Abstract
Background
The perennial O. rufipogon (common wild rice), which is considered to be the ancestor of Asian cultivated rice species, contains many useful genetic resources, including drought resistance genes. However, few studies have identified the drought resistance and tissue-specific genes in common wild rice.
Results
In this study, transcriptome sequencing libraries were constructed, including drought-treated roots (DR) and control leaves (CL) and roots (CR). Using Illumina sequencing technology, we generated 16.75 million bases of high-quality sequence data for common wild rice and conducted de novo assembly and annotation of genes without prior genome information. These reads were assembled into 119,332 unigenes with an average length of 715 bp. A total of 88,813 distinct sequences (74.42% of unigenes) significantly matched known genes in the NCBI NT database. Differentially expressed gene (DEG) analysis showed that 3617 genes were up-regulated and 4171 genes were down-regulated in the CR library compared with the CL library. Among the DEGs, 535 genes were expressed in roots but not in shoots. A similar comparison between the DR and CR libraries showed that 1393 genes were up-regulated and 315 genes were down-regulated in the DR library compared with the CR library. Finally, 37 genes that were specifically expressed in roots were screened after comparing the DEGs identified in the above-described analyses.
Conclusion
This study provides a transcriptome sequence resource for common wild rice plants and establishes a digital gene expression profile of wild rice plants under drought conditions using the assembled transcriptome data as a reference. Several tissue-specific and drought-stress-related candidate genes were identified, representing a fully characterized transcriptome and providing a valuable resource for genetic and genomic studies in plants.
Introduction
Drought is one of the most common abiotic stresses that negatively influence plant growth, biomass production and crop yield [1]. Plants will activate a series of complicated regulatory mechanisms to deal with the unfavorable environment when experiencing drought [2–5]. For example, the regulation of abscisic acid (ABA) and other related transcription factors will work at the molecular and physiological and biochemical level [6–7]. On the other hand, at tissue and cellular level, plants respond to drought environment onset with morphological and cellular changes such as plasma membrane transform, growth retardation, stomatal closure, leaf wax increase, and the accumulation of osmoprotectants [8–12]. During the phenotypic and cell changes process, many genes that regulate metabolism at the physiological and biochemical level are highly expressed to enhance drought resistance [13–15]. Among these genes, several tissue-specific genes can induce morphological changes to address drought conditions. For example, DRO1, a deep-root gene, can induce rice roots to stretch into the deeper, wetter levels of soil to counteract the arid environment at the surface [16]. Overexpression of OsNAC5 enlarges root diameter in rice plants, leading to enhanced drought tolerance [17], and OsGL1-6 is involved in the accumulation of leaf cuticular wax and directly impacts drought resistance in rice [18]. With progress in molecular biology techniques, increasing numbers of genes related to drought have been isolated and identified.
As an important constituent part for plant, root systems have important biological roles in plants’ life cycle. It could anchor the plant, absorb water, inorganic salts and other nutrients, transferring hormones [19]. In recent years, more and more researchers focus on root system researches, including the growth and development of plant roots under drought stress [20]. When plants meet drought stress, responses occur in two different levels. At the morphological level, deep roots can absorb more water and nutrients; and at the molecular level, drought response genes in roots start to express as roots are among the primary sites for stress signal perception [21]. In previous reports, the responses of rice cultivar roots to drought, potassium stress, essential nutrients, structure, heavy metals and plant development have been analyzed on the transcriptomic level [22–26]. In addition, the second generation sequencing technology has been applied to identify stress-inducible transcripts in rice [27,28].
As the ancestor of cultivated rice, common wild rice which undergo a long history of artificial selection has a wide variety of excellent valuable mutated genes which have great potential value for rice breeding [29]. These plants grow in shallow water, small lakes and slow-flowing streams of eutrophication, and they possess the characters of disease and insect resistance and stress tolerance (resisting drought, high salt and low temperature), especially the developed ratoon systems.
Common wild rice grown in its natural habitat shows various resistances to biotic and abiotic stresses, as well as excellent genetic diversity. Previous studies have identified important genes controlling agronomic traits such as insect resistance, cold resistance, and high yield [30–32]. In addition, the PROG1 gene, which is involved in rice domestication [33], and the SH4 and qSH1 genes, which control rice seed shattering [34], have been isolated. Certain genetic resources involved in drought resistance in common wild rice have been analyzed, including 12 drought-tolerance-related QTLs that were identified using introgression lines with the common wild rice Dongxiang accession as a donor [35]. However, the roots and tissue-specific gene expression in common wild rice has not been reported.
