Abstract
Assays that target DNA or RNA (xNA) are highly sensitive, as small amounts of xNA can be amplified by PCR. Unfortunately, PCR is inconvenient in low resource environments, requiring equipment and power that may not be available in these environments. However, isothermal procedures that avoid thermal cycling are often confounded by primer dimers, off-target priming, and other artifacts. Here, we show how a “self avoiding molecular recognition system” (SAMRS) eliminates these artifacts to give clean amplicons in a helicase-dependent isothermal amplification (SAMRS-HDA). We also show that incorporating SAMRS into the 3′-ends of primers facilitates the design and screening of primers for HDA assays. Finally, we show that SAMRS-HDA can be twofold multiplexed, something difficult to achieve with HDA using standard primers. This shows that SAMRS-HDA is a more versatile approach than standard HDA with a broader applicability for xNA-targeted diagnostics and research.
Keywords: helicase dependent isothermal amplification, polymerase chain reaction, primer dimmer, nucleic acids, points-of-care diagnostics
Introduction
The polymerase chain reaction (PCR) is widely used to amplify DNA or RNA (collectively xNA) for research, clinical, and diagnostic applications.[1] However, the cost of PCR instruments and the power required for heating and cooling prevent PCR from being easily used in portable devices, including devices that might detect pathogens in the field, support diagnostics at points-of-care, or allow nucleic acid-targeted tests to be used at home.[2]
For these reasons, methods are sought to amplify xNA targets isothermally.[3] For example, transcription-mediated amplification (TMA)[4] and nucleic acid sequence-based amplification (NASBA)[5] combine reverse transcriptase, RNase H, and RNA polymerase to amplify RNA isothermally. Strand-displacement amplification (SDA)[6] combines restriction endonucleases and strand-displacing DNA polymerases to nick, extend, displace, and amplify DNA targets. Rolling circle amplification (RCA)[7] uses a strand-displacing DNA polymerase to extend primers on circular templates. Recombinase-polymerase amplification (RPA)[8] uses recombinase, single-stranded binding protein (SSB), and strand-displacing polymerase to allow primers to invade duplex DNA. Loop-mediated isothermal amplification (LAMP)[9] uses four primer pairs and a strand-displacing DNA polymerase to allow extension of a primer into a duplex.
Helicase-dependent amplification (HDA) is another isothermal amplification method.[10] Instead of heat, HDA uses helicase to separate strands in a duplex, allowing primers to invade the duplex and be extended by a strand-displacing polymerase. Like PCR, HDA amplifies xNA targets exponentially.[11] Its amplicons can be detected in real-time[12] and hand-held devices are now available for use in the field.[13] Further, when reverse transcriptase is added, HDA can be used to amplify RNA targets.[14]
Unfortunately, HDA has disadvantages, some shared with other isothermal methods, and others unique to the HDA architecture itself. Mixtures of primers at high concentrations in the presence of polymerases and triphosphates are prone to “mischief”. This is especially true during isothermal amplification, as the system is not forced at each cycle to revisit Watson-Crick pairing rules by a heating step.[15] Especially when run at 60 °C (or below), DNA strands can easily generate artifacts, including primer-dimers, off-target hybrids, and non-canonical folds.[16] The consequences include loss of signals, high backgrounds, low sensitivities, and false positives and negatives.
These problems can be mitigated in HDA by careful design of primer sequences. Indeed, those developing classical HDA assays generally must screen many primer combinations to find a pair that can produce a useful amplicon from a particular target.[19] This means that biology cannot, in general, drive selection of HDA primers.
Recognizing the severity of these problems, a blocked-primer helicase-dependent amplification (bpHDA) and quasi-hot start method has recently been developed.[20] In bpHDA, primers are constructed with a single ribonucleotide inserted four nucleotides upstream of a blocked 3′-end. This primer cannot be directly extended. However, after it is hybridized to its target, the RNA linkage is cleaved using a “hot start” RNase H2 over 50 °C, and the cleaved primer can be extended. However, bpHDA cannot amplify RNA targets, including the RNA viruses (such as, Ebola, dengue, chickungunya, and encephalitis) that are currently in the news.
