Abstract
Gadolinium(III) nanoconjugate contrast agents (CAs) have distinct advantages over their small-molecule counterparts in magnetic resonance imaging. In addition to increased Gd(III) payload, a significant improvement in proton relaxation efficiency, or relaxivity (r1), is often observed. In this work, we describe the synthesis and characterization of a nanoconjugate CA created by covalent attachment of Gd(III) to thiolated DNA (Gd(III)–DNA), followed by surface conjugation onto gold nanostars (DNA–Gd@stars). These conjugates exhibit remarkable r1 with values up to 98 mM−1 s−1. Additionally, DNA–Gd@stars show efficient Gd(III) delivery and biocompatibility in vitro and generate significant contrast enhancement when imaged at 7 T. Using nuclear magnetic relaxation dispersion analysis, we attribute the high performance of the DNA–Gd@stars to an increased contribution of second-sphere relaxivity compared to that of spherical CA equivalents (DNA–Gd@spheres). Importantly, the surface of the gold nanostar contains Gd(III)–DNA in regions of positive, negative, and neutral curvature. We hypothesize that the proton relaxation enhancement observed results from the presence of a unique hydrophilic environment produced by Gd(III)–DNA in these regions, which allows second-sphere water molecules to remain adjacent to Gd(III) ions for up to 10 times longer than diffusion. These results establish that particle shape and second-sphere relaxivity are important considerations in the design of Gd(III) nanoconjugate CAs.
Keywords: gadolinium, magnetic resonance, nanostar, relaxivity, second-sphere, contrast agent, nuclear magnetic resonance dispersion
Magnetic resonance imaging (MRI) is considered to be an integral part of medical diagnostic imaging due to its high spatiotemporal resolution, excellent soft tissue contrast, and exceptional safety profile.2–4 Using this technique, high-resolution anatomical images can be obtained without radioactive tracers or ionizing radiation. When native contrast is insufficient for definitive detection, contrast agents (CAs) are used to differentiate among areas where specific anatomical detail is difficult to resolve. To address this shortcoming, paramagnetic chelates of Gd(III) are used to shorten the longitudinal relaxation times (T1) of proximal water protons in regions of agent accumulation and thereby generate positive image contrast.5 However, due to the relatively low sensitivity of clinically approved small-molecule agents, applications are limited because high doses are often required (mM for Gd(III) complexes). To increase the utility of magnetic resonance CAs, current work has focused on developing strategies for specific delivery (greater accumulation of Gd(III) at the site of interest) and greater proton relaxation efficiency (improved r1 relaxivity).
Optimizing CA Performance
The Solomon–Bloembergen–Morgen (SBM) theory of relaxivity describes three primary ways in which a Gd(III) agent can be optimized: (i) changing the rotational correlation time, τr; (ii) increasing the rate of water exchange with the lanthanide (the inverse of the inner-sphere water exchange rate, 1/kex, or τm); and (iii) increasing the number of coordinated water molecules, q.6–8 Despite numerous examples of Gd(III) chelates bearing increased values of q to augment r1, most CA research focuses on complexes wherein q = 1.9,10
Improving r1 by Optimizing τr
Of the aforementioned parameters used to increase r1, the most commonly applied is modulation of τr. Increasing this correlation time (by slowing the Gd(III) reorientation time) is known to improve CA r1 significantly at magnetic field strengths of ≤1.5 T (corresponding to proton Larmor frequencies of ≤64 MHz). The effects of changing this parameter at higher field strengths, however, is unsubstantial or sometimes detrimental to CA performance. 6 Often, increasing τr is achieved by immobilization of Gd(III) to a nanoparticle or macromolecule. This approach is particularly common because nanoconjugate CAs are chemically accessible and couple the tumbling rate of the Gd(III) chelates to that of the much larger species. In addition to increased r1, this strategy is beneficial for increased local concentrations of CA (high Gd(III) payload per particle). For this reason, a variety of Gd(III) nanoconjugate CAs have been described, including agents based on biomacromolecules (proteins and viruses), organic nanoparticles (nanodiamonds, liposomes, and polymers), and inorganic nanoparticles (silica, titanium, and gold).11–21
Recently, Gd(III)–DNA was attached to spherical gold nanoparticles, creating Gd(III)-enriched DNA–gold nanoparticle conjugates (DNA–GdIII@AuNP).22 This nanoconjugate design was particularly effective for cellular MR imaging because DNA–GdIII@AuNPs showed both improved r1 relaxivity per Gd(III) and higher cellular internalization of Gd(III) relative to their small-molecule counterparts. Importantly, the nanoconjugates delivered the expected increase in r1 by lengthening τr, while maintaining the high stability, nuclease resistance, biocompatibility, and efficient cell uptake conferred by the densely packed monolayer of DNA on the gold nanoparticle surface.23–25 Attempts to increase r1 further by increasing the particle size, however, showed only minor improvement.22
Improving r1 by Optimizing τm
Another commonly used approach for improved r1 described by SBM theory focuses on the rapid and efficient exchange of water within the inner-sphere of Gd(III) complexes (τm), a feature which is critical to the generation of high T1 contrast.26–29 However, SBM simulations suggest that a significant enhancement of r1 using this strategy is only probable in CAs bearing near optimal values of τr, particularly at low-field strengths (≤1.5 T).30 By virtue of their size, nanoconjugate CAs inherently possess relatively slow tumbling rates. For this reason improvement of r1 in nanoconjugate systems where q is held constant, is readily accessible through the optimization of τm.
In Gd(III) CA systems, T1 proton relaxation is dominated by a dipole–dipole mechanism.31 Within the equations that describe this behavior, the correlation time parameter, τc, is a field-strength-dependent variable which contains features of Gd(III) complexes that can be affected by the coordination environment of the lanthanide. This relationship is described by eq 1
| (1) |
where T1e is the electronic spin relaxation time, τr is the rotational correlation time, and τm is the inner-sphere water exchange rate. When τc is equal to 2π times the proton Larmor frequency, r1 will be equal to its maximum achievable value at that frequency. Predicting and affecting T1e experimentally is not straightforward, thus, approaches to CA optimization commonly focus on modulation of τr and τm.32 In nanoconjugate CAs, the particle τr often exceeds tens of nanoseconds, and therefore, τc is dominated by the shorter of T1e or τm, or both if of similar length. Typical values of T1e between 0.5 and 1.5 T are approximately 10 ns. In this range of magnetic field strengths, the optimal τm value varies from 20 ns at 0.5 T to 5 ns at 1.5 T, and therefore, an optimized CA will possess a τm within this range.30
Gd(III) Nanoconjugates and Second-Sphere Relaxivity
The water exchange kinetics of lanthanide complexes is a direct result of the metal–ligand coordination environment. Therefore, improvements to τm are readily observed by synthetic modification of the Gd(III)–ligand complex prior to particle conjugation.33–35 Alternately, recent work has suggested that confinement of a CA on or near a protein surface may affect the diffusion of water in the proximity of the CA and thereby impact its performance by providing a significant second- and outer-sphere relaxivity enhancement.36 Although second- and outer-sphere contributions to relaxivity are generally considered insignificant due to the high rate of water diffusion past the Gd(III), this phenomenon is sometimes cited to explain unexpectedly high values of r1 in nanoconjugate systems.6,19,30,37,38
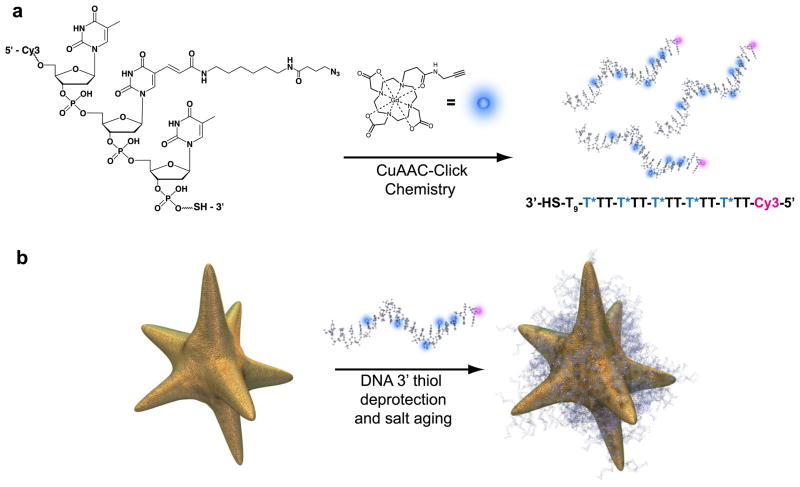
We predicted that this behavior could be affected experimentally by altering the water diffusion adjacent to hydrophilic nanoparticle surfaces, and that this may provide a new design parameter for improved r1 in Gd(III) nanoconjugate CAs. Specifically, we hypothesized that nanoconjugate CA surface dynamics can be affected by variation in particle shape, and that this alteration could be used to augment r1 through the enhancement of second-sphere contributions to CA performance. In support of this rationale, recent work has shown that the surface curvature of irregularly shaped nanoparticles can significantly affect the chemical reactivity of surface ligands.39 In an effort to create a high relaxivity CA, we have examined the effects of particle size and shape within the Gd(III)–DNA gold nanoconjugate platform using gold nanostars to produce DNA–Gd@stars (Scheme 1). Importantly, the nanostar surface contains regions of positive, negative, and neutral curvature, and thus we predict a significant effect on the local environment of the surface conjugated Gd(III)–DNA.