In the present study, common wild rice from the Gaozhou area of Guangdong province in China was used as material for studying root and drought resistance genes. The transcriptomes from different tissues of control and drought-treated common wild rice were sequenced using the Illumina paired-end sequencing technology, the sequencing data were assembled and annotated, and differentially expressed genes (DEGs) were identified. The research is essential and helpful to understand the transcript of the mechanism of different organizations drought in common wild rice, and it will be very useful for gene annotation and discovery and for genomic and transcriptomic assembly in common wild rice.
Materials and Methods
Plant Growth Conditions and Drought Stress Treatment
Common wild rice seeds were collected from Guangzhou Province in China. It was approved by supervision department of Guangdong wild rice protection. The seeds were sowed in Petri dishes at 35°C and then the plants were transferred to pots with water-culture after germination in a growth chamber. The seedlings were grown in four pots (30seedlings/pot) representing two treatments. The composition of the nutrient solution was as described by the International Rice Research Institute [36]. The plants were cultured under the following conditions: 50–70% relative humidity under short-day (8 h light/16 h dark) conditions at 30°C in the light and at 25°C at night.
After the seedlings grew for three weeks, one treatment was performed on the seedling sets using 15% PEG-6000 nutrient solution as the drought treatment for 24 h with the symptoms of wilting, and the second set of seedlings was used as an untreated control. Then, the root and shoot tissues of both control and treated seedlings were harvested and stored at −80°C in preparation for further assays.
RNA Extraction and Transcriptome Sequencing
In this study, the shoot (CL) and root (CR) tissues of control plants and root tissue of the treated group (15% PEG-6000 stress for 24 h) (DR) were harvested for RNA extraction. RNA samples were extracted from five plants, and each sample was analyzed twice. Total RNA was extracted using Trizol reagent (Invitrogen, CA, USA) according to the manufacturer’s instructions, and the RNA quality was verified using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). The RNA Integrity Number (RIN) values for each of the samples were all greater than 8. For transcriptome sequencing libraries construction, NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA) was used. First, mRNA was purified from the pooled total RNA using polyT oligo-attached magnetic beads and fragmented using an RNA fragmentation buffer. First-strand cDNA was synthesized using buffer, dNTPs, random hexamer primers and reverse transcriptase, and the second cDNA strand was synthesized using buffer, dNTPs, RNase H, and DNA polymerase I. Then the double-stranded cDNA was purified using the GeneJET PCR purification kit, then washed with EB buffer and submitted to end repair and single nucleotide adenine addition. Finally, adaptors were added to the fragments, and then were purified with AMPure XP beads and enriched by PCR amplification. After cluster generation, the sequencing libraries were sequenced by Illumina HiSeq 2000 platform.
Data Filtering and De Novo Assembly
Raw sequence data generated from Illumina HiSeq 2000 platform were treated with following steps. First, remove adapters that were added for reverse transcription and sequencing, then sequences containing too many unknown bases (>10%) and low-quality bases (>50% of the bases with a quality score ≤5) were removed. After obtaining clean data for each of the six transcriptome libraries, the clean data were then mixed and assembly was done by using the Trinity software. The parameters were with min_kmer cov set to 2 by default and all other parameters set to the default settings [37]. All of the raw data was submitted to the database with the code Bioproject: SUX1023710 and BioSample: SRS933410.
Unigene Annotation and Functional Classification
For unigene functional annotation, all the assembled unique gene sequences were searched against the NR (http://www.ncbi.nlm.nih.gov/), PFAM (HMMER 3.0 package, hmmscan) Swiss-Prot (http://www.expasy.ch/sprot/), KEGG (http://www.genome.jp/kegg/) and Eukaryotic orthologous groups (KOG) (http://www.ncbi.nlm.nih.gov/cog/) databases using BLASTx algorithms and against the NT database using the BLASTn algorithms, with a threshold of E<1.0E-5. KOG contradistinction algorithms with a threshold of E<1.0E-3 were used to determine the top 10 unigene alignment results. For the NR and PFAM annotations, the Blast2GO program was used to conduct GO annotation of unigenes, and the WEGO software was used to construct GO functional classification [38, 39].