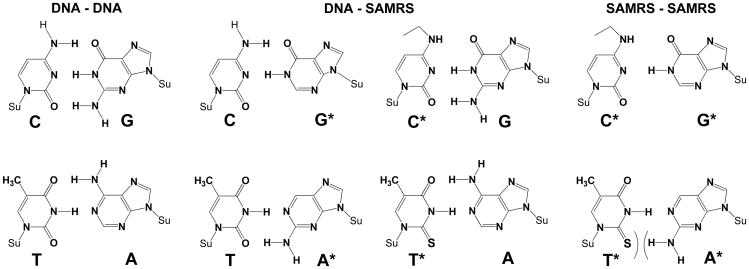
Recently, we introduced a “self-avoiding molecular recognition system” (SAMRS) as a class of DNA analogues designed to support multiplexed PCR.[21] In SAMRS DNA (Scheme 1), the A, T, G and C in natural DNA are replaced by A*, T*, G*, and C*, each with special pairing properties within a Watson-Crick geometry.[22] SAMRS A* pairs with natural T, T* pairs with natural A, G* pairs with natural C, and C* pairs with natural G (Scheme 1, middle), each with two hydrogen bonds. However, A* does not pair with T* and C* does not pair with G* (Scheme 1, right).
Scheme 1.
Chemical structures of the self-avoiding molecular recognition system (SAMRS). Pairs between standard nucleobases (left, DNA-DNA). Pairs between standard nucleobases (C, G, T, and A) and their SAMRS complements (G*, C*, A*, and T*), each joined by two hydrogen bonds in the standard Watson-Crick geometry (middle, DNA-SAMRS). Pairs between SAMRS nucleobases and their formal SAMRS complements; these do not contribute to duplex stability (right, SAMRS-SAMRS). Su = sugar backbone.
These pairing properties (related to a concept known as “pseudocomplementarity”) mean that SAMRS oligonucleotides bind to, and prime on, their natural DNA complements. However, SAMRS molecules do not bind to other SAMRS molecules, even if (formally) 100% complementary. Accordingly, SAMRS components placed near the 3′-ends of primers eliminate “primer-dimers” in standard PCR.[21]
Recently, we noted how SAMRS could improve the performance of recombinase-polymerase amplification (RPA).[23] Here, the long footprint of the primer (∼35 nucleotides must perfectly match) is a disadvantage for targeting rapidly evolving analytes, such as those from RNA viruses. Nevertheless, this success indicated how recombinase systems recognize “alien nucleotides”, and prompted us to ask whether SAMRS could also improve the performance of helicase dependent amplifications (HDA), which have a shorter specificity footprint.
Here, we show that SAMRS components do indeed improve HDA performance. We also show that SAMRS containing primers can be used in HDA to amplify both DNA and RNA targets. These data also suggest that SAMRS-HDA primers can be used without careful design; most sequences that we tried worked. Last, we show that SAMRS supports low-level multiplexing with HDA, something quite difficult for RPA and other isothermal amplification architectures.
Results and Discussion
We began by asking whether the unnatural SAMRS components are accepted by the strand-displacing DNA polymerases and helicase used in commercial HDA kits. Three sets of standard and SAMRS-containing primers (Tables S1) targeting three cancer genes (FLT3, KIT, TSHR) were taken directly from earlier PCR work and tested in HDA assays. With primers built from only standard nucleotides, HDA gave artifactual products with little if any of the desired amplicons, even though we used the most advanced, IsoAmp® III enzyme mix (BioHelix). This failure was observed despite the fact that the control target and primers provided in the BioHelix kit gave the desired amplicon (Figure S1, lane 7), indicating that the assay was working. In contrast, primers containing eight SAMRS nucleotides produced no primer artifacts (Figure S1, lanes 4, 5, and 6).