Scheme 1.
Preparation of DNA–Gd@stars. (a) Cy3-labeled 24-mer poly-dT oligonucleotide is modified via the covalent attachment of Gd(III) to each of five azide-bearing dT bases per strand. (b) Functionalized oligonucleotides are deprotected, revealing the 3′ thiol, and are conjugated to nanostars through a series of increases in salt concentration called salt aging.1
Herein, we present the synthesis of a new τm-optimized, alkyne-bearing Gd(III) chelate to ensure that r1 relaxivity is not limited by inner-sphere water exchange.40 When the Gd(III) complex is conjugated to the surface of gold nanostars, DNA–Gd@stars achieve r1 relaxivities up to 98 mM−1 s−1, a 25-fold greater r1 relaxivity than FDA-approved chelates at 25 °C (32 MHz). Significantly, these values exceed the theoretical maximum relaxivity predicted by SBM theory when a single water molecule is coordinated to the Gd(III) (q = 1).30
Employing detailed nuclear magnetic relaxation dispersion (NMRD) analysis, we compare 15 nm DNA–Gd@spheres with the DNA–Gd@stars and demonstrate that their proton relaxation efficiency is the result of optimized inner-sphere water exchange kinetics and particle surface-mediated elongation of second-sphere water residency lifetimes (and therefore enhanced second-sphere relaxivity).
RESULTS AND DISCUSSION
Synthesis and Characterization of a τm-Optimized Gd(III) Complex
The water exchange kinetics of lanthanide complexes can be affected by interchange of ligand types or synthetic modifications thereof.41 Recent work using the macrocyclic ligand [1,4,7-(tris-tert-butyl acetate)-1,4,7,10-tetraazacyclododecane], or DO3A, has described a modification shown to increase steric crowding around the water coordination site, thereby shortening τm.35,42 Using this previously reported strategy to facilitate an optimal τm in our nanoconjugate system, a new ligand based on DO3A was synthesized (Scheme 2). Importantly, a pendant alkyne is present to facilitate conjugation using click chemistry. Linker arm 1 was synthesized from acryloyl chloride and propargylamine in 72% yield. Attachment of 1 to previously synthesized DO3A macrocycle 2 proceeds via a 1,4 conjugate addition in the presence of base over 3 days.43 Deprotection of compound 3 in trifluoroacetic acid generates the triacetate ligand 4 (Supporting Information Scheme S3), and metalation followed by reverse-phase high-pressure liquid chromatography (RP-HPLC) purification resulted in a 65% yield of the Gd(III) complex 1-(N-(prop-2-yn-1-yl)2-oxopropyl)-4,7, 10-tris(carboxymethyl)-1,4,7,10-tetraazacyclododecyl gadolinium(III) (5).
Scheme 2.
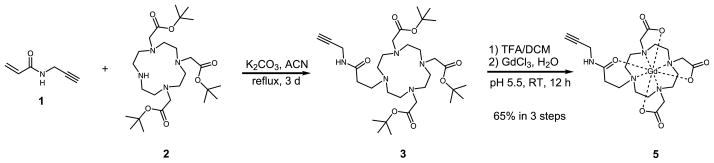
Chemical synthesis of τm-optimized Gd(III) complex 5. Synthesis of 1 proceeds through reaction of acryloyl chloride and propargylamine (Scheme S1). Conjugation of the propionate arm to tert-butyl DO3A (2) is achieved by 1,4 conjugate addition in acetonitrile over 3 days. Deprotection of the macrocycle 4 in trifluoroacetic acid is followed by metalation by GdCl3 at pH 5.5 for 12 h and HPLC purification.
To determine the τm of complex 5, a variable-temperature 17O titration was performed. Results were fitted to the SBM equations, and a τm of 22 ns at 37 °C was observed (Figure S1).31 This result represents an improvement of nearly 680 ns from the previously described Gd(III) complex used in DNA–GdIII@AuNPs (Complex 7, Figure S5).22 Optimization of this parameter is important because when τm ≪ T1 (a scenario known as a “fast exchange” regime), more rapidly exchanging inner-sphere water molecules increase r1 because the Gd(III) can more effectively propagate the effects of proton relaxation to the bulk.30
To verify the number of inner-sphere water molecules (q), the Eu(III) analogue of complex 5 was synthesized (complex 6, Scheme S4). Using the modified Horrocks’ method, equimolar solutions of 6 in water and D2O were prepared, and luminescence lifetimes of each solution were recorded and fitted, resulting in a q = 1.1 ± 0.1 (Figure S2).44,45
Synthesis and Characterization of DNA–Gd@stars
Synthesis of Gd(III)-labeled DNA began by incorporation of a C6-amino-modified deoxythymidine (dT) nucleotide into five positions along the 24-mer poly-dT oligonucleotide sequence (Scheme 1 and Scheme S6). The 3′-thiolated 24-mer (5× amino-modified) poly-dT-Cy3–5′ was reacted with azidobutyrate N-hydroxy succinimide ester in the presence of base to install the azide functionality. Finally, complex 5 was covalently attached to the polyazido DNA via a Cu(I)-catalyzed 1,3 dipolar cyclo-addition (CuAAC, or “click” chemistry). The successful synthesis of the 3′-thiolated Gd(III) poly-dT-Cy3–5′ oligonucleotide was subsequently confirmed by matrix-assisted laser desorption ionization mass spectrometry.
Synthesis of nanostars was performed by the reduction of chloroauric acid by 4-(2-hydroxyethyl)- piperazine-1-ethanesulfonic acid (HEPES) buffer.46,47 DNA–Gd@Star conjugates were synthesized by salt aging purified Gd(III)–DNA in water over 3 days (Scheme 1). Purified particles appear dark green due to the plasmon resonance at 800 nm and are stable in water for >12 weeks when stored at 4 °C.
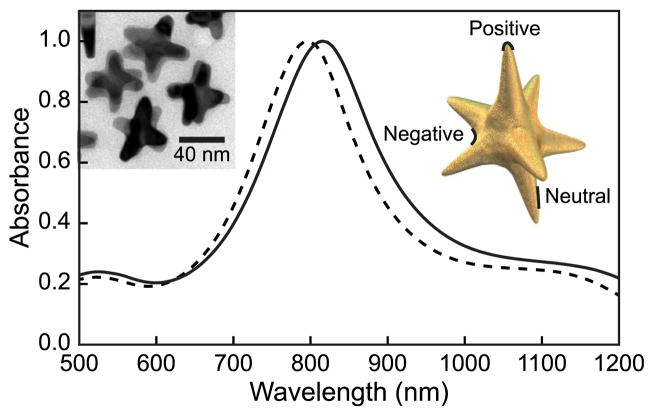
Transmission electron micrographs (TEM) (Figure 1, inset) show that DNA–Gd@stars contain up to 8 branches, have an average tip-to-tip diameter of ca. 50 nm, and an approximate volume of 1.6 × 104 nm3 (Supporting Information). Significantly, individual particles possess regions of positive (branch tips) and negative curvatures (between branches) and flat regions (branch length) (Figure 1). Upon functionalization with Gd(III)–DNA, the average hydrodynamic diameter of nanostars increased from 38.8 ± 0.1 to 63.0 ± 0.7 nm (Table S1) and the maximum surface plasmon absorbance wavelength shifted from 800 to 818 nm, indicating that the Gd(III)–DNA was successfully conjugated to the nanostar surface and that colloidal stability is maintained (Figure 1). To assess the stability of the nanoconjugates, DNA-Gd@stars were subjected to a range of salt (0–450 mM NaCl) and pH (pH 3–11) conditions, and in cell culture media (DMEM + 10% fetal bovine serum) for 24 h (Figure S4). Under all conditions tested, no aggregation of DNA-Gd@stars was observed, which is an important feature for biological applications.
Figure 1.
Absorbance spectra of nanostars (dashed line) and DNA–Gd@stars (solid line) indicate an 18 nm resonance shift after functionalization. Insets are a TEM image of nanostar and scheme indicating nanostar curvature.
For the purpose of comparison, spherical conjugates of 15 and 40 nm diameter were synthesized using the same Gd(III)-DNA (DNA-Gd@ spheres and DNA-Gd@spheres40nm) (Figure S3).1 To quantify DNA loading, inductively coupled plasma mass spectrometry (ICP-MS) was used to determine the ratio of Gd(III) to Au. DNA–Gd@stars contained 1990 ± 450 Gd(III) complexes per nanostar, corresponding to 398 ± 90 DNA strands per nanostar. This loading represents greater than a 3-fold increase in Gd(III) payload relative to DNA–Gd@spheres due to larger particle size and is comparable to the DNA–Gd@spheres40nm (Table S1). The number of gold atoms per particle is approximated by calculating the nanostar volume in >180 particles using TEM and the density of bulk gold (Supporting Information).
