcriptional activation (Chahrour et al., 2008; Uchida et al., 2011). The complexity between the relation between DNA methylation and gene transcription is further realized when one considers that DNA methylation changes do not always result in basal changes in gene expression, but can instead prime transcriptional responses to subsequent stimuli and neural activation (Baker-Andresen, Ratnu, & Bredy, 2013). Members of the Gadd45 (Ma, Guo, Ming, & Song, 2009; Niehrs & Schäfer, 2012) and ten-eleven translocation (TET) (Guo, Su, Zhong, Ming, & Song, 2011; Williams, Christensen, & Helin, 2012) protein families are recently identified enhancers of active DNA demethylation, and the newly discovered 5-hydroxymethylcytosine (5hmC) intermediary (between a methylated and demethylated cytosine) is now commonly considered a sixth base within the genome (Münzel, Globisch, & Carell, 2011; Song & He, 2011). It should be noted that the conventional methods used for mapping 5-mC, such as bisulfite sequencing and methylation-sensitive restriction enzyme-based approaches, do not differentiate it from 5-hmC. As such, although we use the term ‘DNA methylation’ in this review to be consistent with the majority of primary publications to date, the term ‘DNA modification’ may be a more accurate descriptor.
The histone proteins have amino acid tails which protrude beyond the DNA (Figure 1), and these amino acid residues are prone to chemical modifications (Berger, 2007). We mention acetylation and methylation here, since these have been the most studied in terms of plasticity and behavior changes. The addition of an acetyl group (via enzymes called histone acetyltransferases, HATs) neutralizes the positive charge on histones, thereby decreasing the interaction with the negatively charged phosphates of DNA. Histone acetylation is rapid and reversible in an experience-dependent manner, but it too can be long-lived. Histone deacetylases (HDACs) are enzymes that remove the acetyl groups, and because HDACs have much structural diversity they are recognized plausible targets of therapeutic interventions to affect gene activity. Histone methylation too is a crucial regulator of behavioral change and this modification can either suppress or activate gene transcription depending upon which amino acid residue of the histone is targeted and the degree of methylation that occurs. Histone methylation is catalyzed by enzymes called histone methyltransferases (HMTs) while enzymes called histone demethylases (HDMs) catalyze demethylation. Together, histone modifications help regulate gene activity by integrating numerous responses to signal biochemical cascades and repelling/recruiting chromatin remodeling and transcription factors making gene loci either more or less available to transcriptional modulation (Berger, 2007; Kouzarides, 2007).
Increasing evidence is showing that genes, neural plasticity, and behavior can also be epigenetically regulated by noncoding RNAs, or RNA transcripts that have no apparent protein product. For example, microRNAs (miRNAs) are small, single-stranded RNAs with around 22 nucleotides that can silence gene expression through mRNA degradation, inhibition of translation, and destabilization (Bartel, 2009). Small noncoding RNAs, such as Piwi-interacting RNAs (piRNAs) that are slightly larger, around 26-32 nucleotides, have recently been shown to be expressed in neurons and methylate gene targets (Landry, Kandel, & Rajasethupathy, 2013).
Epigenetics modifications associated with plasticity and behavioral change outside of sensitive periods of development
There is a growing consensus that epigenetic regulation of gene transcription is an important component of adulthood cognitive processes. We begin here by summarizing studies with rodents consistent with the notion that environmentally-driven epigenetic tags are able to affect gene activity, creating functional changes in neurons and circuits that facilitate memory formation and prime the genome to respond to stimuli. Next, additional rodent studies are highlighted to illustrate that DNA methylation is also recognized as an epigenetic mediator of the stress response, associated with stress-related changes in behavior. Finally in this section, we discuss observations in humans and rodents that are consistent with the hypothesis that dysregulation of epigenetic mechanisms provides an explanation for symptoms associated with aging and psychiatric disorder.
Neural plasticity and memory
Work with the Marine mollusk Aplysia californica provided some of the first insight that epigenetics play a role in synaptic plasticity (Alberini, Ghirardl, Metz, & Kandel, 1994; Guan et al., 2002), and later experiments using neuronal cultures (Martinowich et al., 2003), brain slices (Levenson et al., 2006) or rodents in a Pavlovian fear conditioning paradigm (Bredy et al., 2007; Lubin, Roth, & Sweatt, 2008; Miller et al., 2010; Mizuno, Dempster, Mill, & Giese, 2012) significantly extended these observations by showing a host of rapid changes in methylation states of memory-linked genes and associated histone changes in the CNS. For example candidate gene approaches in this fashion have revealed concomitant changes in hippocampal Brain-derived neurotrophic factor (Bdnf) DNA methylation and gene expression that facilitate plasticity and memory formation (Lubin, et al., 2008; Mizuno, et al., 2012).
With the growing interest in mechanisms supporting active demethylation of the neuronal genome, several labs have now made the connection between TET proteins and cognition. For example, though able to form a normal associative fear memory, Tet1 knockout mice are impaired in their ability to extinguish the memory (Rudenko et al., 2013). Overexpression of Tet1 (via a viral-mediated approach) leads to increased 5hmC in hippocampal tissue that impairs hippocampal-dependent fear memory formation (Kaas et al., 2013). Additional work in rodents has corroborated the role of changes in DNA methylation in neural processes and epicenters supporting other forms of learning and memory, including novel object recognition (Munoz, Aspe, Contreras, & Palacios, 2010), successful navigation of the Morris water maze (Sultan, Wang, Tront, Liebermann, & Sweatt, 2012), and associative reward learning (Day et al., 2013).
Non-coding RNAs are incredibly responsive to environmental input and have been associated with processes underlying neural plasticity and behavioral change. One of the first such reports found an activity-dependent increase in expression of miR-128b in the infralimbic prefrontal cortex of mice in response to fear extinction training, which is proposed to facilitate extinction by negatively regulating genes associated with retrieval of the original fear memory (Lin et al., 2011). Additional work has shown experience-driven miRNAs in the hippocampus (Kye et al., 2011) and amygdala (Griggs, Young, Rumbaugh, & Miller, 2013) that work to facilitate fear memory formation. A recent and growing body of work on the small noncoding piRNAs has begun to illustrate their role in epigenetic control of memory formation. While miRNAs appear to target facilitators of neural plasticity, piRNAs instead methylate repressors (Landry, et al., 2013; Rajasethupathy et al., 2012).
Stress
Consistent with human physiological and neuroimaging studies, exposing rats to significant stress can produce alterations in stress physiology and modifications in the structure and sensitivity of several brain regions. Changes in hippocampal DNA methylation and histone acetylation have been observed in validated animal models of PTSD (Chertkow-Deutsher, Cohen, Klein, & Ben-Shachar, 2010; Hunter, McCarthy, Milne, Pfaff, & McEwen, 2009; Roth, Zoladz, Sweatt, & Diamond, 2011; Takei et al., 2011), with mounting evidence indicating that epigenetic changes at Bdnf loci not only facilitate fear memory produced by standard Pavlovian conditioning paradigms, but that epigenetic regulation of Bdnf may too be associated with long-lasting memory of traumas associated with PTSD (Roth, et al., 2011; Takei, et al., 2011).
An experimental paradigm commonly used to study the genetic and epigenetic precursors of stress-related psychiatric disorders, particularly depression, is chronic social defeat. In this paradigm, rodents are subjected to repeated aggressive encounters with another individual. The outcome of such a procedure is that this produces avoidance of subsequent social contact in some animals (deemed stress vulnerable) but not in others (resilient animals). Epigenetic regulation of hippocampal Bdnf is likewise altered by defeat stress, with increased repressive histone methylation modifications and concomitant decreases in particular Bdnf transcripts (Tsankova et al., 2006). Regulation of hippocampal Bdnf may also contribute to individual differences in vulnerability to social defeat stress, with epigenetic changes including increased histone acetylation and activation of Bdnf VI protecting against defeat-induced avoidance (Duclot & Kabbaj, 2013). Histone acetylation and the expression of histone modifying enzymes in the hippocampus, medial prefrontal cortex and dorsal raphe nucleus have also been found to correlate with behavioral outcomes associated with chronic social defeat stress (Kenworthy et al., 2013). Of course this form of stress can also have long-term effects on regulation of the hypothalamic-pituitary-adrenal (HPA) axis, and other groups have provided evidence that additional genes associated with HPA regulation are epigenetically modified by stress. Susceptible mice, or mice that spend less time in a social interaction zone after social defeat, have been found to display long-lived demethylation of hypothalamic corticotrophin releasing factor (CRF) gene, which produces an overactive HPA axis and social avoidance behaviors (Elliott, Ezra-Nevo, Regev, Neufeld-Cohen, & Chen, 2010). Resilient mice instead spend more time in the social interaction zone after defeat and do not display the same epigenetic changes.