However, primers containing eight SAMRS nucleotides did not give amplicons either. We therefore asked how many SAMRS components must be put into primers to eliminate HDA artifacts, but still give amplicons. Here, we were guided by the hypothesis that because all SAMRS:standard pairs are joined by just two hydrogen bond equivalents (like the A:T pair), eight SAMRS components in the primers might destabilize the primer/target duplex too much for the enzyme mix in Biohelix kit. According to this hypothesis and independent observations, we reasoned that we could improve the SAMRS primer:template duplex stability while retaining the ability to avoid artifacts with primers that contained fewer SAMRS components. As an alternative hypothesis, different strand-displacing polymerases might accept SAMRS better than those in the IsoAmp® III kit.
Therefore, we tested the ability of different mutants of Bst and other strand-displacing DNA polymerases to amplify DNA with primers containing eight or four SAMRS components (Table 1 and Table S1). As shown in Figure S2, standard primers gave largely “primer-dimer” for all polymerases and conditions tested. In contrast, primers containing both four and eight SAMRS components gave no primer artifacts. Further, trace amounts of product were produced with primers containing four SAMRS components using GspSSD DNA polymerase (Figure S2, lane 16); no products were generated with primers containing eight SAMRS components regardless of the polymerase or conditions (Figure S2).
Table 1.
| Name | Sequence[a] |
|---|---|
| KIT-F-Std: | 5′-ACAAAGATTTGTGATTTTGGTCTAGCCAG-3′ |
| KIT-R-Std: | 5′-GGACTGTCAAGCAGAGAATGGGTACTCAC-3′ |
| KIT-F-SAMRS4: | 5′-ACAAAGATTTGTGATTTTGGTCTAGCCAG-3′ |
| KIT-R-SAMRS4: | 5′-GGACTGTCAAGCAGAGAATGGGTACTCAC-3′ |
| rpoB-Std-I-F: | 5′-GTCGCCGCGATCAAGGAGTTCTTCG-3′ |
| rpoB-Std-I-R: | 5′-TCAACCCCGACAGCGGGTTGTTCTG-3′ |
| rpoB-SAMRS-I-F: | 5′-GTCGCCGCGATCAAGGAGTTCTTCG-3′ |
| rpoB-SAMRS-I-R: | 5′-TCAACCCCGACAGCGGGTTGTTCTG-3′ |
| rpoB-Std-II-F: | 5′-GTCGCCGCGATCAAGGAGTTCTTCG-3′ |
| rpoB-Std-II-R: | 5′-GACAGTCGGCGCTTGTGGGTCAACC-3′ |
| rpoB-SAMRS-II-F: | 5′-GTCGCCGCGATCAAGGAGTTCTTCG-3′ |
| rpoB-SAMRS-II-R: | 5′-GACAGTCGGCGCTTGTGGGTCAACC-3′ |
Nucleotides in bold and underlined are SAMRS components.
The amounts of amplicons observed with primers containing four SAMRS components could be increased by adjusting the concentrations of MgSO4. In particular, higher MgSO4 (5 mM) produced sufficient amounts of amplicons with both Bst2.0 and GspSSD DNA polymerases (Figure S3).
To determine the sensitivity and limit of detection (LOD) of SAMRS-HDA with this optimization, we used the KIT cancer gene in human genomic DNA as a target. Here, ten-fold serial dilutions of human genomic DNA were amplified by HDA using both SAMRS-containing primers and standard primers with the GspSSD polymerase added to the commercial IsoAmp® II enzyme mix (BioHelix), recognizing that the IsoAmp® II kit has a higher amplification efficiency over the IsoAmp III kit. The isothermal amplification assay was monitored in real-time (Figure S4); formation of the desired amplicon was confirmed by agarose gel electrophoresis (Figure 1).
Figure 1.