Molar Relaxivity of DNA–Gd@stars
To assess the performance of DNA–Gd@stars, the r1 of the nanoconjugates was determined by taking the slope of the linear plot of 1/T1 versus Gd(III) concentration (Table S3 and Figure S5). The surprising value of 54.7 mM−1 s−1 per Gd(III) in water at 60 MHz and 37 °C was obtained (the average of multiple batches, Table S2). This value is among the highest reported for a Gd(III) nanoconjugate (for one inner-sphere water molecule, or q = 1) and is in the same range as supramolecular- or protein bound Gd(III) complexes. Similarly high values of r1 are observed for Gd(III) conjugated to viral capsids, non-covalently bound to human serum albumin (MS-325), or entrapped in apoferritin.15,48,49
To investigate the source of such high r1 values, the relaxivities of the unconjugated Gd(III) complex (Scheme 2) and DNA-bound Gd(III) were examined separately. The r1 values of complex 5 and unconjugated Gd(III)–DNA were found to be 3.8 and 9.5 mM−1 s−1, respectively (Table 1). Though incrementally higher than equivalent values using the previously described Gd(III) complex 7, the relaxivities observed were unremarkable.22 In order to validate the high relaxivity of the assembled nanostar constructs, five replicate batches of DNA–Gd@stars were synthesized and measured, showing relaxivities between 42.3 and 69.1 mM−1 s−1 (37 °C, 60 MHz) and Gd(III) loading between 1231 and 2473 Gd(III) per particle (Table S2). When examined at low-field strength (where differences in r1 are most evident, here 1.41 T),30 r1 relaxivities measured only 14.6 and 16.8 mM−1 s−1, respectively. Considering the product of the particle loading (Table S1) and high r1 per Gd(III), DNA–Gd@stars exhibit a 3-fold greater relaxivity per particle relative to DNA-Gd@ spheres40nm, despite comparable Gd(III) payload. These results (Table 1) indicate that the shape, not the size of the nanostar conjugates, is the cause of the large r1 in this system.
TABLE 1.
Relaxivities of DNA–Gd@stars and DNA–Gd@ spheres at 60 and 300 MHza
|
r1 relaxivity (mM−1 s−1)
|
||
|---|---|---|
| 60 MHzb | 300 MHzc | |
| ProHance | 3.0d | 4.0 |
| complex 5 | 3.8 | NM |
| Gd(III)–DNA | 9.5 | NM |
| DNA–Gd@stars/ionic | 54.7 | 9.4 |
| DNA–Gd@stars/particle | 108850 | 19345 |
| DNA–Gd@spheres/ionic | 14.6 | 5.8 |
| DNA–Gd@spheres/particle | 8230 | 3270 |
| DNA–Gd@spheres40nm/ionic | 16.8 | NM |
| DNA–Gd@spheres40nm/particle | 34520 | NM |
“Ionic” r1 refers to the contribution of each individual Gd(III) complex to proton relaxation, whereas “particle” describes the product of each particle’s payload and ionic r1.
Measured at 37 °C in water plus 0.01% Tween 20.
Measured at 25 °C in water plus 0.01% Tween 20.
As previously reported;50
NM = not measured.
High-Field r1 of DNA–Gd@stars
As the demand for improved contrast-to-noise ratios and image resolution continues to grow, research has trended toward the use of higher field strength MR instruments.3 As a consequence, Gd(III) contrast agent performance at high field is becoming increasingly important. To demonstrate the ability of DNA–Gd@stars to increase T1 contrast at high magnetic fields, solution phantoms were acquired on a 7 T Bruker Pharma Scan (300 MHz). DNA–Gd@star samples were imaged at varying concentrations, and the T1 relaxation times of each tube were used to calculate high-field relaxivity. DNA–Gd@ stars generated an r1 of 9.4 mM−1 s−1, while the r1 of the commercially available CA, ProHance, was only 4.0 mM−1 s−1 at 25 °C in water (Figure 2). Solutions of ProHance at the concentrations examined (equimolar in Gd(III) with DNA–Gd@stars) showed contrast enhancement indistinguishable from the water control (Figure 2 and Supporting Information Figure S12).
Figure 2.
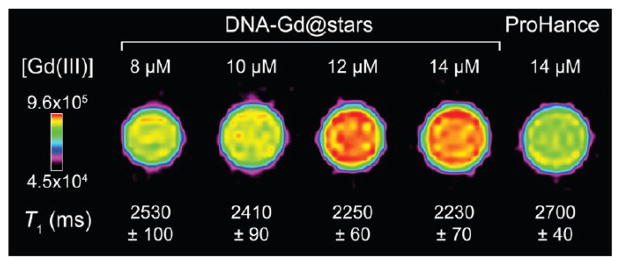
High-field MR solution phantoms (25 °C, 7 T) of aqueous solutions of DNA–Gd@stars vs equimolar concentrations of the clinical CA ProHance (standardized to [Gd(III)]). Approximate diameter of capillary tubes = 1 mm. Calibration bar is signal intensity.
NMRD Acquisition
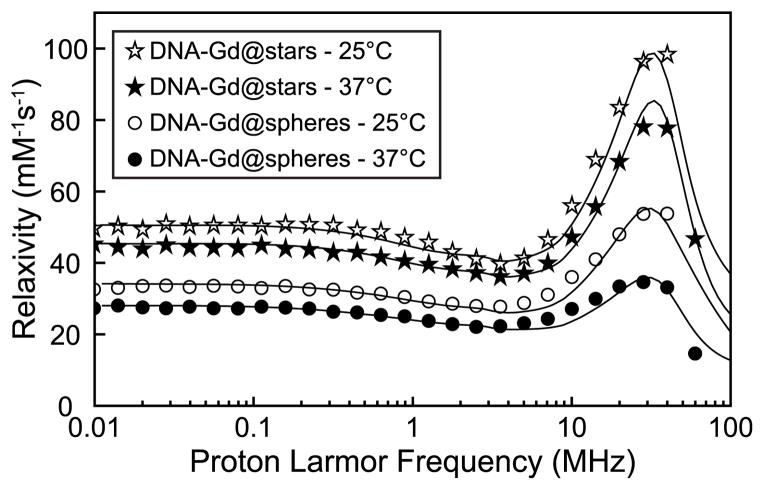
For a detailed mechanistic investigation into the cause of the very high relaxivities observed, DNA–Gd@stars and DNA–Gd@spheres were subjected to a NMRD analysis. Due to the small difference in r1 observed between the different sizes of spherical nanoconjugates at 1.41 T, only the 15 nm DNA–Gd@spheres (which most closely resemble the DNA–gold nanoparticle conjugates in literature) were taken forward for NMRD comparison to DNA–Gd@ stars.1,22–25,51–53 Using this technique, proton T1 relaxation rates of water are observed in the presence of paramagnetic species across a range of magnetic field strengths. With the acquired data, proton relaxivity profiles are created and fitted using SBM theory or other more recently developed approaches such as the modified Florence model.54–56 The analysis of relaxivity profiles provides a means of extracting valuable mechanistic information, including dynamic parameters such as τr and τm.31,57–59
The NMRD profiles of the nanostar and 15 nm spherical nanoparticle conjugates were measured at 25 and 37 °C in water. After acquisition, spectra were normalized to the Gd(III) concentration, and the resulting relaxivity profiles were analyzed using the modified Florence model and the independently measured values for τm and q (Figure 3).54,55,60,61
Figure 3.
NMRD profiles for water solutions of DNA–Gd@stars and DNA–Gd@spheres. Best fit parameters for the profiles are reported in Supporting Information Tables S12 and S13.
NMRD Analysis
The NMRD profile of DNA–Gd@stars shows a maximum relaxivity peak of 98 mM−1 s−1 at 25 °C, centered at approximately 32 MHz. This very large value exceeds the theoretical limit that can be calculated with SBM theory, including that calculated with the modified Florence model, when only one water molecule is coordinated to the Gd(III) ion (Figure S13).30
As a consequence, this profile could not be reproduced by constraining q = 1. Even when including contributions from the outer coordination sphere (water molecules freely diffusing up to a distance of closest approach of 3.6 Å), the data could not be fit.62,63 Allowing the analysis to consider a value of q > 1 was observed to provide the best fit of the NMRD data. This is a physically unlikely scenario, however, because neither the attachment of the Gd(III) complex to the DNA nor conjugation to the gold nanoparticle is expected to affect the coordination environment of the Gd(III) and affect q.50 To rationalize these results, we once again constrained the analysis to require that q = 1, retained the previously applied outer-sphere contribution, and included significant contributions from second coordination sphere water molecules.64,65 In this analysis, the τr of the DNA–Gd@stars is assumed to be larger than T1e and τm (τr ≥ 1 μs) and thus beyond the ability to affect r1 significantly. Using these parameters, four second-sphere water molecules (qSS = 4) at a Gd(III)–proton distance of 3.5 Å were required to fit the data successfully. This interpretation required an exceptionally large contribution to r1 from the second-sphere water molecules, which is unusual because in most Gd(III) complexes this contribution is <10% of total r1.30 Due to the covariance between qSS and the Gd(III)–proton distance, larger values of qSS are feasible for even larger proton distances.