Recent experimental work in laboratory settings has begun to illustrate the ability of mildly stressful experiences to evoke rapid epigenetic changes in the human genome. Participants following the Trier Social Stress Test (TSST) have been reported to show a short-lived increase in methylation of the oxytocin receptor gene (Unternaehrer et al., 2012). Consistent with the fact that the response to the TSST is known to differ for male and females, another study has reported greater methylation of the Nr3c1 gene after the TSST in females compared to males, which coincided with a decrease in salivary cortisol released during the TSST (Edelman et al., 2012). Other reports helping to experimentally establish a link between epigenetic patterns and human brain function include one demonstrating that greater stress and lower methylation of catechol-O-methyltransferase Val158 allele are correlated with more inefficient prefrontal activity (Ursini et al., 2011), and a second showing that DNA methylation of the gene encoding the oxytocin receptor is associated with individual variability in neural responses within brain regions supporting social perception (Jack, Connelly, & Morris, 2012).
Finally, we highlight a growing body of literature demonstrating the ability of parental traumatic exposure (as adults) to be inherited transgenerationally. Paternal transmission of stress-related behaviors induced by social defeat has been demonstrated (Dietz et al., 2011). Specifically, adult male mice that were exposed to chronic social defeat stress as well as control mice were bred with female mice that had never experienced any type of stress. Offspring were then assessed for anxiety- and depressive-like behaviors. Not only did chronic exposure to social defeat produce social avoidance behavior in fathers, both their male and female offspring too showed greater amounts of social avoidance behavior. Offspring of defeated fathers also showed reduced preference for sucrose and decreased latencies in immobility in the forced swim test, suggestive of depressive-like behavior. Remarkably, some of the transgenerational effects could even be replicated with in vitro fertilization experiments utilizing the father’s sperm.
Disruptions in social hierarchy in adolescence mice (modelled through repeatedly changing a rodent’s cagemate) has been shown to increase anxiety-like behaviors in both mothers and first generation females (Saavedra-Rodríguez & Feig, 2013). Fathers from this paradigm also appear able to transmit the anxiety and defective social interaction phenotypes to F2 and F3 daughters (Saavedra-Rodríguez & Feig, 2013). Exposure of adolescent female mice to an enriched environment on the other hand, consisting of novel objects, exercise, and increased capacity for social interaction, is known to have a beneficial effect on LTP induction and learning ability in her offspring (Arai, Li, Hartley, & Feig, 2009). The final study that we highlight here to demonstrate that environmental information experienced later in life can be inherited is one demonstrating that subjecting mice to fear conditioning with a novel odor before conception can alter behavioural sensitivity to that same odor (but not other odors) in F1 and F2 generation offspring (Dias & Ressler, 2014). Demonstrating an epigenetic influence independent of social transmission, the authors found differences in methylation of DNA associated with a specific olfactory receptor gene (Olfr151) that was present in both the fathers and offspring.
Cognitive dysfunction and psychiatric disorder
Over the last several years, the cognitive symptoms associated with aging and psychiatric disorders have begun to receive an epigenetic explanation. In regards to the cognitive decline associated with aging and Alzheimer’s disease, some very early work provided the first glimpses that there are age-dependent changes in methylation, particularly methylation associated with the amyloid precursor protein gene (Tohgi et al., 1999; West, Lee, & Maroun, 1995). Empirical studies have continued to provide support linking aging, epigenetic dysregulation, and learning and memory deficits. For example, one study utilized a mouse model of Alzheimer’s disease, and showed that increased histone acetylation achieved through HDAC inhibition increased dendrite sprouting and synapse formation, and enhanced Morris water maze performance (Fischer, Sananbenesi, Wang, Dobbin, & Tsai, 2007). In a second exemplary study, Activity regulated cytoskeletal-associated protein (Arc, a synaptic plasticity and memory-linked gene) transcripts were found downregulated in the hippocampus of aged rats (24–32 months) in comparison to adult rats (9–12 months), an effect attributed to aberrant DNA methylation of the Arc gene (Penner et al., 2011). Increased DNA methylation has been reported for several plasticity-related genes whose expression correlates with spatial behaviour and decreases with age (Haberman, Quigley, & Gallagher, 2012). Additional work with humans shows a dramatic change in the epigenetic landscape of the CNS with age (Lister, et al., 2013; Numata et al., 2012). Together data are consistent with the notion that the aged brain is characterized by accumulating epigenetic modifications, which can alter the expression or responsiveness of plasticity-related and memory-linked genes, with implications for brain and behavioral plasticity.
Nature vs. nurture questions have long plagued scientists in understanding mechanisms responsible for behavioral development and the etiology of psychiatric disorders. Though it has been difficult to link any one specific gene with their pathophysiology, numerous studies have provided compelling evidence for the contribution of gene-environment interactions. With the revolution of behavioural epigenetics, investigators then turned to epigenetic mechanisms as a plausible route for facilitating this interaction and whether these mechanisms may play a role in processes that contribution to the pathophysiology of psychiatric disorders.
An early hypothesis that emerged regarding Schizophrenia was that epigenetic regulation of developmental and plasticity-related genes was a significant contributing factor in the development of this disorder. Early post-mortem and animal model work focused on understanding the neurobiological underpinnings of Schizophrenia had long-suggested that deficiencies in the extracellular matrix protein reelin and GABA synthesis enzyme GAD67 play a significant role in the etiology of this disorder. When investigators began examining whether there was a link between epigenetic mechanisms and these events, they found that deficits in reelin GAD67 protein levels paralleled significant methylation alterations within the promoter regions of these genes (Abdolmaleky et al., 2005; Connor & Akbarian, 2008; Grayson et al., 2005; Huang & Akbarian, 2007). Genome-wide epigenetic approaches since have suggested there are hundreds of gene loci with altered DNA methylation in Schizophrenia, including other gene families related to GABAergic and neurotrophic function (Connor & Akbarian, 2008; Mill et al., 2008).
Epigenetic phenomena have similarly been associated with suicide and depression. Dnmt mRNA alterations (Poulter et al., 2008), increased Bdnf DNA methylation (Keller, Sarchiapone, Zarrilli, & et al., 2010), and altered methylation patterns of numerous genes that play a role in neuronal growth, development, and plasticity (Sabunciyan et al., 2012) have been found in the brains (within the frontal cortex, amygdala, and PVN) of individuals who committed suicide and/or had been diagnosed with major depression. Altered levels of Dnmt mRNA (Higuchi et al., 2011) and Bdnf DNA methylation have likewise been found in peripheral measures in patients with major depression (Fuchikami et al., 2011; Kang et al., 2013). Other findings in depressed patients include aberrant methylation of genes involved in cardiovascular health and regulation of the immune system (Uddin et al., 2011; Zill et al., 2012).
As PTSD, by definition, requires exposure to a traumatic event, and because genes within the CNS are exquisitely sensitive to stress, epigenetic alterations have received attention as possible contributors to the etiology and maintenance of PTSD. Indeed, some of the earliest work utilizing peripheral measures of methylation revealed strong associations between child abuse, total life stress, methylation of DNA associated with genes related to serotonin function (Koenen et al., 2011), immune regulation and plasticity, (Smith et al., 2011; Uddin et al., 2010) and the diagnosis of PTSD. Additional studies since have revealed an interaction between trauma and methylation status of other gene loci, including genomic repetitive elements (Rusiecki et al., 2013; Rusiecki et al., 2012), and genes involved in regulation of the HPA axis (Klengel et al., 2013; Yehuda et al., 2013a) and dopamine regulation and fear inhibition (Norrholm et al., 2013).