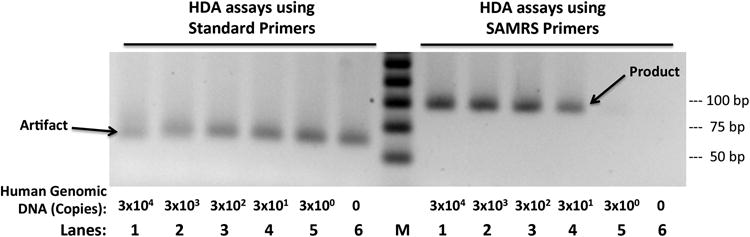
Agarose gel (2.5%) resolving amplicons from HDA assays to estimate the limit of detection of KIT gene in human genomic DNA. (Left) With standard primers, HDA assays produce only artifacts. (Right) With SAMRS-containing primers, HDA assays produce the desired amplicon products. M is a 25 bp DNA ladder.
Here, the impact of SAMRS was striking. HDA with standard primers gave only artifacts. In contrast, HDA with primers containing four SAMRS components gave the desired amplicon. Further, the dynamic range was impressive. As few as 30 copies of genomic DNA were detected by SAMRS-HDA, with the assay responding quantitatively over four orders of magnitude in target concentration (Figure 1).
To further evaluate and compare the standard primers with analogous SAMRS-containing primers, two sets of standard HDA primers (Table 1) were designed to meet the parameter settings provided by BioHelix. These targeted the core region of the beta subunit of the RNA polymerase (rpoB) gene from Mycobacterium tuberculosis; this genetic region includes mutations that confer multidrug-resistance. Two sets of SAMRS-containing primers (rpoB-SAMRS-I and rpoB-SAMRS-II, Table 1) were derived from the “well-designed” standard primers (rpoB-Std-I and rpoB-Std-II, Table 1). Simulants of the rpoB gene were amplified by standard primers and SAMRS-containing primers. Standard primers produced the desired amplicons in the presence of 1 ng of target DNA (Figure 2, left, lanes 1 and 4).
Figure 2.
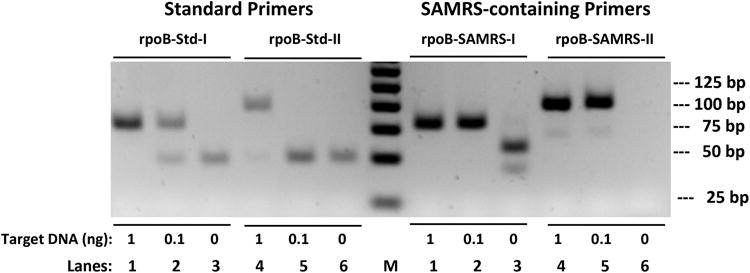
HDA assays using standard primers (left) or the analogous SAMRS-containing primers (right) to amplify the rpoB gene. Lanes 1 and 4: 1 ng (2.6 × 108 copies) of input target DNA. Lanes 2 and 5: 0.1 ng (2.6 × 107 copies) of input target DNA. Amplicons from HDA assays were resolved on agarose gel (2.5%) and stained with ethidium bromide. M indicates 25 bp DNA ladder.
However, as the amounts of target DNA were reduced to 0.1 ng, the amount of amplicon decreased and artifacts emerged with standard primers. As shown in Figure 2 (left, lanes 2 and 5), standard rpoB-Std-I primers produced both desired amplicon and primer-dimer in a ratio of 6:4; standard rpoB-Std-II primers produced no amplicon, only artifact. The concentrations of standard primers (from 0.05 to 0.2 μM), MgSO4 (4 - 6 mM), and additives (betaine) were tested as well, however, none of these conventional means had the capability to fully avoid the artifacts (data not shown).
In contrast, both SAMRS-containing primer sets gave the desired amplicons for all target concentrations (Figure 2, right, lanes 1, 2, 4, and 5), with essentially no artifact if any template at all present. As few as ∼300 copies of the target were detected in SAMRS-HDA under conditions where standard primers gave no products at all, only artifacts (data not shown).
Given the success of SAMRS-HDA for amplifying DNA targets, we asked if the SAMRS-HDA platform could be adapted to RNA targets. This requires, of course, a moderately thermostable reverse transcriptase (RT) that initially converts the RNA target to cDNA using SAMRS-containing primers, followed by amplification of the resulting cDNA using SAMRS-HDA.