By definition, the magnitude of second-sphere effects on r1 is determined by the number of second-sphere water protons, their distance from, and residency lifetimes adjacent to the metal center. In most cases, the second-sphere water molecule lifetimes (τmSS) are sufficiently short that their contributions to the bulk paramagnetic relaxivity is small, and therefore, these effects are accounted for by combining the second-sphere contribution with the outer-sphere relaxation (when their residence times adjacent to the metal are at or near the diffusional residence time). In order to obtain the large contribution from the second-sphere water molecules in the DNA–Gd@stars, τmSS is requisitely assigned relatively long values (0.1–1 ns) compared to diffusion (tens of picoseconds). Based on this analysis, we hypothesize that the shape of the DNA–Gd@stars provides a unique, hydrophilic environment where an extended network of transiently associated second-sphere water molecules can reside adjacent to the Gd(III). Many recent examples of nanoconjugate CAs appear to support this hypothesis, though in many cases we find that it is not described explicitly.15,19,48,49
While seldom investigated in mechanistic detail, nanoconjugate CAs that exhibit unusually high r1 relaxivities often mention the chemical environment of the Gd(III) as a possible explanation. In particular, for CAs that bear extended hydrophilic polymer or protein surfaces, the chemical environment surrounding the lanthanide center is believed to slow water diffusion in the second-sphere of the Gd(III) complexes.15,19,37,48,49 The result of this hydrogen-bonding-rich environment is that each metal center has incrementally longer contact with second-sphere water molecules. In extensive work to study the effects of second-sphere relaxivity (r1SS), chelates of Gd(III) were synthesized using pendant phosphate groups.64,66,67 Importantly, Gd(III) complexes were presented wherein the inner-sphere water was displaced by steric crowding (q = 0), which allowed for direct experimental determination of second- and outer-sphere relaxivities compared to q = 1 controls. In further work, q = 0 chelates of Gd(III) were bound to serum albumin (in order to lengthen τr and thereby optimize r1) and could achieve second- and outer-sphere relaxivities as high as 12 mM−1 s−1 at 20 MHz and 25 °C. This work showed that using pendant hydrogen-bonding functional groups and an adjacent hydrophilic protein surface could facilitate contributions from the second-sphere that are quite significant.68
The precedent studies and our current results suggest that a sizable contribution to r1 must be provided by second-sphere water molecules present in the dense monolayer of DNA on the surface of DNA–Gd@stars. This sterically crowded, hydrogen-bonding-rich environment likely plays a major role in helping to slow the diffusion of passing water molecules in the proximity of the Gd(III).
Investigating r1SS in DNA–Gd@stars
For additional evidence to support the second-sphere relaxivity hypothesis, the NMRD profile was examined for detailed information about the on-particle water exchange dynamics by observing the change in relaxivities with respect to temperature. A decrease in r1 with increasing temperature is observed in the DNA–Gd@stars, in agreement with the “fast exchange” behavior of inner-sphere water molecules observed in the exchange dynamics of 5.69 Formally, fast exchange behavior is manifest by a decrease in τc with increasing temperature. From the NMRD fit, the T1e at very low fields increases with increasing temperature and is about 0.5–1 ns, whereas τr is maintained at values larger than tens of nanoseconds. Therefore, we conclude that the decrease in r1 with increasing temperature is the result of a faster τm.
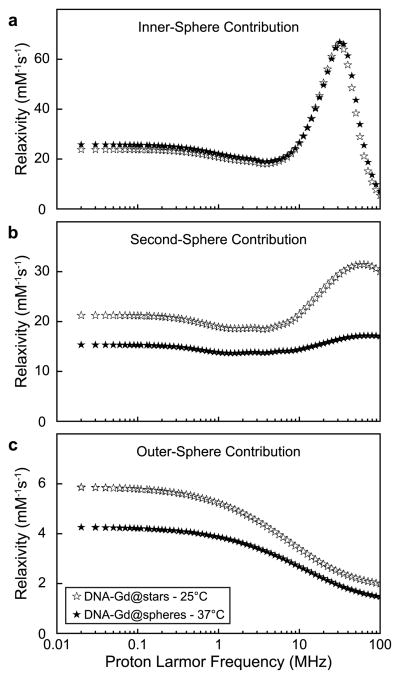
In the NMRD fit, the τm was fixed to the previously measured values of 39 and 22 ns at 25 and 37 °C, respectively. The τmSS, however, was left to vary. After fitting, the respective inner-, second-, and outer-sphere components of r1 were plotted and analyzed (Figure 4 and Supporting Information Table S11). The results showed that the contribution to r1 from inner-sphere water molecules was almost independent of temperature, despite the nearly 2-fold difference in τm between the two temperatures. This indicates that the inner-sphere residency time of coordinated water molecules is not the major determinant of τc for the dipole–dipole interaction with the Gd(III).
Figure 4.
Simulated deconvolution of DNA–Gd@stars NMRD profiles into component inner- (a), second- (b), and outer-sphere (c) contributions.
The τmSS values calculated from the NMRD fitting were 0.6 and 0.3 ns at temperatures of 25 and 37 °C (Table S12) and were found to be the predominant cause of the observed decrease in r1 between the two temperatures (Figure 4b). This change in τmSS represents a similar 2-fold change in the exchange rate between the two temperatures, but on a time scale nearly 2 orders of magnitude more rapidly than the inner-sphere exchange rate (although still slow compared to diffusion). Here, we show that the 44% decrease of r1 with increasing temperature is largely determined by second-sphere water proton residence lifetimes (Table S11). Specifically, at the clinically relevant field strength of 1.5 T (64 MHz), we find an r1SS of 31.5 mM−1 s−1 of a total 54.8 mM−1 s−1 at 25 °C and 17.2 mM−1 s−1 of a total 44.3 mM−1 s−1 at 37 °C (57.4 and 38.8% second-sphere contribution, respectively). Based on this analysis, the r1SS contribution provides a significant increase in the overall performance of the DNA–Gd@stars relative to other Gd(III) nanoconjugates.
NMRD Analysis of DNA–Gd@spheres
To verify that the irregular shape of the nanostars is responsible for the high relaxivities observed, the 15 nm DNA–Gd@ spheres synthesized using the same Gd(III)–DNA were studied by NMRD (Figure 3). The spherical particles show a similar behavior with respect to temperature (a fast exchange regime) but generate a considerably lower r1. The NMRD profile from the spherical nanoparticle system could be fitted using q = 1, where most of r1 can be attributed to inner-sphere relaxation (with only minor contributions from the second- and outer sphere). The temperature-dependent difference in r1 is characteristic of less optimal reorientation times resulting from increased local mobility, which is included in the analysis by applying the parameter τfast (Table S13).15,70 The effects of local mobility were not included in the analysis of the DNA–Gd@stars because this treatment would result in even larger r1SS (due to faster Gd(III) reorientation times and thus decreased inner-sphere relaxivity, r1IS).
By comparing the NMRD data for the DNA–Gd@spheres and the DNA–Gd@stars, we observe that even without the large second-sphere contribution to r1, the Gd(III) nanostar conjugates generate an r1IS of 66.9 mM−1 s−1 at 32 MHz and 37 °C. This value is approximately twice the total r1 of the DNA–Gd@sphere conjugates at the same field and temperature. This suggests that the shape of DNA–Gd@stars provides a unique environment capable of delivering high relaxivities even in the absence of r1SS. Furthermore, when comparing the r1 of the 15 nm DNA–Gd@spheres with that of the 40 nm equivalent at 1.41 T (64 MHz), only a minor improvement was observed (Table 1). We believe that this evidence further supports our hypothesis that the particle shape is a critical factor for the high performance of the DNA–Gd@stars.
DNA–Gd@star Cell Uptake and Biocompatibility
Due to the impressive r1 observed for DNA–Gd@stars and the precedent that other oligonucleotide-coated nanoparticles are able to efficiently enter cells without transfection agents, we believe DNA–Gd@stars are promising CAs for applications such as cellular tracking or cancer diagnosis.22,51 For this reason, we investigated the ability of DNA–Gd@stars to deliver Gd(III) to cancer cells in vitro. To examine if DNA–Gd@stars maintain the desired cellular uptake and biocompatibility of small spherical nanoconjugate platforms that are well-characterized in the literature, we did a comparison with the 15 nm DNA–Gd@spheres.22–24,68 We also compared DNA–Gd@stars to a commercially available CA (ProHance).
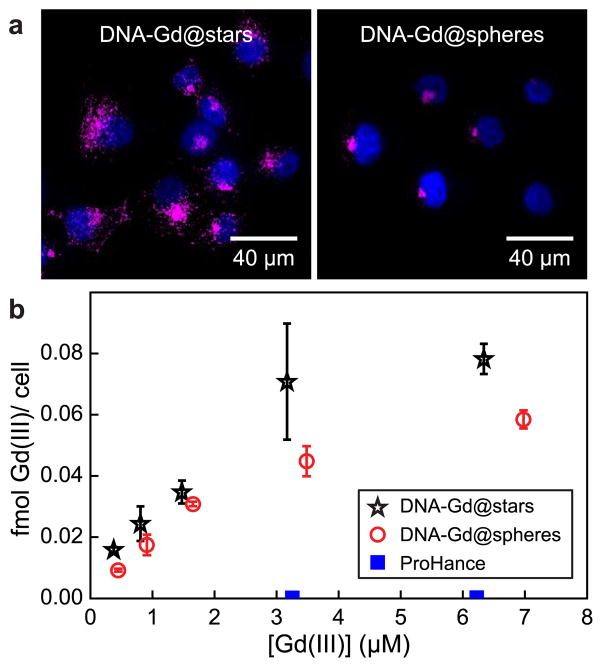
In cellular uptake studies, pancreatic cancer cells (PANC-1) were incubated with increasing Gd(III) concentrations of DNA–Gd@stars, DNA–Gd@spheres, or ProHance for 24 h. To remove noninternalized CAs, cells were washed three times with ice-cold phosphate buffered saline (PBS) prior to trypsinization and then washed an additional two times by centrifugation and resuspension in fresh buffer. Finally, cells were counted and digested in a 1:1 mixture of HNO3 and HCl for metals analysis using ICP-MS. Results indicated that the Gd(III) payload delivered per cell by incubation with DNA–Gd@stars was 50 times higher than incubation with ProHance and ca. 1.4 times higher than that by DNA–Gd@spheres (Figure 5b).