DGE Library Preparation, Sequencing and Mapping Analysis
Total RNAs from six samples, including shoot (CL) and root (CR) tissues of control plants and root tissue of the treated group (DR), two replicates each, were used for DGE library sequencing. For each sample, three micrograms of RNA was used for the DGE library construction. The process of the DGE sequencing library construction was same as those of the transcriptome sequencing libraries.
After getting the raw sequence data and performing the data-processing steps, the clean reads were then mapped to the assembly transcriptome reference sequences using the RSEM software [40]. Mismatches of no more than 2 bases were allowed in the alignment, and read counts for each gene were obtained from the mapping results.
Identification of Differentially Expressed Unigenes
For calculating the gene expression level, the FPKM method was used [41], then the DEGs were screened by comparing the gene expression levels. The DESeq R package (1.10.1) [42] was used to analyze the DEGs between the two conditions. In order to control the false discovery rate, the resulting P values were adjusted using the Benjamini and Hochberg’s approach. In this study, genes with an adjusted P-value < 0.05 for CR vs CL with 1.5 times changes of gene expression and an adjusted P-value < 0.1 for DR vs CR with 1.5 times changes of gene expression were used for identifying the DEGs.
Quantitative Real-Time PCR Analysis
To confirm the DEG results, quantitative real-time reverse transcription PCR (qRT-PCR) was performed on eighteen unigenes that were randomly chosen from the three libraries. qRT-PCR was implemented using the SYBR premix Ex Taq kit (TaKaRa, Dalian, China) on an ABI 7500 Real-Time System (Applied Biosystems). The relative expression value was calculated using the 2-ΔΔCt method [43], with actin (Os03g0718100) as an internal control. The six known drought response genes, including OsMYB2, SNAC1, OsNADPH-1, OsMAPK5 and OsAPX2 were also choosed for qRT-PCR analysis. In addition to these 18 unigenes, another four drought-induced unigenes were selected for expression analysis after different times of drought treatment. The gene-specific primers used in the qRT-PCR analysis are listed in S1 Table. The RNA pools used in the qRT-PCR analyses were extracted from three independent samples that were different from those used for RNA-seq. All reactions were performed using one biological sample and three technical replicates.
Development and Detection of EST-SSR Markers
Besides screening DEG information, microsatellites distribution was also detected in the assemblied transcriptomes. The MISA software (http://pgrc.ipk-gatersleben.de/misa/misa.html) was used to identify microsatellites. EST-SSRs were considered to contain motifs consisting of one to six nucleotides.
Primers for each SSR were designed using Primer3 software (http://primer3.ut.ee).
Results
Illumina Paired-End Sequencing and De Novo Assembly
Six cDNA libraries were generated from the seedling stage (shoot and root of control, root-treated) of common wild rice using Illumina deep sequencing. After removing adapters and filtering the low-quality sequences, 6.42 million, 5.74 million, and 4.59 million clean 100-bp reads were generated for the control shoot (CL1, CL2), control root (CR1, CR2), and drought-treated root (DR1, DR2) libraries, respectively (Table 1). All of the high-quality reads from these six libraries were mixed to do transcriptome assembly by using the Trinity software [37]. Based on the overlapping information provided by the high-quality assembly, 201,352 transcripts, with a mean length of 1189 bp and an N50 of 2159 bp, were generated. After extracting the longest transcript for each gene to serve as the unigene, 119,332 unigenes were obtained. The average length was 715 bp, and approximately 38.88% of the unigenes were at least 500 bp long (Table 2, S1 Fig).
Table 1. Summary of data output quality of the various libraries.
| Library | Raw Reads | Clean reads | Error(%) | Q20(%) | GC(%) |
|---|---|---|---|---|---|
| CL1 | 36,316,581 | 33,680,504 | 0.05 | 95.00 | 53.45 |
| CL2 | 32,904,668 | 30,527,534 | 0.05 | 95.01 | 53.84 |
| CR1 | 33,161,644 | 30,846,212 | 0.04 | 95.34 | 51.95 |
| CR2 | 28,544,583 | 26,630,389 | 0.04 | 95.31 | 51.90 |
| DR1 | 25,417,458 | 23,716,321 | 0.04 | 95.37 | 52.16 |
| DR2 | 26,938,151 | 25,164,179 | 0.04 | 95.38 | 52.20 |
Table 2. Summary of the common wild rice transcriptome.