For RT-SAMRS-HDA, several thermostable RTs were tested, including ThermoScript RT, Enhanced Avian RT, AffinityScript RT, SuperScript III RT, Maxima RT, and RevertAid Premium RT. The target was a conserved region in the HIV gag gene in a sample of HIV-1 “armored RNA” (Asuragen).
Again, primers contained four SAMRS components near their 3′-ends (Table S2). Most of the reverse transcriptases tested in RT-SAMRS-HDA worked well, regardless of the target. The detailed performance of the RT-SAMRS-HDA depended, however, on both the type and amount of reverse transcriptase used. Enhanced Avian RT proved to be the preferred choice for both high efficiency and specificity. As shown in Figure S5, amplicons from 2000 and 200 copies of HIV-1 armored RNA were easily detected when ThermoScript and Enhanced Avian RTs were used.
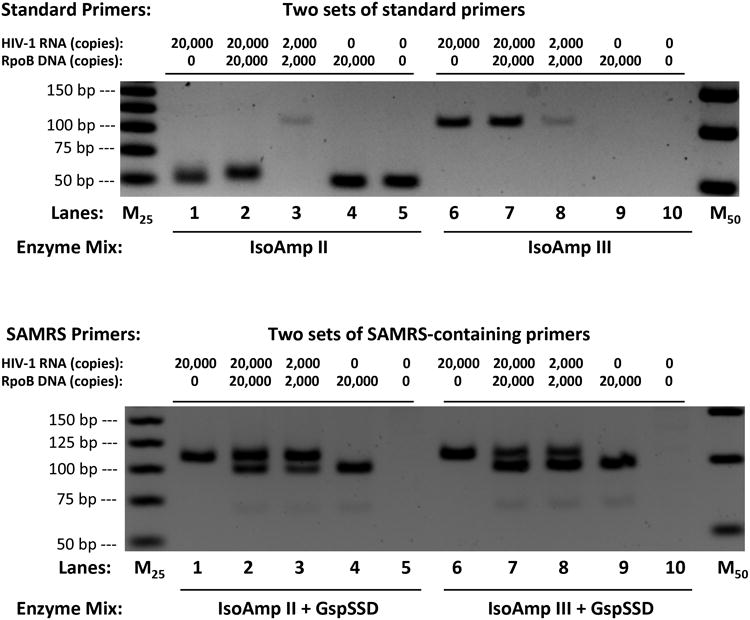
To extend RT-SAMRS-HDA to target complete genomic HIV-1 viral RNA, six primer pairs were designed to cover conserved regions and regions that give rise to drug resistance within three genes (gag, env, and pol). SAMRS-containing primer pairs were chosen for these biological reasons; four standard nucleotides near their 3′-ends were replaced by SAMRS components (Table S2). Both standard and SAMRS-containing primer pairs were tested to amplify 20,000 copies of complete HIV-1 viral RNA under identical conditions using the IsoAmp II enzyme mix with Enhanced Avian RT and GspSSD DNA polymerase (Figure 3).
Figure 3.