Figure 5.
(a) Confocal images of DNA–Gd@stars and DNA–Gd@spheres ([Gd(III)] = 1.5 μM) taken up by PANC-1 cells. Magenta signal represents Cy3-labeled nanoconjugates imaged under identical microscope conditions (without postprocessing), and blue signal represents DAPI-stained nuclei. (b) Femtomoles of Gd(III) taken up per cell, quantified by ICP-MS.
Previous reports suggest that between 107 and 109 Gd(III) ions per cell are required to produce detectable contrast in MRI.22 Thus, DNA–Gd@stars delivered Gd(III) payloads that are expected to generate visible contrast using only low micromolar incubation concentrations (4.5 × 107 ions per cell with 6 μM Gd(III) incubation concentration, batch 5 in Table S2). To check for detectable contrast in cell pellet images, PANC-1 cells were incubated with 0, 3, and 6 μM Gd(III) concentrations of DNA–Gd@stars or the commercial CA ProHance for 24 h. After being washed, cell pellets were added to glass capillary tubes and imaged using a 7 T Bruker PharmaScan (Figure S14). Analysis of the resulting images confirmed that the T1 of cells incubated with DNA–Gd@stars were reduced compared to untreated cells and ProHance (p < 0.02). These results highlight that, in addition to significantly improving r1, DNA–Gd@stars efficiently penetrate cells, enabling detectable contrast in cell pellets using as low as 3 μM Gd(III) incubation concentrations.
Confocal fluorescence micrographs were obtained to visualize the uptake and intracellular localization of DNA–Gd@stars and 15 nm DNA–Gd@spheres in PANC-1 cells (24 h incubation, [Gd(III)] = 1.5 μM). The fluorescence from the Cy3-labeled conjugates was obtained under identical microscope conditions and was not post processed for a more accurate visual comparison (Figure 5a). The Cy3 signal of DNA–Gd@stars appears brighter than that of DNA–Gd@spheres, consistent with the quantitative results gathered by ICP-MS.
We attribute the difference in Gd(III) delivery primarily to greater loading as a result of the higher surface area of the gold nanostars compared to the 15 nm spheres. Interestingly, we noticed a difference in the intracellular localization of the two conjugates. DNA–Gd@stars are distributed through the cytoplasm of the cells, while the DNA–Gd@spheres are primarily localized in the perinuclear region, consistent with the previous generation of DNA–GdIII@AuNPs.22 Although beyond the scope of the current study, this result suggests that differences may exist in the uptake or intracellular trafficking (e.g., endosomal–lysosomal pathway) behavior between the star- and spherical-shaped conjugates and warrants further investigation into the effects of the nanostar shape and size on cellular interactions.
Finally, the toxicity of the DNA–Gd@stars and DNA–Gd@spheres conjugates was evaluated using the Cell Titer 96 MTS assay. After a 24 h incubation with up to 6 μM Gd(III) concentrations, no cytotoxicity was observed for either conjugate (Figure S15). When we consider the exceptionally high r1 of DNA–Gd@stars, these in vitro results demonstrate that this platform is an effective and biocompatible CA for cellular MR imaging that requires only low micromolar Gd(III) incubation concentrations to generate detectable image contrast.
CONCLUSION
The use of MRI for longitudinal, in vivo studies presents clear advantages over competing modalities for its safety, soft tissue contrast, and limitless depth penetration. As a result, the investigation of new Gd(III) CAs has been of great interest to the research community. Herein we report a new class of gold nanoconjugates that exhibit exceptionally high relaxivities at both low- and high-field strengths. Using NMRD analysis, we have shown that nanoparticle shape and surface curvature affect the organization of conjugated DNAs on the particle surface and that this plays a meaningful role in sequestration of water molecules in the proximity of the Gd(III) complexes. Upon incubation with pancreatic cancer cells, DNA–Gd@stars improve Gd(III) delivery and maintain biocompatibility when compared side-by-side with DNA–Gd@spheres and commercial CAs. These results indicate the strategic value of designing the Gd(III) chelate into a dense, hydrophilic microenvironment to facilitate longer access to second-sphere water molecules and thereby generate contrast enhancement greater than can be achieved using small-molecule CAs alone. These results will enable future nanoconjugate CA development by consideration of particle shape and surface chemistry as vital parameters in the proton relaxation mechanism of Gd(III)-based agents.
MATERIALS AND METHODS
Chemical Synthesis and Characterization
All reagents and solvents were purchased from Sigma-Aldrich unless otherwise noted. All chemical syntheses were performed under ambient conditions unless described explicitly. Chemical characterization was achieved using a Varian 500 MHz NMR and a Bruker AutoFlex III MALDI spectrometer. Ligand and Gd(III) complex purification was performed using a Varian Prostar 500 HPLC using a Waters 4.6 × 250 mm 5 μm Atlantis C18 column and mobile phases of Millipore water, 0.05% trifluoroacetic acid in Millipore water, and acetonitrile. Nanoparticle characterization was performed on JEOL 1230 and Hitachi HD7700 TEMs. UV/vis/NIR spectra of colloidal solutions were collected on a LAMBDA 1050 spectrophotometer (PerkinElmer). DLS and ζ-potential measurements were done with a Brookhaven ZetaPals ζ-potential and particle size analyzer.
Oligonucleotides were synthesized on solid-phase-controlled pore glass beads (CPGs) by standard techniques on a MerMade automated synthesizer. All reagents and protected 3′ thiol modifier CPGs and C6 amino modifier dT-modified bases were purchased from Glen Research (Sterling, VA). Oligonucelotides were deprotected from the solid phase using AMA conditions [(1:1 methylamine/ammonium hydroxide (saturated)] for 1 h. Purification of oligonucleotides was performed using reverse-phase high-performance liquid chromatography on a Varian HPLC using a mobile phase consisting of 30 mM triethylammonium acetate buffer pH 7 and acetonitrile. Separation was achieved using a gradient of 75% acetonitrile over 45 min, as monitored by backbone and Cy3 wavelengths at 254 and 546 nm, respectively. Purified oligonucleotides were lyophilized and stored at −20 °C until particle synthesis. Inorganic Gd(III) complex 5 was synthesized using standard organic chemistry techniques.
ICP-MS was performed on either a computer-controlled (Plasmalab software) Thermo (Thermo Fisher Scientific, Waltham, MA) PQ ExCell ICP-MS equipped with a CETAC 500 autosampler or a computer-controlled (Plasmalab software) Thermo X series II ICP-MS equipped with an ESI (Omaha, NE, USA) SC-2 autosampler.
Synthesis of Gold Nanostars
Gold nanostars were synthesized by reducing chloroauric acid in HEPES buffer to create biocompatible, surfactant-free gold nanoparticles.46 Specifically, chloroauric acid (0.2 mM final concentration) was added to HEPES buffer (100 mM), shaken for 1 min, then left to grow for at least 30 min. The resonance wavelength of the nanostars was measured using UV/vis spectroscopy.
Synthesis of Spherical Gold Nanoparticles
Gold nanoparticles were synthesized by published procedures. The plasmon resonance wavelength was observed by UV/vis spectroscopy, and size was confirmed by DLS and TEM (Figure S3b). Unconjugated gold colloids (40 nm) were purchased from Ted Pella and characterized by DLS and TEM (Figure S3c).
Relaxivity (r1)
A stock solution of DNA–Gd@star conjugates was made (700 μL). This stock was serially diluted four times for a total of five solutions. Solutions were heated to 37 °C, and 200 μL of each concentration was placed into a Bruker minispec mq60 NMR spectrometer (60 MHz) for measurement of T1 relaxation time. Data were collected using an inversion recovery pulse sequence using four averages, a 15 s repetition time, and 10 data points. The remaining volumes of each solution were utilized for ICP analysis of [Gd(III)]. The inverse of the longitudinal relaxation time (1/T1, s−1) was plotted versus the Gd(III) concentration (mM). When a linear fit was applied to these data, the slope that is generated is defined as the relaxivity of the agent (mM−1 s−1). Relaxivities of DNA–Gd@spheres, Gd(III)–DNA, and complex 5 were collected by the analogous procedure.