Compelling evidence is emerging that having a so-called risk allele and aberrant DNA methylation may be a better predictor of PTSD. For example, 9-repeat allele carriers of the gene encoding the dopamine transporter (SLC6A3) gene show an increased risk of lifetime PTSD when in conjunction with high methylation present in the SLC6A3 promoter (Chang et al., 2012). Methylation of and SNP variants of genes within the pituitary adenylate cyclase-activating polypeptide (PACAP) system, a system responsive to cellular stress and implicated in neurotrophic function (Ressler et al., 2011) or the dopamine regulator catechol-O-methyltransferase (Norrholm, et al., 2013), also appears to predict PTSD diagnosis or symptomatology. Finally, the risk of suffering from PTSD is significantly increased by exposure to early trauma in FKBP5 (a gene whose product is important in modulating the stress response) risk allele carriers with concomitant demethylation of cytosines within the FKBP5 gene (Klengel, et al., 2013). Together, observations have been consistent with the hypothesis that epigenetic marking of genes could underlie aspects of neuropsychiatric disorders that can be associated with environmental factors and abnormal brain function. Our current understanding is that epigenetic processes, acting either separately or in conjunction with genetic polymorphisms, serve as risk or protective factors responsible for long-term and even multi-generational trajectories in the development of psychiatric disorders.
Epigenetics modifications associated with sensitive periods of development
The maternal environment exerts a profound mediating role between environmental exposures and the neurodevelopmental plasticity that shapes behavioral outcomes. In mammals, this mediation can occur via alterations of the placenta at the maternal-fetal interface, alterations in maternal physiology pre- and post-natally affecting for example nutrition or circulating hormones, and changes in mother-offspring interactions during early postnatal life. A prominent and well-studied feature of maternal effects on neurodevelopment in offspring is its influence on the HPA axis, a major regulator of the endocrine response to environmental challenges. The regulation of circulating glucocorticoids maintains homeostatic energy balance across the circadian cycle (Landys, Ramenofsky, & Wingfield, 2006) and mediates physiological and behavioral responses to stress (Breuner, Patterson, & Hahn, 2008). Output from the stress axis begins with sensory input from environmental variation that initiates a cascade of endocrine responses from the hypothalamus, culminating with the release of glucocorticoids in the form of cortisol or corticosterone that feedback on a variety of neural circuitry (Love, McGowan, & Sheriff, 2012; McEwen, 2012).
Ecologists have long recognized that chronic stressors play key organizing roles in ecosystems via their actions on HPA activity. Indeed, the function of the HPA axis is highly conserved across vertebrate taxa, underscoring the biological importance of optimal glucocorticoid regulation (Breuner, et al., 2008). A diverse array of stressors can induce relatively permanent changes in the HPA axis of offspring via exposure to maternal stress during pre- and post-natal development, for example predation threat, the quality of the rearing environment, and the unpredictability of the social environment (Love, et al., 2012). The relative permanence of such changes in an ecological (natural world) context suggests that the effects of stress on HPA are adaptive responses that prepare offspring for environments where similar stressors are likely to be encountered (Clinchy et al., 2010; Love, et al., 2012).
The focus of ecologists on adaptive responses related to maternal stress (and perhaps a more obvious relationship to measures of fitness) is somewhat distinct from those of many human and laboratory animal studies that have focused on the role of early stress in psychopathology. Indeed, more is known about the pathways altered by adversity than other forms of early social experience. Large numbers of human epidemiological studies have indicated that early life experiences have enduring consequences for health in middle and later adulthood – including physical and mental health – as a consequence of establishing long-term health trajectories (Hertzman & Boyce, 2010; Sperry & Widom, 2013; Widom, Czaja, Bentley, & Johnson, 2012). For example, early adverse experiences such as physical abuse or neglect are well-known risk factors for mental health problems later in life (Sperry & Widom, 2013; Turecki, Ernst, Jollant, Labonte, & Mechawar, 2012). Childhood physical and sexual abuse impair intellectual function and increase the risk of affective disorders and suicide (Gould et al., 2012; Mann & Currier, 2010; Nemeroff, 2004; Nikulina & Widom, 2013). It has been proposed that adverse environmental experiences such as these during early life exert an enhanced impact on health trajectories in part because early postnatal development is a time of enhanced plasticity (Hanson, Godfrey, Lillycrop, Burdge, & Gluckman, 2010).
Elucidating the biological mechanisms underlying effects of stress and adverse experiences during development on later mental health is challenging in humans for reasons that include limited access to relevant biological material known to be affected by alterations in HPA function. However, studies in animal models have suggested that early-life stress directly impairs neuroplasticity in brain regions such as the hippocampus and has a lasting impact on endocrine systems underlying the response to psychosocial stress (McEwen, 2012; Meaney, 2001). In this section, we will focus on plasticity associated with the HPA axis, and highlight several studies of laboratory animals and humans that indicate a profound effect of parental care early in life on the epigenetic programming of genes sensitive to the effects of early care and stress-associated behaviors. In these studies, laboratory rodent models have been particularly useful in identifying mechanisms of epigenetic regulation in the brain that have then been used to generate hypotheses in humans.
Several decades of research in animal models has established that variations in maternal care induce long-term changes in gene expression in the brain of offspring. A variety of paradigms have been used to examine these effects, including experimenter-induced separation of pups and dams for varying lengths of time and monitoring the natural variation in maternal care exhibited by dams towards their offspring (Meaney, 2001). This research has found that early postnatal life – during approximately the first week of life in the rat – is a period sensitive to the effects of stress on long-term stress-related behavior and HPA function. The offspring of dams who naturally exhibit high levels of care show elevated levels of glucocorticoid receptors (GR) in the hippocampus, enhanced negative feedback sensitivity and a more modest response to stressors in adulthood (Liu et al., 1997). Cross fostering studies showed that this phenotype is directly attributable to maternal behavior rather than factors related to the prenatal environment, as offspring phenotype typically matches that of an adoptive dams rather than that of the biological dam (Francis, Diorio, Liu, & Meaney, 1999).
A series of landmark studies were initiated to examine putative epigenetic mechanisms involved in this long-term programming of gene expression. These studies indicated that the accompanying change in GR expression was regulated by DNA methylation of the GR17 splice variant in the hippocampus (Weaver et al., 2004; Weaver, Meaney, & Szyf, 2006). In vitro studies showed that site-specific DNA methylation inhibited the binding of NGFI-A, a transcription factor that drives GR expression, to its canonical recognition site (Weaver et al., 2007). GR17 is one of at least 11 untranslated first exons of the GR gene. Though GR is expressed in virtually all cell types, GR exon 1 splice variants regulate levels of expression in a tissue-specific manner (this is also true for the human GR exon 1 splice variants, as will be discussed later) (McCormick et al., 2000; Turner & Muller, 2005). In the hippocampus, GR17 was previously shown to vary in expression as a function of the average level of maternal care provided to a litter during early postnatal life (McCormick, et al., 2000). Interestingly, offspring of dams providing relatively high levels of maternal care showed demethylation of this promoter during the first week of life, while relatively high levels of DNA methylation persisted among the offspring of low maternal care dams, whereas coinciding with emergence of differences in maternal care between the two litter types. The results implied that DNA demethylation leads to an increased number of GRs and an attenuated response to stress, however the molecular mechanisms regulating site-specific DNA demethylation of the GR promoter remain unknown. DNA methylation differences were stable throughout adulthood in these animals, but were reversible by intra-cerebral infusion of Trichostatin A (TSA), a histone deacetylase inhibitor, which was also associated with increased gene expression in hundreds of other genes (Weaver, et al., 2006). In this study, the epigenomic response to TSA infusion was not examined. However, additional experiments indicated that the enzymes responsible for DNA methylation may be poised to act in the adult brain in response to methyl donor availability, as higher levels of DNA methylation of the GR17 promoter were observed among the offspring of high maternal care mothers given central infusions of the methyl donor l-methionine (Weaver, et al., 2006).
In a recent study, stress leading to altered NGFI-A levels was found not to alter DNA methylation of the NGFI-A response element in GR17, though other CG sites within the promoter were found differentially methylated (Witzmann, Turner, Meriaux, Meijer, & Muller, 2012). These data indicate that other factors in addition to NGFI-A may play a role in targeting DNA methylation/demethylation to the GR17 NGFI-A response element. It is likely that DNA methylation of GR17 gene expression involves the binding of additional transcription factors and/or is context and brain region specific. It is also likely that the GR17 is itself part of a response mechanism that involves additional splice variants of GR and other transcription factors.