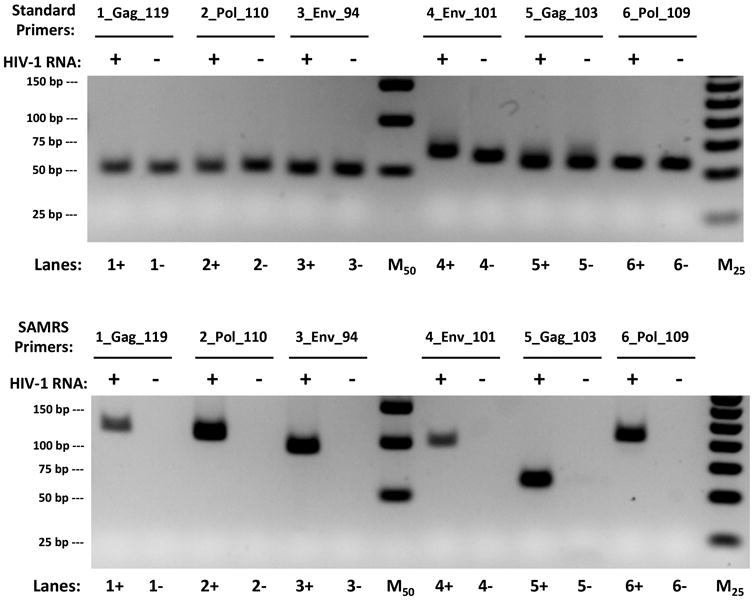
Reverse transcriptase helicase dependent amplification (RT-HDA) assays using standard primers (top) and SAMRS primers (bottom) to amplify gag, pol, and env genes in complete HIV-1 viral RNA. The desired amplicon size for each pair of primer is 119bp, 110bp, 94bp, 101bp, 103bp, and 109bp, respectively. M50 and M25 are 50 bp and 25 bp ladders.
The contrast between the results obtained with standard primers and SAMRS primers was again striking. For all six standard primer pairs, only artifacts were seen, both in the presence and absence of HIV-1 RNA target (Figure 3, top). However, all SAMRS primer pairs, except (5_Gag_103), gave desired products with HIV-1 RNA target (Figure 3, bottom).
In addition, a “hot start” IsoAmp III enzyme mix was also tested with both standard and SAMRS-containing primers (Figure S6). For all six standard primer pairs, only two primer pairs (2_Pol_110 and 6_Pol_109) gave the desired amplicons using “hot start” enzyme, the rest of four primer pairs generated either primer artifacts or no products at all (Figure S6, top). In contrast, four SAMRS-primer pairs generated expected amplicons, one SAMRS-primer pair (1_Gag_119) gave no product and one (5_Gag_103) gave an artifact (Figure S6, bottom).
To understand these results, we predicted the secondary structures of the various RNA targets. This analysis found that the RNA target (5_Gag_103) has the strongest hairpin structure of the targets (as judged by predicted free energies of formation). This seems to be a reasonable explanation for these results. Thus, while SAMRS appears to solve some problems in isothermal HDA amplification (e.g. preventing primer-dimers from arising via 3′-overlap), it does not solve all problems, especially those intrinsic in the target.
We then asked whether SAMRS-HDA could be multiplexed. This is generally challenging with isothermal systems, since the increased complexity of a multiplexed system offers still more opportunities for xNA species to go astray.
Here, twofold multiplexed HDA assays were tested to amplify both HIV-1 viral RNA and rpoB DNA targets. For two sets of standard primers, only primer artifacts were generated using IsoAmp II enzyme mix (Figure 4, top, left panel) and single product (HIV target) was produced using “hot start” IsoAmp III enzyme mix (Figure 4, top, right panel). However, for two sets of SAMRS-containing primers, both RNA and DNA targets were amplified and all desired products were generated using both enzyme mix (IsoAmp II and III) with additional GspSSD DNA polymerase (Figure 4, bottom). These results further demonstrated that SAMRS-containing primers support twofold multiplexed HDA assays when standard primers fail even with “hot start” approach.
Figure 4.
Amplification of HIV-1 viral RNA and rpoB DNA using twofold multiplexed RT-HDA with standard primers (top) and SAMRS-containing primers (bottom).
Lanes 1 and 6: Only HIV-1 RNA target.
Lanes 2, 3, 7, and 8: Both HIV-1 RNA and rpoB DNA as targets.
Lanes 4 and 9: Only rpoB DNA target.
Lanes 5 and 10: No target control.
M25 and M50 indicate 25bp and 50bp DNA ladders.
To test the effect of SAMRS components at different positions of primers, four SAMRS components were placed one, two, or three standard nucleotides away from their 3′-ends (Table S3) to amplify DNA targets. As shown in Figure S7, primers containing four SAMRS components and one standard nucleotide at their 3′-ends (KIT-SAMRS4) gave desired product without any artifacts. In contrast, SAMRS-containing primers with two standard nucleotides at their 3′-ends (KIT-SAMRS4-2N) produced both product and artifact, with three standard nucleotides at 3′-ends (KIT-SAMRS4-3N) gave only artifacts (Figure S7). These results define the optimal number and placement of SAMRS components in HDA primers.