Metals Analysis by ICP-MS
Quantitation of metal concentration was performed by acid digestion of nanoconjugate samples, followed by ICP-MS analysis. Gadolinium and gold analyses were prepared by different dilution factors such that they were within the range of the selected standard concentrations. Specifically, Gd analyses were digested by addition of 20 μL of nanoconjugate sample into 120 μL of 1:1 concentrated nitric acid/concentrated hydrochloric acid (TraceSelect Nitric acid, >69%; TraceSelect HCl, fuming 37%) and mixed thoroughly. Au analyses were made by addition of 5 μL of a nanoconjugate sample to 500 μL of 1:1 HNO3/HCl as above and mixed thoroughly. Millipore water and multielement internal standard (CLISS-1, Spex Certiprep, Metuchen, NJ, USA) containing Li, Sc, Y, In, Ho, and Bi were added to produce a solution of 2% nitric acid (v/v), 2% HCl (v/v), and 5.0 ng/mL internal standard up to a total sample volume of 3 mL (Gd) and 10 mL (Au) after 20-fold dilution of the original aliquot. Individual Au and Gd elemental standards were prepared at 0, 0.78125, 1.5625, 3.125, 6.25, 12.5, 25.0, 50.0, 100, and 200 ng/mL concentrations with 2% nitric acid (v/v), 2% HCl (v/v), and 5.0 ng/mL internal standards up to a total sample volume of 5 mL. Each sample was acquired using one survey run (10 sweeps) and three main (peak jumping) runs (100 sweeps). The isotopes selected were 197Au, 156,157Gd and 115In, 165Ho, and 209Bi (as internal standards for data interpolation and instrument stability).
NMRD Data
Water proton relaxation rates of solutions containing nanostar and nanosphere conjugates were measured from 0.01 to 40 MHz proton Larmor frequency using a fast-field cycling Stelar relaxometer. The relaxivity profiles of the DNA–Gd@stars and DNA–Gd@spheres were obtained after the subtraction of the diamagnetic 24-mer poly-dT gold nanoconjugate equivalents and normalized to 1 mM Gd(III) concentration.
Cell Culture
Pancreatic cancer cells (PANC-1, ATCC) were maintained in Dulbecco’s modified Eagle medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (Invitrogen). Cells were grown in T25 flasks (VWR) at 37 °C and 5% CO2.
Quantification of Gd(III) Uptake in Cells
Thirty thousand PANC-1 cells were seeded in each well of a 24-well plate overnight. The cell culture medium was replaced with varying concentrations (in triplicate) of DNA–Gd@stars, DNA–Gd@spheres, or Pro-Hance suspended in the serum-containing growth medium and incubated for 24 h at 37 °C. The CA-doped medium was removed, and the wells were washed three times with ice-cold PBS followed by trypsinization. The cells were then washed twice more via centrifugation (900g, 5 min) and resuspended in PBS. Finally, cells were counted with a hemacytometer and digested in 120 μL of 1:1 concentrated nitric acid/concentrated hydrochloric acid overnight. ICP-MS samples and standards were prepared as described above.
Confocal Imaging of Cy3-Labeled DNA–Gd@stars and DNA–Gd@ spheres
PANC-1 cells were grown on collagen-coated coverslips and incubated for 24 h with 1.5 μM [Gd(III)] of DNA–Gd@stars and DNA–Gd@spheres in growth medium. Cells were washed three times with ice-cold PBS, fixed in 4% paraformaldehyde, and mounted on a glass slide with a drop of ProLong gold antifade reagent (Invitrogen). The fixed samples were imaged on an inverted Zeiss 510 confocal microscope using a 40× water immersion objective and a 533 nm HeNe laser to excite the Cy3-labeled nanoconjugates. All microscope parameters (pinhole size, gain, offset, laser intensity) were kept constant between imaging DNA–Gd@star and DNA–Gd@sphere samples. The Cy3 channel was not post processed. The blue DAPI channel was processed (despeckle, Gausian blur, adjust min/max) with FIJI to enhance visualization of the cell nuclei.
Supplementary Material
Acknowledgments
We gratefully acknowledge support by NIH Grant R01EB005866, NIH Grant P01HL108795, the National Cancer Institute Center for Cancer Nanotechnology Excellence initiative at Northwestern University Award No. U54CA151880, and the National Institutes of Health (NIH) Director’s Pioneer Award DP1OD003899. We acknowledge government support under FA9550-11-C-0028 and awarded by the Department of Defense, Air Force Office of Scientific Research, National Defense Science and Engineering Graduate (NDSEG) Fellowship, 32 CFR 168a (K.S.B.C.). NMRD acquisition and analysis was performed at the Magnetic Resonance Center (CERM) at the University of Florence and was supported by the Ente Cassa di Risparmio di Firenze, MIUR PRIN 2012SK7ASN, EC Contract pNMR No. 317127. The Center for Advanced Molecular Imaging (CAMI) provided MR imaging, and the Quantitative Bioelemental Imaging Center (QBIC) at Northwestern provided confocal microscopy and ICP-MS analysis and was supported by National Science Foundation CHE-9810378/005 and NASA Ames Research Center NNA06CB93G. Compound purification and characterization was performed at the Integrated Molecular Structure Education and Research Center (IMSERC). M.W.R. would like to thank the Mirkin Lab at Northwestern for the gracious use of lab equipment for DNA synthesis and purification and for the supply of helpful advice over many years. The authors would also like to thank Raymond C. Lee for synthesis of the gold nanostar stock solutions. TEM imaging work was performed at the Northwestern University Biological Imaging Facility (BIF) and was generously supported by the NU Office for Research. Electron microscopy was performed on a JEOL 3200 FETEM purchased with the support of NCRR 1S10RR025092. UV–vis spectroscopic measurements were performed at the NU Keck Biophysics Facility supported by Cancer Center Support Grant (NCI CA060553). This work made use of the EPIC facility (NUANCE Center-Northwestern University), which has received support from the MRSEC program (NSF DMR-1121262) at the Materials Research Center; the Nanoscale Science and Engineering Center (NSF EEC-0647560) at the International Institute for Nanotechnology; and the State of Illinois, through the International Institute for Nanotechnology. Cell viability assays were carried out in the High Throughput Analysis (HTA) Laboratory at Northwestern University. The authors would like to thank Northwestern Visualization and the NUIT Academic & Research Technologies for preparation of the nanoconjugate graphics herein.
Footnotes
Conflict of Interest: The authors declare no competing financial interest.
Supporting Information Available: Details of synthesis, purification, and chemical characterization of small molecules, nanoparticle synthesis, characterization, raw data for calculations of τm and q NMRD fitting parameters, additional TEM images, relaxometry measurements, ICP-MS analysis, nanoconjugate colloidal stability, cell pellet images, and additional confocal images are available. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES AND NOTES
- 1.Hurst SJ, Lytton-Jean AKR, Mirkin CA. Maximizing DNA Loading on a Range of Gold Nanoparticle Sizes. Anal Chem. 2006;78:8313–8318. doi: 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dempsey MF, Condon B, Hadley DM. MRI Safety Review. Semin Ultrasound CT MR. 2002;23:392–401. doi: 10.1016/s0887-2171(02)90010-7. [DOI] [PubMed] [Google Scholar]
- 3.Willinek WA, Schild HH. Clinical Advantages of 3.0 T MRI over 1.5 T. Eur J Radiol. 2008;65:2–14. doi: 10.1016/j.ejrad.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Shellock FG, Spinazzi A. MRI Safety Update 2008: Part 2, Screening Patients for MRI. Am J Roentgenol. 2008;191:1140–1149. doi: 10.2214/AJR.08.1038.2. [DOI] [PubMed] [Google Scholar]
- 5.Shellock FG, Spinazzi A. MRI Safety Update 2008: Part 1, MRI Contrast Agents and Nephrogenic Systemic Fibrosis. Am J Roentgenol. 2008;191:1129–1139. doi: 10.2214/AJR.08.1038.1. [DOI] [PubMed] [Google Scholar]