We examined DNA methylation, H3K9 acetylation, and gene expression in a 7 million base pair region containing the GR gene in the rat hippocampus (McGowan et al., 2011). Epigenetic differences in adulthood that were associated with early maternal care occurred in statistically related clusters of up to 100KB but were nonetheless exquisitely patterned, whereby increased transcription was associated with hyperacetylated and hypermethylated exons, and hypomethylated promoters. We found epigenetic differences in association with altered transcription as a function of maternal care across several GR1 splice variants. Large epigenetic differences were noted in proximity to the transcription start site of GR, within the first coding exon (exon 2) and within GR introns, suggesting there may be additional regions of GR regulation via yet-to-be-identified non-coding RNAs within the GR locus. These data were the first to link epigenetic changes across both coding and non-coding regions in the mammalian brain, and implicate a non-random ‘epigenetic programming’ across large-scale loci in response to differences in early care. Accumulating evidence indicates that additional genes in neural pathway mediating the stress response are epigenetically regulated by DNA methylation of gene regulatory elements in association with early life stress, for example arginine vasopressin in the hypothalamus (Murgatroyd et al., 2009), Bdnf in the hippocampus (Roth, Lubin, Funk, & Sweatt, 2009) and GAD67 in the prefrontal cortex (Zhang et al., 2010).
These postnatal programming effects appear to derive from environmentally induced alterations of maternal-neonatal interactions, involving systems that determine the methylation patterns of GR gene promoter sequences and additional loci. It will be important to understand the precise nature of the maternal-neonatal interactions that mediate these changes. For example, there is evidence that artificial stimulation of pups with a paint-brush as a substitute for maternal licking can alter DNA methylation of a promoter region of the estrogen receptor alpha gene in the preoptic area of the hypothalamus (Kurian, Olesen, & Auger, 2010). These data have important implications for studies of transgenerational epigenetic effects of maternal care, via behavioral mechanism of inheritance rather than gametic inheritance, as maternal behavior is associated with levels of maternal care provided by offspring to their progeny (Champagne, Francis, Mar, & Meaney, 2003). Such transgenerational effects may be associated with adaptive functions of epigenetic programming, and may therefore be highly important source of transgenerational programming of behavioral and neural plasticity (Daxinger & Whitelaw, 2012). Collaborations among ecologists and neurobiologists will be important in addressing these questions in future studies. Nevertheless, there is mounting evidence that epigenetic mechanisms coordinate wide spread changes in gene expression in response to differences in early maternal care or adversity.
Human studies of epigenetic programming of the HPA and its consequences for plasticity and psychopathology rely on obtaining relevant tissue susceptible to epigenetic variation as a function of HPA dysregulation. There is evidence that some peripheral tissues may be informative in this regard. For example, recent research has identified DNA methylation of GR1F promoter, the human equivalent of the GR17 variant in rodents, in lymphocytes as a predictor of treatment outcome in PTSD patients (Yehuda et al., 2013b). These data suggest that GR promoter methylation in lymphocytes is under epigenetic control as a function of factors that alter HPA function.
We examined postmortem brain tissue from adults with well-characterized life histories to investigate the influence of early life adversity on GR DNA methylation in adults with a history of trauma. Our focus was on individuals with a history of severe physical or sexual abuse or neglect during childhood, which is common among suicide victims, and is an important risk factor for suicide (Turecki, et al., 2012). We examined the GR1F promoter in the hippocampus of human suicide victims and controls (McGowan et al., 2009). Family dysfunction and childhood adversity are linked to altered HPA stress responses and an increased risk for suicide. The promoter region we examined is upstream of one of several untranslated exon 1 splice variants that are known to regulate tissue-specific expression of GR, akin to the function that the GR exon 1 splice variants serve in the rodent (Turner & Muller, 2005). The study included three conditions: (1) suicide completers with a history of childhood abuse or severe neglect, (2) suicide completers without a history of childhood abuse or neglect and (3) individuals who has neither committed suicide nor had a history of childhood abuse or neglect. A fourth group of non-suicide victims with a history of abuse or neglect was not available, partly due to the fact that tissues from such a ‘control’ group are exceedingly rare, and were unavailable for our study. In this study, we found that the GR gene was differentially methylated among suicide victims with a history of abuse in childhood, but not among suicide victims with a negative history of childhood abuse, compared to control individuals without a history of suicide.
The data suggest that epigenetic processes might mediate the effects of the social environment during childhood on hippocampal gene expression and that stable epigenetic marks such as DNA methylation might then persist into adulthood and influence vulnerability for psychopathology through effects on intermediate levels of function such as activity of the HPA axis that regulates the stress response. However, it is still unclear whether the epigenetic aberrations were present in the germ line, whether they were introduced during embryogenesis, or whether they were truly changes occurring during early childhood. We also do not yet know the extent to which parental factors per se play a role in this phenotype. Despite these important caveats, these data were the first to link the early life environment to changes in the GR gene in humans. The data parallel that in the rodent study mentioned above, though in a very different context.
We have applied high throughput approaches to examining DNA methylation, chromatin modifications, and mRNA expression in gene regulatory, coding, intragenic and intergenic regions in humans in a study that paralleled that described above in rats. We analyzed the GR gene locus by interrogating a 7MB region containing the GR gene in hippocampi of adult suicide victims who were abused early in life compared to controls using high-throughput DNA microarray (Suderman et al., 2012). The GR gene locus shows substantial conservation with the same locus in rodents, with an almost identical order or orthologous genes across the locus. Like the study describe above in the rat (McGowan, et al., 2011), methylation levels were non-randomly distributed across the locus, indicating that stochastic processes are unlikely to account for the range of variation that we observed in this study. Proximal to the GR gene itself, we found a large region hypermethylated in suicide completers relative to controls within the first coding exon of the GR gene and its proximal promoters, extending previous observations of hypermethylation of the GR1F promoter among suicide victims with a history of abuse (McGowan, et al., 2009). This analysis also revealed differences in DNA methylation in intragenic regions of the GR gene.
At this time, we can only speculate that unrecognized non-coding RNAs may reside within this region and affect GR expression. Other differences were discovered within coding regions and the 3′ UTR of the GR gene. These data suggest that GR is epigenetically plastic in response to the early life social environment in both rodents and humans, though the specific alterations that we observed are not identical in both species (Suderman, et al., 2012). Nevertheless, the data indicate that the animal model of parental care may have broad applicability for translational studies aimed at understanding the consequences of epigenetic modification of GR in humans.
Future directions for the study of epigenetics in plasticity
Studies in a range of organisms have linked early life events to changes in neuroplasticity that have a lasting impact of endocrine systems mediating the response to stress (McEwen, 2012). However, significant challenges remain in linking studies of epigenetic mechanisms in laboratory animal models to translational human studies and to ecological studies examining ultimate explanations of epigenetic plasticity in the life history of the species. In this section, we will highlight several issues for future research relevant for this explanatory interplay.
First, mechanistic studies in animal models are hampered by a limited ability to target epigenetic modifications to select loci, although there has been progress in this regard (de Groote, Verschure, & Rots, 2012). In addition, knowledge about how specific environmental factors target select gene sequences remains poor, though we have discussed one such example in the effects of maternal care on the regulation of the GR17 promoter obtained from studies of the effects of maternal care in rodents. Enzymes that participate in DNA methylation and demethylation are non-specific, and must be directed to particular regions of the genome. Precisely how this occurs remains a significant challenge for the field. Transcriptional enhancers and repressors are known to recruit non-specific histone modifying enzymes to specific genomic loci and target specific genes (Jenuwein & Allis, 2001). For example, DNMT3a is known to interact with EZH2, which targets the DNA methylation-histone modification multi-protein complexes to specific sequences in DNA (Vire et al., 2005). These factors recognize specific cis-acing sequences in genes, bind to these sequences and attract specific chromatin modifying enzymes to genes through protein-protein interactions. Specific transacting factors are responsive to cellular signaling pathways that are activated by cell-surface receptors, and could thus serve as conduits for epigenetic change linking an environmental or physiological trigger at cell surface receptors with gene-specific chromatin alterations and the reprogramming of gene activity. Likewise, factors that interfere with the signaling pathway may result in chromatin alterations.
Second, a challenge in translating mechanistic results from animal studies to humans concerns access to relevant tissues. Tissue types are known to be sensitive to differences in constituent cell numbers, which could bias results (Lam et al., 2013; Suderman et al., 2013). Analysis of whole blood (Borghol et al., 2012; Naumova et al., 2012) and lymphocyte (Beach et al., 2013; Vijayendran, Beach, Plume, Brody, & Philibert, 2012) samples from individuals exposed to various forms of early-life adversity have consistently revealed aberrant methylation patterns that are present on a genome-wide scale. Peripheral cells such as lymphocytes do offer an avenue to examine the HPA, as lymphocytes are sensitive to endocrine modulation of HPA (e.g. (de Kloet et al., 2006). The most commonly available tissue however for human epigenetic studies is buccal cells from mouth swabs or saliva. Intriguingly, there is evidence that this tissue is responsive to early-life adversity (Essex et al., 2013; Yang et al., 2013). Buccal cells complement studies of adversity in neurons in the sense that they do represent cells with a common embryonic origin. Studies across tissue types in humans and animal models will provide a valuable means of identifying epigenetically plastic regions of the genome across cell-types in response to environmental factors.