Conclusions
These results show that self-avoiding molecular recognition systems (SAMRS) can eliminate primer interaction and artifacts that propagate in HDA, with a shorter specificity footprint than with RPA. Indeed, the versatility of SAMRS-HDA is quite remarkable. By improving the statistics of success in SAMRS-HDA when HDA with standard primers and “hot start” approaches fail, SAMRS makes the assays easier to design. Further, both DNA and RNA can be targeted. Its ability to support low level multiplexing (here, for two pathogens that often go together in immunosuppressed patients) adds still more to the applicability of this procedure. This makes SAMRS-HDA a more attractive architecture for isothermal amplification, especially in resource limited environments, at points-of-care, and in the developing world.
Experimental Section
Materials
Standard oligonucleotides were obtained from Integrated DNA Technologies (IDT; Coralville, IA). Oligonucleotides containing SAMRS nucleotides were synthesized using published methods.[21] Human genomic DNA male was from Promega (G1471, WI, USA). The plasmid for RpoB in Topo 2.1 was a gift from the Ellington laboratory (University of Texas; Austin, TX). HIV-1 armored RNA was from Asuragen (42103, TX, USA). HIV-1B/US1 purified RNA was from SeraCare Life Sciences (500411, MD, USA). The IsoAmp® enzyme mix kit was ordered from BioHelix (Beverly, MA, USA). Gsp DNA polymerase variants (GspM and GspM2.0) and GspSSD were ordered from OptiGene (Horsham, UK). Bst2.0 DNA polymerase was from New England Biolabs (NEB, MA, USA). ThermoScript reverse transcriptase was from Invitrogen (12236-014). Enhanced Avian reverse transcriptase was from Sigma Aldrich (A4464-1KU). EvaGreen was obtained from Biotium (Hayward, CA, USA).
Experimental Details are described in Supporting Information.
Supplementary Material
Acknowledgments
This work was supported by the Defense Threat Reduction Agency (DTRA1-13-1-0004), the Defense Advanced Research Project Administration (DARPA ADEPT, HR0011-11-2-0018), the National Institute of Allergies and Infectious Disease (NIAID, R01AI098616), Nucleic Acids Licensing LLC, and Firebird Biomolecular Sciences LLC under a grant from the Department of Defense W911NF-12-C-0059.
Contributor Information
Dr. Zunyi Yang, Email: zyang@ffame.org, Foundation for Applied Molecular Evolution (FfAME), 720 SW 2nd Avenue, Suite 201, Gainesville, FL 32601.
Chris McLendon, Foundation for Applied Molecular Evolution (FfAME), 720 SW 2nd Avenue, Suite 201, Gainesville, FL 32601.
Daniel Hutter, Firebird Biomolecular Sciences LLC, 13709 Progress Blvd, N112, Alachua, FL 32615. Fax: (+1)352-271-7076; Homepage: (http://ffame.org).
Kevin M. Bradley, Foundation for Applied Molecular Evolution (FfAME), 720 SW 2nd Avenue, Suite 201, Gainesville, FL 32601
Shuichi Hoshika, Foundation for Applied Molecular Evolution (FfAME), 720 SW 2nd Avenue, Suite 201, Gainesville, FL 32601.
Carole Frye, Firebird Biomolecular Sciences LLC, 13709 Progress Blvd, N112, Alachua, FL 32615. Fax: (+1)352-271-7076; Homepage: (http://ffame.org).
Steven A. Benner, Email: sbenner@ffame.org, Foundation for Applied Molecular Evolution (FfAME), 720 SW 2nd Avenue, Suite 201, Gainesville, FL 32601; Firebird Biomolecular Sciences LLC, 13709 Progress Blvd, N112, Alachua, FL 32615. Fax: (+1)352-271-7076; Homepage: (http://ffame.org).