- 6.Caravan P, Ellison JJ, McMurry TJ, Lauffer RB. Gadolinium(III) Chelates as MRI Contrast Agents: Structure, Dynamics, and Applications. Chem Rev. 1999;99:2293–2352. doi: 10.1021/cr980440x. [DOI] [PubMed] [Google Scholar]
- 7.Solomon I. Relaxation Processes in a System of Two Spins. Phys Rev. 1955;99:559–565. [Google Scholar]
- 8.Bloembergen N, Morgan LO. Proton Relaxation Times in Paramagnetic Solutions: Effects of Electron Spin Relaxation. J Chem Phys. 1961;34:842–850. [Google Scholar]
- 9.Aime S, Calabi L, Cavallotti C, Gianolio E, Giovenzana GB, Losi P, Maiocchi A, Palmisano G, Sisti M. [Gd-AAZTA]−: A New Structural Entry for an Improved Generation of MRI Contrast Agents. Inorg Chem. 2004;43:7588–7590. doi: 10.1021/ic0489692. [DOI] [PubMed] [Google Scholar]
- 10.Cohen SM, Xu J, Radkov E, Raymond KN, Botta M, Barge A, Aime S. Syntheses and Relaxation Properties of Mixed Gadolinium Hydroxypyridinonate MRI Contrast Agents. Inorg Chem. 2000;39:5747–5756. doi: 10.1021/ic000563b. [DOI] [PubMed] [Google Scholar]
- 11.Villaraza AJ, Bumb A, Brechbiel MW. Macromolecules, Dendrimers, and Nanomaterials in Magnetic Resonance Imaging: The Interplay between Size, Function, and Pharmacokinetics. Chem Rev. 2010;110:2921–2959. doi: 10.1021/cr900232t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michael AB, Xin Y, Nicole FS. Engineering Gd-Loaded Nanoparticles To Enhance MRI Sensitivity via T1 Shortening. Nanotechnology. 2013;24:462001. doi: 10.1088/0957-4484/24/46/462001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prasuhn JDE, Yeh RM, Obenaus A, Manchester M, Finn MG. Viral MRI Contrast Agents: Coordination of Gd by Native Virions and Attachment of Gd Complexes by Azide–Alkyne Cycloaddition. Chem Commun. 2007:1269–1271. doi: 10.1039/b615084e. [DOI] [PubMed] [Google Scholar]
- 14.Garimella PD, Datta A, Romanini DW, Raymond KN, Francis MB. Multivalent, High-Relaxivity MRI Contrast Agents Using Rigid Cysteine-Reactive Gadolinium Complexes. J Am Chem Soc. 2011;133:14704–14709. doi: 10.1021/ja204516p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caravan P, Parigi G, Chasse JM, Cloutier NJ, Ellison JJ, Lauffer RB, Luchinat C, McDermid SA, Spiller M, McMurry TJ. Albumin Binding, Relaxivity, and Water Exchange Kinetics of the Diastereoisomers of MS-325, a Gadolinium(III)-Based Magnetic Resonance Angiography Contrast Agent. Inorg Chem. 2007;46:6632–6639. doi: 10.1021/ic700686k. [DOI] [PubMed] [Google Scholar]
- 16.Manus LM, Mastarone DJ, Waters EA, Zhang XQ, Schultz-Sikma EA, MacRenaris KW, Ho D, Meade TJ. Gd(III)–Nanodiamond Conjugates for MRI Contrast Enhancement. Nano Lett. 2010;10:484–489. doi: 10.1021/nl903264h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kielar F, Tei L, Terreno E, Botta M. Large Relaxivity Enhancement of Paramagnetic Lipid Nanoparticles by Restricting the Local Motions of the Gd(III) Chelates. J Am Chem Soc. 2010;132:7836–7837. doi: 10.1021/ja101518v. [DOI] [PubMed] [Google Scholar]
- 18.Lee SM, Song Y, Hong BJ, MacRenaris KW, Mastarone DJ, O’Halloran TV, Meade TJ, Nguyen ST. Modular Polymer-Caged Nanobins as a Theranostic Platform with Enhanced Magnetic Resonance Relaxivity and pH-Responsive Drug Release. Angew Chem. 2010;122:10156–10160. doi: 10.1002/anie.201004867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Courant T, Roullin VG, Cadiou C, Callewaert M, Andry MC, Portefaix C, Hoeffel C, de Goltstein MC, Port M, Laurent S, et al. Hydrogels Incorporating Gd-DOTA: Towards Highly Efficient Dual T1/T2 MRI Contrast Agents. Angew Chem, Int Ed. 2012;51:9119–9122. doi: 10.1002/anie.201203190. [DOI] [PubMed] [Google Scholar]
- 20.Endres PJ, Paunesku T, Vogt S, Meade TJ, Woloschak GE. DNA–TiO2 Nanoconjugates Labeled with Magnetic Resonance Contrast Agents. J Am Chem Soc. 2007;129:15760–15761. doi: 10.1021/ja0772389. [DOI] [PubMed] [Google Scholar]
- 21.Heffern MC, Matosziuk LM, Meade TJ. Lanthanide Probes for Bioresponsive Imaging. Chem Rev. 2014;114:4496–4539. doi: 10.1021/cr400477t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song Y, Xu X, MacRenaris KW, Zhang XQ, Mirkin CA, Meade TJ. Multimodal Gadolinium-Enriched DNA–Gold Nanoparticle Conjugates for Cellular Imaging. Angew Chem, Int Ed. 2009;48:9143–9147. doi: 10.1002/anie.200904666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Oligonucleotide-Modified Gold Nanoparticles for Intracellular Gene Regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 24.Giljohann DA, Seferos DS, Patel PC, Millstone JE, Rosi NL, Mirkin CA. Oligonucleotide Loading Determines Cellular Uptake of DNA-Modified Gold Nanoparticles. Nano Lett. 2007;7:3818–3821. doi: 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seferos DS, Prigodich AE, Giljohann DA, Patel PC, Mirkin CA. Polyvalent DNA Nanoparticle Conjugates Stabilize Nucleic Acids. Nano Lett. 2009;9:308–311. doi: 10.1021/nl802958f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helm L, Merbach AE. Water Exchange on Metal Ions: Experiments and Simulations. Coord Chem Rev. 1999;187:151–181. [Google Scholar]
- 27.Manus LM, Strauch RC, Hung AH, Eckermann AL, Meade TJ. Analytical Methods for Characterizing Magnetic Resonance Probes. Anal Chem. 2012;84:6278–6287. doi: 10.1021/ac300527z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Micskei K, Helm L, Brucher E, Merbach AE. Oxygen-17 NMR Study of Water Exchange on Gadolinium Polyaminopolyacetates [Gd(DTPA)(H2O)]2− and [Gd(DOTA)(H2O)]− Related to NMR Imaging. Inorg Chem. 1993;32:3844–3850. [Google Scholar]
- 29.Aime S, Barge A, Borel A, Botta M, Chemerisov S, Merbach AE, Müller U, Pubanz D. A Multinuclear NMR Study on the Structure and Dynamics of Lanthanide(III) Complexes of the Poly(amino carboxylate) EGTA4− in Aqueous Solution. Inorg Chem. 1997;36:5104–5112. [Google Scholar]
- 30.Caravan P, Farrar CT, Frullano L, Uppal R. Influence of Molecular Parameters and Increasing Magnetic Field Strength on Relaxivity of Gadolinium- and Manganese-Based T1 Contrast Agents. Contrast Media Mol Imaging. 2009;4:89–100. doi: 10.1002/cmmi.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toth E, Helm L, Merbach AE. Relaxivity of Gadolinium (III) Complexes: Theory and Mechanism. In: Toth E, Merbach AE, editors. Chemistry of Contrast Agents in Medical Magnetic Resonance Imaging. John Wiley & Sons Ltd; New York: 2001. pp. 45–119. [Google Scholar]
- 32.Clarkson RB, Smirnov AI, Smirnova TI, Belford RL. Multi-frequency and High-Frequency EPR Methods in Contrast Agent Research: Examples from Gd3+ Chelates. In: Toth E, Merbach AE, editors. Chemistry of Contrast Agents in Medical Magnetic Resonance Imaging. John Wiley & Sons Ltd; New York: 2001. pp. 383–415. [Google Scholar]
- 33.Ruloff R, Toth E, Scopelliti R, Tripier R, Handel H, Merbach AE. Accelerating Water Exchange for Gd(III) Chelates by Steric Compression around the Water Binding Site. Chem Commun. 2002:2630–2631. doi: 10.1039/b207713b. [DOI] [PubMed] [Google Scholar]
- 34.Laus S, Ruloff R, Tóth É, Merbach AE. Gd(III) Complexes with Fast Water Exchange and High Thermodynamic Stability: Potential Building Blocks for High-Relaxivity MRI Contrast Agents. Chem—Eur J. 2003;9:3555–3566. doi: 10.1002/chem.200204612. [DOI] [PubMed] [Google Scholar]
- 35.Jaszberenyi Z, Sour A, Toth E, Benmelouka M, Merbach AE. Fine-Tuning Water Exchange on Gd(III) Poly(amino carboxylates) by Modulation of Steric Crowding. Dalton Trans. 2005:2713–2719. doi: 10.1039/b506702b. [DOI] [PubMed] [Google Scholar]
- 36.Tóth É, Pubanz D, Vauthey S, Helm L, Merbach AE. The Role of Water Exchange in Attaining Maximum Relaxivities for Dendrimeric MRI Contrast Agents. Chem—Eur J. 1996;2:1607–1615. [Google Scholar]
- 37.Strauch RC, Mastarone DJ, Sukerkar PA, Song Y, Ipsaro JJ, Meade TJ. Reporter Protein-Targeted Probes for Magnetic Resonance Imaging. J Am Chem Soc. 2011;133:16346–16349. doi: 10.1021/ja206134b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moriggi LC, Cannizzo C, Dumas E, Mayer CDR, Ulianov A, Helm L. Gold Nanoparticles Functionalized with Gadolinium Chelates as High-Relaxivity MRI Contrast Agents. J Am Chem Soc. 2009;131:10828–10829. doi: 10.1021/ja904094t. [DOI] [PubMed] [Google Scholar]