Third, identifying the effects of specific environmental conditions on the range of epigenetic plasticity and neurobehavioral outcomes may shed light on the reasons for which particular regions of the genome respond to the environment in early life. For example, Barker’s hypothesis (Hales & Barker, 1992), the proposal that pathological outcomes resulted from reduced fetal growth, stimulated research on a variety of health-related conditions arising from early environmental exposures (Low, Gluckman, & Hanson, 2011). This research revealed that nutrition and parental care can alter health trajectories in a manner consistent with that of an adaptive response, as both early undernutrition and overnutrition can lead to the same pathological outcomes (i.e. metabolic syndrome, cardiovascular disorders; (Low, et al., 2011)). Thus, the range of responses to early adversity suggests instead that pathology may arise as a function of ‘mismatch’ between the early-life environment and the later-life environment rather than as a consequence of early-life dysfunction. This distinction is potentially important, because it implies that for animal and human studies, specific postnatal environmental conditions may exist in which pathological responses may instead confer an apparently adaptive advantage (see (Champagne et al., 2008)). Studies in wild animals existing in the context in which they have evolved will be particularly useful in understanding the ultimate causes of epigenetic plasticity (Clinchy, et al., 2010; Love, et al., 2012).
Finally, we point our here that we have only discussed epigenetic modifications throughout this review in the context of the nuclear genome. There is increasing evidence however that CNS mitochondrial DNA is also subject to methylation and hydroxymethylation (Chen, Dzitoyeva, & Manev, 2012; Dzitoyeva, Chen, & Manev, 2012; Iacobazzi, Castegna, Infantino, & Andria, 2013; Shock, Thakkar, Peterson, Moran, & Taylor, 2011). Though little attention has been given to these phenomena to date, the recent discovery of DNA methylation regulatory enzymes and proteins inside mitochondria (Chestnut et al., 2011; Dzitoyeva, et al., 2012; Shock, et al., 2011) has now led investigators to question whether mitochondrial DNA methylation changes are present under a variety of conditions (for example in aging (Dzitoyeva, et al., 2012) or in response to valproic acid (Chen, et al., 2012)). This has led to the emergence of a new field of mitochondrial epigenetics, and further research is warranted to explore whether environmentally-induced changes in mitochondrial DNA methylation play a role in the relationship between early-life adversity and psychopathology.
Concluding remarks
Since the birth of behavioral epigenetics research, we have gained fascinating insight into the link between regulation of chromatin structure and plasticity. Studies have revealed that environmental adversity, for example in the form of social stress or traumatic experiences, can become encoded within epigenetic factors that control gene activity. Together, it has become clear that epigenetic mechanisms are poised to facilitate gene-environment communication throughout our lifespan. Epigenetic effects may also have implications for the stress susceptibility and well-being of future generations, providing a molecular mechanism to explain the transgenerational continuity of the effects of, for example, abuse and trauma. We certainly still lack a complete understanding of the cause-and-effect role of epigenetic mechanisms in brain development, function, and plasticity, but continued exploration of the regulatory role of epigenetic processes in aspects of normal and abnormal brain and behavior development will continue to be an informative approach for understanding the biology of risk and resilience for cognitive dysfunction and psychiatric disorders.
Acknowledgments
Preparation of this review was supported in part by The National Institute of General Medical Sciences (1P20GM103653) to TLR and the Natural Sciences and Engineering Research Council of Canada (NSERC) to POM.
References
- Abdolmaleky HM, Cheng KH, Russo A, Smith CL, Faraone SV, Wilcox M, et al. Hypermethylation of the reelin (reln) promoter in the brain of schizophrenic patients: A preliminary report. American Journal of Medical Genetics. 2005;134:60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- Alberini CM, Ghirardl M, Metz R, Kandel ER. C/ebp is an immediate-early gene required for the consolidation of long-term facilitation in aplysia. Cell. 1994;76:1099–1114. doi: 10.1016/0092-8674(94)90386-7. [DOI] [PubMed] [Google Scholar]
- Arai JA, Li S, Hartley DM, Feig LA. Transgenerational rescue of a genetic defect in long-term potentiation and memory formation by juvenile enrichment. The Journal of Neuroscience. 2009;29:1496–1502. doi: 10.1523/JNEUROSCI.5057-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker-Andresen D, Ratnu VS, Bredy TW. Dynamic DNA methylation: A prime candidate for genomic metaplasticity and behavioral adaptation. Trends in neurosciences. 2013;36:3–13. doi: 10.1016/j.tins.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Bartel DP. Micrornas: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach SRH, Brody GH, Lei MK, Gibbons FX, Gerrard M, Simons RL, et al. Impact of child sex abuse on adult psychopathology: A genetically and epigenetically informed investigation. Journal of Family Psychology. 2013;27:3–11. doi: 10.1037/a0031459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [10.1038/nature05915] [DOI] [PubMed] [Google Scholar]
- Borghol N, Suderman M, McArdle W, Racine A, Hallett M, Pembrey M, et al. Associations with early-life socio-economic position in adult DNA methylation. International journal of epidemiology. 2012;41:62–74. doi: 10.1093/ije/dyr147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredy TW, Wu H, Crego C, Zellhoefer J, Sun YE, Barad M. Histone modifications around individual bdnf gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learning & Memory. 2007;14:268–276. doi: 10.1101/lm.500907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuner CW, Patterson SH, Hahn TP. In search of relationship between the acute adrenocortical response and fitness. General and Comparative Endocrinology. 2008;157:288–295. doi: 10.1016/j.ygcen.2008.05.017. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong STC, Qin J, et al. Mecp2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne DL, Bagot RC, van Hasselt F, Ramakers G, Meaney MJ, de Kloet ER, et al. Maternal care and hippocampal plasticity: Evidence for experience-dependent structural plasticity, altered synaptic functioning, and differential responsiveness to glucocorticoids and stress. Journal of Neuroscience. 2008;28:6037–6045. doi: 10.1523/JNEUROSCI.0526-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne FA, Francis DD, Mar A, Meaney MJ. Variations in maternal care in the rat as a mediating influence for the effects of environment on development. Physiolgy and Behavior. 2003;79:359–371. doi: 10.1016/s0031-9384(03)00149-5. [DOI] [PubMed] [Google Scholar]
- Chang S-C, Koenen KC, Galea S, Aiello AE, Soliven R, Wildman DE, et al. Molecular variation at the <italic>slc6a3</italic> locus predicts lifetime risk of ptsd in the detroit neighborhood health study. PLoS ONE. 2012;7:e39184. doi: 10.1371/journal.pone.0039184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Dzitoyeva S, Manev H. Effect of valproic acid on mitochondrial epigenetics. European Journal of Pharmacology. 2012;690:51–59. doi: 10.1016/j.ejphar.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertkow-Deutsher Y, Cohen H, Klein E, Ben-Shachar D. DNA methylation in vulnerability to post-traumatic stress in rats: Evidence for the role of the post-synaptic density protein dlgap2. The International Journal of Neuropsychopharmacology. 2010;13:347–359. doi: 10.1017/S146114570999071X. [DOI] [PubMed] [Google Scholar]
- Chestnut BA, Chang Q, Price A, Lesuisse C, Wong M, Martin LJ. Epigenetic regulation of motor neuron cell death through DNA methylation. The Journal of Neuroscience. 2011;31:16619–16636. doi: 10.1523/JNEUROSCI.1639-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchetti D. Developmental psychopathology: Reactions, reflections, projections. Developmental Review. 1993;13:471–502. [Google Scholar]
- Cicchetti D. Development and psychopathology. In: Cicchetti D, Cohen D, editors. Developmental psychopathology. 2nd ed Vol. Vol 1: Theory and Method. Wiley; New York: 2006. pp. 1–23. [Google Scholar]
- Clinchy M, Schulkin J, Zanette LY, Sheriff MJ, McGowan PO, Boonstra R. The neurological ecology of fear: Insights neuroscientists and ecologists have to offer one another. Frontiers in Behaviornal Neuroscience. 2010;4:21. doi: 10.3389/fnbeh.2011.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor CM, Akbarian S. DNA methylation changes in schizophrenia and bipolar disorder. Epigenetics. 2008;3:55–58. doi: 10.4161/epi.3.2.5938. [DOI] [PubMed] [Google Scholar]
- Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nature Reviews Genetics. 2012;13:153–162. doi: 10.1038/nrg3188. [DOI] [PubMed] [Google Scholar]
- Day JJ, Childs D, Guzman-Karlsson MC, Kibe M, Moulden J, Song E, et al. DNA methylation regulates associative reward learning. Nature Neuroscience. 2013;16:1445–1452. doi: 10.1038/nn.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groote ML, Verschure PJ, Rots MG. Epigenetic editing: Targeted rewriting of epigenetic marks to modulate expression of selected target genes. Nucleic Acids Research. 2012;40:10596–10613. doi: 10.1093/nar/gks863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kloet CS, Vermetten E, Geuze E, Kavelaars A, Heijnen CJ, Westenberg HG. Assessment of hpa-axis function in posttraumatic stress disorder: Pharmacological and non-pharmacological challenge tests, a review. Journal of Psychiatric Research. 2006;40:550–567. doi: 10.1016/j.jpsychires.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Dias BG, Ressler KJ. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nature Neuroscience, advance online publication. 2014;17:89–96. doi: 10.1038/nn.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz DM, LaPlant Q, Watts EL, Hodes GE, Russo SJ, Feng J, et al. Paternal transmission of stress-induced pathologies. Biological Psychiatry. 2011;70:408–414. doi: 10.1016/j.biopsych.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclot F, Kabbaj M. Individual differences in novelty seeking predict subsequent vulnerability to social defeat through a differential epigenetic regulation of brain-derived neurotrophic factor expression. The Journal of Neuroscience. 2013;33:11048–11060. doi: 10.1523/JNEUROSCI.0199-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzitoyeva S, Chen H, Manev H. Effect of aging on 5-hydroxymethylcytosine in brain mitochondria. Neurobiology of Aging. 2012;33:2881–2891. doi: 10.1016/j.neurobiolaging.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman S, Shalev I, Uzefovsky F, Israel S, Knafo A, Kremer I, et al. Epigenetic and genetic factors predict women’s salivary cortisol following a threat to the social self. PLoS ONE. 2012;7:e48597. doi: 10.1371/journal.pone.0048597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott E, Ezra-Nevo G, Regev L, Neufeld-Cohen A, Chen A. Resilience to social stress coincides with functional DNA methylation of the crf gene in adult mice. Nature neuroscience. 2010;13:1351–1353. doi: 10.1038/nn.2642. [10.1038/nn.2642] [DOI] [PubMed] [Google Scholar]
- Essex MJ, Boyce WT, Hertzman C, Lam LL, Armstrong JM, Neumann SM, et al. Epigenetic vestiges of early developmental adversity: Childhood stress exposure and DNA methylation in adolescence. Child Development. 2013;84:58–75. doi: 10.1111/j.1467-8624.2011.01641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai L-H. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [10.1038/nature05772] [DOI] [PubMed] [Google Scholar]
- Francis D, Diorio J, Liu D, Meaney MJ. Nongenomic transmission across generations of maternal behavior and stress responses in the rat. Science. 1999;286:1155–1158. doi: 10.1126/science.286.5442.1155. [DOI] [PubMed] [Google Scholar]
- Fuchikami M, Morinobu S, Segawa M, Okamoto Y, Yamawaki S, Ozaki N, et al. DNA methylation profiles of the brain-derived neurotrophic factor (bdnf) gene as a potent diagnostic biomarker in major depression. PLoS ONE. 2011;6:e23881. doi: 10.1371/journal.pone.0023881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould F, Clarke J, Heim C, Harvey PD, Majer M, Nemeroff CB. The effects of child abuse and neglect on cognitive functioning in adulthood. Journal of Psychiatric Research. 2012;46:500–506. doi: 10.1016/j.jpsychires.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson D, Jia X, Chen Y, Sharma RP, Mitchell C, Guidotti A, et al. Reelin promoter hypermethylation in schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griggs EM, Young EJ, Rumbaugh G, Miller CA. Microrna-182 regulates amygdala-dependent memory formation. The Journal of Neuroscience. 2013;33:1734–1740. doi: 10.1523/JNEUROSCI.2873-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Giustetto M, Lomvardas S, Kim JH, Miniaci MC, Schwartz JH, et al. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 2002;111:483–493. doi: 10.1016/s0092-8674(02)01074-7. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming G.-l., Song H. Emerging roles of tet proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle. 2011;10:2662–2668. doi: 10.4161/cc.10.16.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberman RP, Quigley CK, Gallagher M. Characterization of cpg island DNA methylation of impairment-related genes in a rat model of cognitive aging. Epigenetics. 2012;7:1008–1019. doi: 10.4161/epi.21291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales CN, Barker DJ. Type 2 (non-insulin-dependent) diabetes mellitus: The thrifty phenotype hypothesis. Diabetologia. 1992;35:595–601. doi: 10.1007/BF00400248. [DOI] [PubMed] [Google Scholar]
- Hanson M, Godfrey KM, Lillycrop KA, Burdge GC, Gluckman PD. Developmental plasticity and developmental origins of non-communicable disease: Theoretical considerations and epigenetic mechanisms. Progress in Biophysics and Molecular Biology. 2010;106:272–280. doi: 10.1016/j.pbiomolbio.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Hertzman C, Boyce T. How experience gets under the skin to create gradients in developmental health. Annual Review of Public Health. 2010;31:329–347. doi: 10.1146/annurev.publhealth.012809.103538. [DOI] [PubMed] [Google Scholar]
- Higuchi F, Uchida S, Yamagata H, Otsuki K, Hobara T, Abe N, et al. State-dependent changes in the expression of DNA methyltransferases in mood disorder patients. Journal of Psychiatric Research. 2011;45:1295–1300. doi: 10.1016/j.jpsychires.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Huang H-S, Akbarian S. Gad1 mrna expression and DNA methylation in prefrontal cortex of subjects with schizophrenia. PLoS ONE. 2007;2:e809. doi: 10.1371/journal.pone.0000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter RG, McCarthy KJ, Milne TA, Pfaff DW, McEwen BS. Regulation of hippocampal h3 histone methylation by acute and chronic stress. Proceedings of the National Academy of Sciences. 2009;106:20912–20917. doi: 10.1073/pnas.0911143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobazzi V, Castegna A, Infantino V, Andria G. Mitochondrial DNA methylation as a next-generation biomarker and diagnostic tool. Molecular Genetics and Metabolism. 2013;110:25–34. doi: 10.1016/j.ymgme.2013.07.012. [DOI] [PubMed] [Google Scholar]
- Jack A, Connelly JJ, Morris JP. DNA methylation of the oxytocin receptor gene predicts neural response to ambiguous social stimuli. Frontiers in Human Neuroscience. 2012;6 doi: 10.3389/fnhum.2012.00280. [Original Research] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kaas Garrett A., Zhong C, Eason Dawn E., Ross Daniel L., Vachhani Raj V., Ming G.-l., et al. Tet1 controls cns 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79:1086–1093. doi: 10.1016/j.neuron.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H-J, Kim J-M, Lee J-Y, Kim S-Y, Bae K-Y, Kim S-W, et al. Bdnf promoter methylation and suicidal behavior in depressive patients. Journal of Affective Disorders. 2013;151:679–685. doi: 10.1016/j.jad.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Keller S, Sarchiapone M, Zarrilli F, et al. Increased bdnf promoter methylation in the wernicke area of suicide subjects. Archives of General Psychiatry. 2010;67:258–267. doi: 10.1001/archgenpsychiatry.2010.9. [DOI] [PubMed] [Google Scholar]
- Kenworthy CA, Sengupta A, Luz SM, Ver Hoeve ES, Meda K, Bhatnagar S, et al. Social defeat induces changes in histone acetylation and expression of histone modifying enzymes in the ventral hippocampus, prefrontal cortex, and dorsal raphe nucleus. Neuroscience. 2013 doi: 10.1016/j.neuroscience.2013.01.024. [DOI] [PubMed] [Google Scholar]
- Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM, et al. Allele-specific fkbp5 DNA demethylation mediates gene-childhood trauma interactions. Nature Neuroscience. 2013;16:33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenen KC, Uddin M, Chang S-C, Aiello AE, Wildman DE, Goldmann E, et al. Slc6a4 methylation modifies the effect of the number of traumatic events on risk for posttraumatic stress disorder. Depression and Anxiety. 2011;28:639–647. doi: 10.1002/da.20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Snapshot: Histone-modifying enzymes. Cell. 2007;131:822–822.e821. doi: 10.1016/j.cell.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Kurian JR, Olesen KM, Auger AP. Sex differences in epigenetic regulation of the estrogen receptor-alpha promoter within the developing preoptic area. Endocrinology. 2010;151:2297–2305. doi: 10.1210/en.2009-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kye MJ, Neveu P, Lee Y-S, Zhou M, Steen JA, Sahin M, et al. Nmda mediated contextual conditioning changes mirna expression. PLoS One. 2011;6:e24682. doi: 10.1371/journal.pone.0024682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam LL, Emberly E, Fraser HB, Neumann SM, Chen E, Miller GE, et al. Reply to suderman et al.: Importance of accounting for blood cell composition in epigenetic studies. Proceedings of the National Academy of Sciences. 2013;110:E1247. doi: 10.1073/pnas.1222104110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CD, Kandel ER, Rajasethupathy P. New mechanisms in memory storage: Pirnas and epigenetics. Trends in neurosciences. 2013;36:535–542. doi: 10.1016/j.tins.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Landys MM, Ramenofsky M, Wingfield JC. Actions of glucocorticoids at a seasonal baseline as compared to stress-related levels in the regulation of periodic life processes. General and Comparative Endocrinology. 2006;148:132–149. doi: 10.1016/j.ygcen.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, et al. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. Journal of Biological Chemistry. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Lin Q, Wei W, Coelho CM, Li X, Baker-Andresen D, Dudley K, et al. The brain-specific microrna mir-128b regulates the formation of fear-extinction memory. Nature Neuroscience. 2011;14:1115–1117. doi: 10.1038/nn.2891. [DOI] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341 doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Diorio J, Tannenbaum B, Caldji C, Francis D, Freedman A, et al. Maternal care, hippocampal glucocorticoid receptors, and hypothalamic-pituitary-adrenal responses to stress. Science. 1997;277:1659–1662. doi: 10.1126/science.277.5332.1659. [DOI] [PubMed] [Google Scholar]
- Love OP, McGowan PO, Sheriff MJ. Maternal adversity and ecological stressors in natural populations: The role of stress axis programming in individuals, with implications for populations and communities. Functional Ecology. 2012 [Google Scholar]
- Low FM, Gluckman PD, Hanson MA. Developmental plasticity and epigenetic mechanisms underpinning metabolic and cardiovascular diseases. Epigenomics. 2011;3:279–294. doi: 10.2217/epi.11.17. [DOI] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of bdnf gene transcription in the consolidation of fear memory. The Journal of Neuroscience. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Guo JU, Ming G.-l., Song H. DNA excision repair proteins and gadd45 as molecular players for active DNA demethylation. Cell Cycle. 2009;8:1526–1531. doi: 10.4161/cc.8.10.8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann JJ, Currier DM. Stress, genetics and epigenetic effects on the neurobiology of suicidal behavior and depression. European Psychiatry. 2010;25:268–271. doi: 10.1016/j.eurpsy.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, et al. DNA methylation-related chromatin remodeling in activity-dependent bdnf gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- McCormick JA, Lyons V, Jacobson MD, Noble J, Diorio J, Nyirenda M, et al. 5′-heterogeneity of glucocorticoid receptor messenger rna is tissue specific: Differential regulation of variant transcripts by early-life events. Molecular Endocrinology. 2000;14:506–517. doi: 10.1210/mend.14.4.0438. [DOI] [PubMed] [Google Scholar]
- McEwen BS. Brain on stress: How the social environment gets under the skin. Proceedings of the National Academy of Sciences. 2012;109:17180–17185. doi: 10.1073/pnas.1121254109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonte B, Szyf M, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nature Neuroscience. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Suderman M, Sasaki A, Huang TC, Hallett M, Meaney MJ, et al. Broad epigenetic signature of maternal care in the brain of adult rats. PLoS ONE. 2011;6:e14739. doi: 10.1371/journal.pone.0014739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ. Maternal care, gene expression, and the transmission of individual differences in stress reactivity across generations. Annual Review of Neuroscience. 2001;24:1161–1192. doi: 10.1146/annurev.neuro.24.1.1161. [DOI] [PubMed] [Google Scholar]
- Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. The American Journal of Human Genetics. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, et al. Cortical DNA methylation maintains remote memory. Nature Neuroscience. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Dempster E, Mill J, Giese KP. Long-lasting regulation of hippocampal bdnf gene transcription after contextual fear conditioning. Genes, Brain and Behavior. 2012;11:651–659. doi: 10.1111/j.1601-183X.2012.00805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz PC, Aspe MA, Contreras LS, Palacios AG. Correlations of recognition memory performance with expression and methylation of brain-derived neurotrophic factor in rats. Biological Research. 2010;43:251–258. [PubMed] [Google Scholar]
- Münzel M, Globisch D, Carell T. 5-hydroxymethylcytosine, the sixth base of the genome. Angewandte Chemie International Edition. 2011;50:6460–6468. doi: 10.1002/anie.201101547. [DOI] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, Fischer D, et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nature Neuroscience. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Naumova OY, Lee M, Koposov R, Szyf M, Dozier M, Grigorenko EL. Differential patterns of whole-genome DNA methylation in institutionalized children and children raised by their biological parents. Development and Psychopathology. 2012;24:143–155. doi: 10.1017/S0954579411000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeroff CB. Neurobiological consequences of childhood trauma. Journal of Clinical Psychiatry. 2004;65:18–28. [PubMed] [Google Scholar]
- Niehrs C, Schäfer A. Active DNA demethylation by gadd45 and DNA repair. Trends in cell biology. 2012;22:220–227. doi: 10.1016/j.tcb.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Nikulina V, Widom CS. Child maltreatment and executive functioning in middle adulthood: A prospective examination. Neuropsychology. 2013;27:417–427. doi: 10.1037/a0032811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrholm SD, Jovanovic T, Smith A, Binder EB, Klengel T, Conneely K, et al. Differential genetic and epigenetic regulation of catechol-o-methyl-transferase (comt) is associated with impaired fear inhibition in posttraumatic stress disorder. Frontiers in Behavioral Neuroscience. 2013;7 doi: 10.3389/fnbeh.2013.00030. [Original Research] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata S, Ye T, Hyde Thomas M., Guitart-Navarro X, Tao R, Wininger M, et al. DNA methylation signatures in development and aging of the human prefrontal cortex. American Journal of Human Genetics. 2012;90:260–272. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penner MR, Roth TL, Chawla MK, Hoang LT, Roth ED, Lubin FD, et al. Age-related changes in arc transcription and DNA methylation within the hippocampus. Neurobiology of Aging. 2011;32:2198–2210. doi: 10.1016/j.neurobiolaging.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter MO, Du L, Weaver ICG, Palkovits M, Faludi G, Merali Z, et al. Gabaa receptor promoter hypermethylation in suicide brain: Implications for the involvement of epigenetic processes. Biological Psychiatry. 2008;64:645–652. doi: 10.1016/j.biopsych.2008.05.028. [DOI] [PubMed] [Google Scholar]
- Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, et al. A role for neuronal pirnas in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149:693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler KJ, Mercer KB, Bradley B, Jovanovic T, Mahan A, Kerley K, et al. Post-traumatic stress disorder is associated with pacap and the pac1 receptor. Nature. 2011;470:492–497. doi: 10.1038/nature09856. [10.1038/nature09856] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the bdnf gene. Biological Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Zoladz PR, Sweatt JD, Diamond DM. Epigenetic modification of hippocampal bdnf DNA in adult rats in an animal model of post-traumatic stress disorder. Journal of Psychiatric Research. 2011;45:919–926. doi: 10.1016/j.jpsychires.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko A, Dawlaty Meelad M., Seo J, Cheng Albert W., Meng J, Le T, et al. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79:1109–1122. doi: 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiecki JA, Byrne C, Galdzicki Z, Srikantan V, Chen L, Poulin M, et al. Ptsd and DNA methylation in select immune function gene promoter regions: A repeated measures case-control study of u.S. Military service members. Frontiers in Psychiatry. 2013;4 doi: 10.3389/fpsyt.2013.00056. [Original Research] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiecki JA, Chen L, Srikantan V, Zhang L, Yan L, Polin ML, et al. DNA methylation in repetitive elements and post-traumatic stress disorder: A case–control study of us military service members. Epigenomics. 