References
- 1.Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad F, Hashsham SA. Anal Chim Acta. 2012;733:1–15. doi: 10.1016/j.aca.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 3.Asiello PJ, Baeumner AJ. Lab Chip. 2011;11:1420–1430. doi: 10.1039/c0lc00666a. [DOI] [PubMed] [Google Scholar]
- 4.Hofmann WP, Dries V, Herrmann E, Gartner B, Zeuzem S, Sarrazin C. J Clin Virol. 2005;32:289–293. doi: 10.1016/j.jcv.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Vandervliet GME, Schukkink RAF, Vangemen B, Schepers P, Klatser PR. J Gen Microbiol. 1993;139:2423–2429. doi: 10.1099/00221287-139-10-2423. [DOI] [PubMed] [Google Scholar]
- 6.Walker GT, Nadeau JG, Spears PA, Schram JL, Nycz CM, Shank DD. Nucleic Acids Res. 1994;22:2670–2677. doi: 10.1093/nar/22.13.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inoue J, Shigemori Y, Mikawa T. Nucleic Acids Res. 2006;34 doi: 10.1093/nar/gkl350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piepenburg O, Williams CH, Stemple DL, Armes NA. Plos Biol. 2006;4:1115–1121. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mori Y, Kanda H, Notomi T. J Infect Chemother. 2013;19:404–411. doi: 10.1007/s10156-013-0590-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vincent M, Xu Y, Kong HM. Embo Rep. 2004;5:795–800. doi: 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.An LX, Tang W, Ranalli TA, Kim HJ, Wytiaz J, Kong HM. J Biol Chem. 2005;280:28952–28958. doi: 10.1074/jbc.M503096200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tong Y, Tang W, Kim HJ, Pan X, Ranalli TA, Kong H. Biotechniques. 2008;45:543–557. doi: 10.2144/000112959. [DOI] [PubMed] [Google Scholar]
- 13.Goldmeyer J, Li HJ, McCormac M, Cook S, Stratton C, Lemieux B, Kong F, Tang W, Tang YW. J Clin Microbiol. 2008;46:1534–1536. doi: 10.1128/JCM.02234-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldmeyer J, Kong H, Tang W. J Mol Diagn. 2007;9:639–644. doi: 10.2353/jmoldx.2007.070012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gill P, Ghaemi A. Nucleos Nucleot & Nucleic Acids. 2008;27:224–243. doi: 10.1080/15257770701845204. [DOI] [PubMed] [Google Scholar]
- 16.Das S, Mohapatra SC, Hsu JT. Biotechnol Tech. 1999;13:643–646. [Google Scholar]
- 17.Paul N, Shum J, Le T. In: RT-PCR Protocols. Second. King N, editor. Vol. 630. 2010. pp. 301–318. [Google Scholar]
- 18.Fan JB, Chee MS, Gunderson KL. Nat Rev Genet. 2006;7:632–644. doi: 10.1038/nrg1901. [DOI] [PubMed] [Google Scholar]
- 19.Lemieux B, Li Y, Kong H, Tang YW. Expert rev mol diagn. 2012;12:437–443. doi: 10.1586/erm.12.34. [DOI] [PubMed] [Google Scholar]
- 20.Denys GA. Expert rev mol diagn. 2014;14:17–26. doi: 10.1586/14737159.2014.864239. [DOI] [PubMed] [Google Scholar]
- 21.Hoshika S, Chen F, Leal NA, Benner SA. Angew Chem Int Ed. 2010;49:5554–5557. doi: 10.1002/anie.201001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kutyavin IV, Rhinehart RL, Lukhtanov EA, Gorn Vv, Meyer RB, Gamper HB. Biochemistry. 1996;35:11170–11176. doi: 10.1021/bi960626v. [DOI] [PubMed] [Google Scholar]
- 23.Sharma N, Hoshika S, Hutter D, Bradley KM, Benner SA. ChemBioChem. 2014;15:2268–2274. doi: 10.1002/cbic.201402250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.