- 39.Walker DA, Leitsch EK, Nap RJ, Szleifer I, Grzybowski BA. Geometric Curvature Controls the Chemical Patchiness and Self-Assembly of Nanoparticles. Nat Nanotechnol. 2013;8:676–681. doi: 10.1038/nnano.2013.158. [DOI] [PubMed] [Google Scholar]
- 40.Ferreira MF, Mousavi B, Ferreira PM, Martins CIO, Helm L, Martins JA, Geraldes CFGC. Gold Nanoparticles Functionalised with Stable, Fast Water Exchanging Gd3+ Chelates as High Relaxivity Contrast Agents for MRI. Dalton Trans. 2012;41:5472–5475. doi: 10.1039/c2dt30388d. [DOI] [PubMed] [Google Scholar]
- 41.Graeppi N, Powell DH, Laurenczy G, Zékány L, Merbach A. Coordination Equilibria and Water Exchange Kinetics of Lanthanide(III) Propylenediaminetetraacetates and Other Magnetic Resonance Imaging Related Complexes. Inorg Chim Acta. 1995;235:311–326. [Google Scholar]
- 42.Tei L, Gugliotta G, Baranyai Z, Botta M. A New Bifunctional Gd(III) Complex of Enhanced Efficacy for MR-Molecular Imaging Applications. Dalton Trans. 2009:9712–9714. doi: 10.1039/b917566k. [DOI] [PubMed] [Google Scholar]
- 43.Axelsson O, Olsson A. Synthesis of Cyclen Derivatives. WO/2006/112723. Patent Appl. 2006
- 44.Supkowski RM, Horrocks WD., Jr On the Determination of the Number of Water Molecules, q,Coordinatedto Europium(III) Ions in Solution from Luminescence Decay Lifetimes. Inorg Chim Acta. 2002;340:44–48. [Google Scholar]
- 45.Beeby A, Clarkson IM, Dickins RS, Faulkner S, Parker D, Royle L, de Sousa AS, Williams JAG, Woods M. Non-radiative Deactivation of the Excited States of Europium, Terbium and Ytterbium Complexes by Proximate Energy-Matched OH, NH and CH Oscillators: An Improved Luminescence Method for Establishing Solution Hydration States. J Chem Soc, Perkin Trans. 1999;2:493–504. [Google Scholar]
- 46.Xie J, Lee JY, Wang DIC. Seedless, Surfactantless, High-Yield Synthesis of Branched Gold Nanocrystals in HEPES Buffer Solution. Chem Mater. 2007;19:2823–2830. [Google Scholar]
- 47.Dam DHM, Lee JH, Sisco PN, Co DT, Zhang M, Wasielewski MR, Odom TW. Direct Observation of Nanoparticle–Cancer Cell Nucleus Interactions. ACS Nano. 2012;6:3318–3326. doi: 10.1021/nn300296p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Min J, Jung H, Shin HH, Cho G, Cho H, Kang S. Implementation of P22 Viral Capsids as Intravascular Magnetic Resonance T1 Contrast Conjugates via Site-Selective Attachment of Gd(III)-Chelating Agents. Biomacromolecules. 2013;14:2332–2339. doi: 10.1021/bm400461j. [DOI] [PubMed] [Google Scholar]
- 49.Aime S, Frullano L, Geninatti Crich S. Compartmentalization of a Gadolinium Complex in the Apoferritin Cavity: A Route To Obtain High Relaxivity Contrast Agents for Magnetic Resonance Imaging. Angew Chem, Int Ed. 2002;41:1017–1019. doi: 10.1002/1521-3773(20020315)41:6<1017::aid-anie1017>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 50.Mastarone DJ, Harrison VSR, Eckermann AL, Parigi G, Luchinat C, Meade TJ. A Modular System for the Synthesis of Multiplexed Magnetic Resonance Probes. J Am Chem Soc. 2011;133:5329–5337. doi: 10.1021/ja1099616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patel PC, Giljohann DA, Daniel WL, Zheng D, Prigodich AE, Mirkin CA. Scavenger Receptors Mediate Cellular Uptake of Polyvalent Oligonucleotide-Functionalized Gold Nanoparticles. Bioconjugate Chem. 2010;21:2250–2256. doi: 10.1021/bc1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prigodich AE, Lee OS, Daniel WL, Seferos DS, Schatz GC, Mirkin CA. Tailoring DNA Structure To Increase Target Hybridization Kinetics on Surfaces. J Am Chem Soc. 2010;132:10638–10641. doi: 10.1021/ja104859j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jensen SA, Day ES, Ko CH, Hurley LA, Luciano JP, Kouri FM, Merkel TJ, Luthi AJ, Patel PC, Cutler JI, et al. Spherical Nucleic Acid Nanoparticle Conjugates as an RNAi-Based Therapy for Glioblastoma. Sci Transl Med. 2013;5:209ra152. doi: 10.1126/scitranslmed.3006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bertini I, Galas O, Luchinat C, Parigi G. A Computer Program for the Calculation of Paramagnetic Enhancements of Nuclear-Relaxation Rates in Slowly Rotating Systems. J Magn Reson Ser A. 1995;113:151–158. [Google Scholar]
- 55.Bertini I, Kowalewski J, Luchinat C, Nilsson T, Parigi G. Nuclear Spin Relaxation in Paramagnetic Complexes of S = 1: Electron Spin Relaxation Effects. J Chem Phys. 1999;111:5795–5807. [Google Scholar]
- 56.Kowalewski J, Luchinat C, Nilsson T, Parigi G. Nuclear Spin Relaxation in Paramagnetic Systems: Electron Spin Relaxation Effects under Near-Redfield Limit Conditions and Beyond. J Phys Chem A. 2002;106:7376–7382. [Google Scholar]
- 57.Bertini I, Luchinat C, Parigi G. Advances in Inorganic Chemistry. Vol. 57. Academic Press; New York: 2005. 1H NMRD Profiles of Paramagnetic Complexes and Metalloproteins; pp. 105–172. [Google Scholar]
- 58.Preslar AT, Parigi G, McClendon MT, Sefick SS, Moyer TJ, Haney CR, Waters EA, MacRenaris KW, Luchinat C, Stupp SI, et al. Gd(III)-Labeled Peptide Nanofibers for Reporting on Biomaterial Localization in Vivo. ACS Nano. 2014;8:7325–7332. doi: 10.1021/nn502393u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hung AH, Duch MC, Parigi G, Rotz MW, Manus LM, Mastarone DJ, Dam KT, Gits CC, MacRenaris KW, Luchinat C, et al. Mechanisms of Gadographene-Mediated Proton Spin Relaxation. J Phys Chem C. 2013;117:16263–16273. doi: 10.1021/jp406909b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lauffer RB. Paramagnetic Metal Complexes as Water Proton Relaxation Agents for NMR Imaging: Theory and Design. Chem Rev. 1987;87:901–927. [Google Scholar]
- 61.Helm L. Relaxivity in Paramagnetic Systems: Theory and Mechanisms. Prog Nucl Magn Reson Spectrosc. 2006;49:45–64. [Google Scholar]
- 62.Hwang LP, Freed JH. Dynamic Effects of Pair Correlation Functions on Spin Relaxation by Translational Diffusion in Liquids. J Chem Phys. 1975;63:4017–4025. [Google Scholar]
- 63.Freed JH. Dynamic Effects of Pair Correlation Functions on Spin Relaxation by Translational Diffusion in Liquids. II. Finite Jumps and Independent T1 Processes. J Chem Phys. 1978;68:4034–4037. [Google Scholar]
- 64.Botta M. Second Coordination Sphere Water Molecules and Relaxivity of Gadolinium(III) Complexes: Implications for MRI Contrast Agents. Eur J Inorg Chem. 2000;2000:399–407. [Google Scholar]
- 65.Bertini I, Fragai M, Luchinat C, Parigi G. Solvent 1H NMRD Study of Hexaaquochromium(III): Inferences on Hydration and Electron Relaxation. Inorg Chem. 2001;40:4030–4035. doi: 10.1021/ic010161j. [DOI] [PubMed] [Google Scholar]
- 66.Geraldes CFGC, Brown RD, Cacheris WP, Koenig SH, Sherry AD, Spiller M. Evaluation of Polyaza Macrocyclic Methylene Phosphonate Chelates of Gd3+ Ions as MRI Contrast Agents. Magn Reson Med. 1989;9:94–104. doi: 10.1002/mrm.1910090111. [DOI] [PubMed] [Google Scholar]
- 67.Aime S, Botta M, Terreno E, Anelli PL, Uggeri F. Gd(DOTP)5− Outer-Sphere Relaxation Enhancement Promoted by Nitrogen Bases. Magn Reson Med. 1993;30:583–591. doi: 10.1002/mrm.1910300509. [DOI] [PubMed] [Google Scholar]
- 68.Aime S, Batsanov AS, Botta M, Dickins RS, Faulkner S, Foster CE, Harrison A, Howard JAK, Moloney JM, Norman TJ, et al. Nuclear Magnetic Resonance, Luminescence and Structural Studies of Lanthanide Complexes with Octadentate Macrocyclic Ligands Bearing Benzylphosphinate Groups. J Chem Soc, Dalton Trans. 1997:3623–3636. [Google Scholar]
- 69.Aime S, Botta M, Fasano M, Terreno E. Prototropic and Water-Exchange Processes in Aqueous Solutions of Gd(III) Chelates. Acc Chem Res. 1999;32:941–949. [Google Scholar]
- 70.Alhaique F, Bertini I, Fragai M, Carafa M, Luchinat C, Parigi G. Solvent 1H NMRD Study of Biotinylated Paramagnetic Liposomes Containing Gd-Bis-SDA-DTPA or Gd-DMPE-DTPA. Inorg Chim Acta. 2002;331:151–157. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.