2012;4:29–40. doi: 10.2217/epi.11.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra-Rodríguez L, Feig LA. Chronic social instability induces anxiety and defective social interactions across generations. Biological Psychiatry. 2013;73:44–53. doi: 10.1016/j.biopsych.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabunciyan S, Aryee MJ, Irizarry RA, Rongione M, Webster MJ, Kaufman WE, et al. Genome-wide DNA methylation scan in major depressive disorder. PLoS ONE. 2012;7:e34451. doi: 10.1371/journal.pone.0034451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirane K, Toh H, Kobayashi H, Miura F, Chiba H, Ito T, et al. Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-cpg methylation and role of DNA methyltransferases. PLoS Genet. 2013;9:e1003439. doi: 10.1371/journal.pgen.1003439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proceedings of the National Academy of Sciences. 2011;108:3630–3635. doi: 10.1073/pnas.1012311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AK, Conneely KN, Kilaru V, Mercer KB, Weiss TE, Bradley B, et al. Differential immune system DNA methylation and cytokine regulation in post-traumatic stress disorder. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2011;156:700–708. doi: 10.1002/ajmg.b.31212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C-X, He C. The hunt for 5-hydroxymethylcytosine: The sixth base. Epigenomics. 2011;3:521–523. doi: 10.2217/epi.11.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry DM, Widom CS. Child abuse and neglect, social support, and psychopathology in adulthood: A prospective investigation. Child Abuse & Neglect. 2013;37:415–425. doi: 10.1016/j.chiabu.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sroufe LA, Rutter M. The domain of developmental psychopathology. Child Development. 1984;55:17–29. [PubMed] [Google Scholar]
- Suderman M, Borghol N, Pappas JJ, McArdle W, Racine A, Hallett MT, et al. Epigenomic socioeconomic studies more similar than different. Proceedings of the National Academy of Sciences. 2013;110:E1246. doi: 10.1073/pnas.1221019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suderman M, McGowan PO, Sasaki A, Huang TC, Hallett MT, Meaney MJ, et al. Conserved epigenetic sensitivity to early life experience in the rat and human hippocampus. Proceedings of the National Academy of Sciences. 2012;109:17266–17272. doi: 10.1073/pnas.1121260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan FA, Wang J, Tront J, Liebermann DA, Sweatt JD. Genetic deletion of gadd45b, a regulator of active DNA demethylation, enhances long-term memory and synaptic plasticity. The Journal of Neuroscience. 2012;32:17059–17066. doi: 10.1523/JNEUROSCI.1747-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei S, Morinobu S, Yamamoto S, Fuchikami M, Matsumoto T, Yamawaki S. Enhanced hippocampal bdnf/trkb signaling in response to fear conditioning in an animal model of posttraumatic stress disorder. Journal of Psychiatric Research. 2011;45:460–468. doi: 10.1016/j.jpsychires.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Tohgi H, Utsugisawa K, Nagane Y, Yoshimura M, Ukitsu M, Genda Y. The methylation status of cytosines in a τ gene promoter region alters with age to downregulate transcriptional activity in human cerebral cortex. Neuroscience Letters. 1999;275:89–92. doi: 10.1016/s0304-3940(99)00731-4. [DOI] [PubMed] [Google Scholar]
- Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [10.1038/nn1659] [DOI] [PubMed] [Google Scholar]
- Turecki G, Ernst C, Jollant F, Labonte B, Mechawar N. The neurodevelopmental origins of suicidal behavior. Trends in Neuroscience. 2012;35:14–23. doi: 10.1016/j.tins.2011.11.008. [DOI] [PubMed] [Google Scholar]
- Turner JD, Muller CP. Structure of the glucocorticoid receptor (nr3c1) gene 5′ untranslated region: Identification, and tissue distribution of multiple new human exon 1. Journal of Molecular Endocrinology. 2005;35:283–292. doi: 10.1677/jme.1.01822. [DOI] [PubMed] [Google Scholar]
- Uchida S, Hara K, Kobayashi A, Otsuki K, Yamagata H, Hobara T, et al. Epigenetic status of gdnf in the ventral striatum determines susceptibility and adaptation to daily stressful events. Neuron. 2011;69:359–372. doi: 10.1016/j.neuron.2010.12.023. [DOI] [PubMed] [Google Scholar]
- Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de los Santos R, et al. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proceedings of the National Academy of Sciences. 2010;107:9470–9475. doi: 10.1073/pnas.0910794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Koenen KC, Aiello AE, Wildman DE, de los Santos R, Galea S. Epigenetic and inflammatory marker profiles associated with depression in a community-based epidemiologic sample. Psychological Medicine. 2011;41:997–1007. doi: 10.1017/S0033291710001674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unternaehrer E, Luers P, Mill J, Dempster E, Meyer AH, Staehli S, et al. Dynamic changes in DNA methylation of stress-associated genes (oxtr, bdnf) after acute psychosocial stress. Translational Psychiatry. 2012;2:e150. doi: 10.1038/tp.2012.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursini G, Bollati V, Fazio L, Porcelli A, Iacovelli L, Catalani A, et al. Stress-related methylation of the catechol-o-methyltransferase val158 allele predicts human prefrontal cognition and activity. The Journal of Neuroscience. 2011;31:6692–6698. doi: 10.1523/JNEUROSCI.6631-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayendran M, Beach S, Plume JM, Brody G, Philibert R. Effects of genotype and child abuse on DNA methylation and gene expression at the serotonin transporter. Frontiers in Psychiatry. 2012;3 doi: 10.3389/fpsyt.2012.00055. [Original Research] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The polycomb group protein ezh2 directly controls DNA methylation. Nature. 2005 doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, et al. Epigenetic programming by maternal behavior. Nature Neuroscience. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Weaver IC, D’Alessio AC, Brown SE, Hellstrom IC, Dymov S, Sharma S, et al. The transcription factor nerve growth factor-inducible protein a mediates epigenetic programming: Altering epigenetic marks by immediate-early genes. Journal of Neuroscience. 2007;27:1756–1768. doi: 10.1523/JNEUROSCI.4164-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Meaney MJ, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proceedings of the National Academy of Sciences. 2006;103:3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R, Lee J, Maroun L. Hypomethylation of the amyloid precursor protein gene in the brain of an alzheimer’s disease patient. Jouranl of Molecular Neuroscience. 1995;6:141–146. doi: 10.1007/BF02736773. [DOI] [PubMed] [Google Scholar]
- Widom CS, Czaja SJ, Bentley T, Johnson MS. A prospective investigation of physical health outcomes in abused and neglected children: New findings from a 30-year follow-up. American Journal of Public Health. 2012;102:1135–1144. doi: 10.2105/AJPH.2011.300636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Helin K. DNA methylation: Tet proteins-guardians of cpg islands? EMBO Reports. 2012;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzmann SR, Turner JD, Meriaux SB, Meijer OC, Muller CP. Epigenetic regulation of the glucocorticoid receptor promoter 1(7) in adult rats. Epigenetics. 2012;7:1290–1301. doi: 10.4161/epi.22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B-Z, Zhang H, Ge W, Weder N, Douglas-Palumberi H, Perepletchikova F, et al. Child abuse and epigenetic mechanisms of disease risk. American Journal of Preventive Medicine. 2013;44:101–107. doi: 10.1016/j.amepre.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R, Daskalakis NP, Desarnaud F, Makotkine I, Lehrner A, Koch E, et al. Epigenetic biomarkers as predictors and correlates of symptom improvement following psychotherapy in combat veterans with ptsd. Frontiers in Psychiatry. 2013a;4 doi: 10.3389/fpsyt.2013.00118. [Original Research] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R, Daskalakis NP, Desarnaud F, Makotkine I, Lehrner AL, Koch E, et al. Epigenetic biomarkers as predictors and correlates of symptom improvement following psychotherapy in combat veterans with ptsd. Front Psychiatry. 2013b;4:118. doi: 10.3389/fpsyt.2013.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TY, Hellstrom IC, Bagot RC, Wen X, Diorio J, Meaney MJ. Maternal care and DNA methylation of a glutamic acid decarboxylase 1 promoter in rat hippocampus. Journal of Neuroscience. 2010;30:13130–13137. doi: 10.1523/JNEUROSCI.1039-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zill P, Baghai TC, Schüle C, Born C, Früstück C, Büttner A, et al. DNA methylation analysis of the angiotensin converting enzyme (ace) gene in major depression. PLoS ONE. 2012;7:e40479. doi: 10.1371/journal.pone.0040479. [DOI] [PMC free article] [PubMed] [Google Scholar]