| Category | Number | Total number | Mean length (bp) | N50 (bp) | Total nucleotides | |||
|---|---|---|---|---|---|---|---|---|
| 200-500bp | 500-1kb | 1k-2kb | >2kb | |||||
| Transcripts | 84,613 | 39,012 | 38,705 | 39,022 | 201,352 | 1,189 | 2,159 | 239,341,036 |
| Unigenes | 72,936 | 24,321 | 13,563 | 8,512 | 119,332 | 715 | 1,147 | 85,352,510 |
Functional Annotation and Classification of the Unigenes
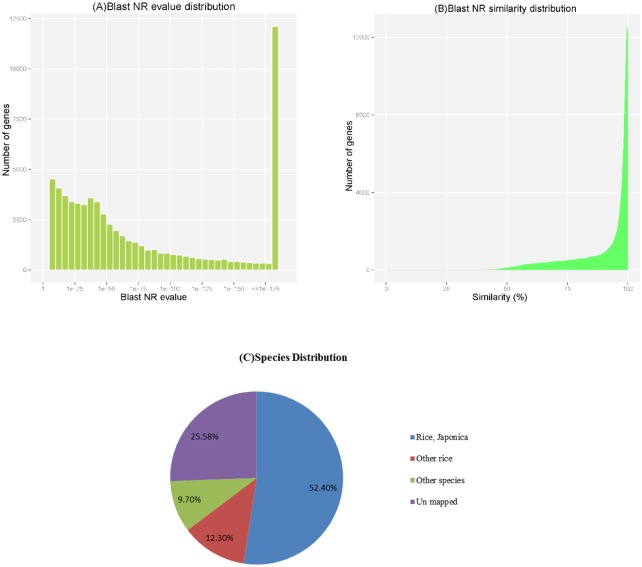
After getting the sequence of assembled unigenes, the sequences were searched against the SwissProt protein and NCBI non-redundant (NR) databases using the BLAST 2.2.28+ program (with E-value threshold of 1E-5). Of the 119,332 unique sequences, 111,171 unigenes (93.16%) had at least one significant match to an existing gene model. A total of 88,813 distinct sequences (74.42% of the unigenes) significantly matched known genes in the NT database (Table 3). Of the matched unigenes, approximately 18.87% (12424/65856) matched the NR database with an E value smaller than 1E-175 (Fig 1A), and approximately 71.71% (47226/65856) shared more than 90% similarity with an established sequence (Fig 1B). The majority of sequences (64.76%) were homologous to genes of Oryza sativa, and of those, 52.4% of the unigenes were best matched to sequences from Oryza sativa Japonica, followed by 12.3% from other Oryza sativa species and 9.7% from other species (Fig 1C). In the absence of matching to the NT database of 25.58% genes in 2572 genes have higher expression level, and 8161 genes were not matched to any database(S2 Table).
Table 3. Summary of functional annotation of the assembled unigenes.
| Public database | Number of Unigenes | Percentage (%) |
|---|---|---|
| Annotated in NR | 65,856 | 55.18 |
| Annotated in NT | 88,813 | 74.42 |
| Annotated in KO | 18,286 | 15.32 |
| Annotated in SwissProt | 38,097 | 31.92 |
| Annotated in PFAM | 42,906 | 35.95 |
| Annotated in GO | 55,318 | 46.35 |
| Annotated in KOG | 23,905 | 20.03 |
| Annotated in all Databases | 8400 | 7.03 |
| Annotated in at least one Database | 111,171 | 93.16 |
| Total Unigenes | 119,332 | 100 |
Fig 1. Characteristics of the BLAST matches of the transcriptome unigenes.
(A) E-value and (B) Similarity distribution of the top BLAST hits for each unigene in the NR database, with a cut-off E-value of 1.0E-5. (C) Species distribution of the BLAST hits for each unigene in the NT database.
Functional Classification by GO and KOG
Gene Ontology (GO) analysis was performed based on the NR annotation. Among the 65,856 annotated unigenes, 55,318 sequences were assigned to 56 GO functional groups terms (Table 2, S3 Table). Among the three main categories the biological process category was the most (147,154), following with the cellular components category (124,519) and molecular function category (69,458) (Fig 2A). Within the biological processes category, related to metabolic processes (21.44%) and cellular processes (22.21%) were the most enriched. Meanwhile, genes encoding binding proteins (45.42%) and proteins related to catalytic activity (37.99%) were enriched in the molecular function category. In the cellular components category, cell (22.38%), cell part (22.38%) and organelle (17.70%) were the largest proportion represented GO terms.
Fig 2. Functional classification of the assembled unigenes.
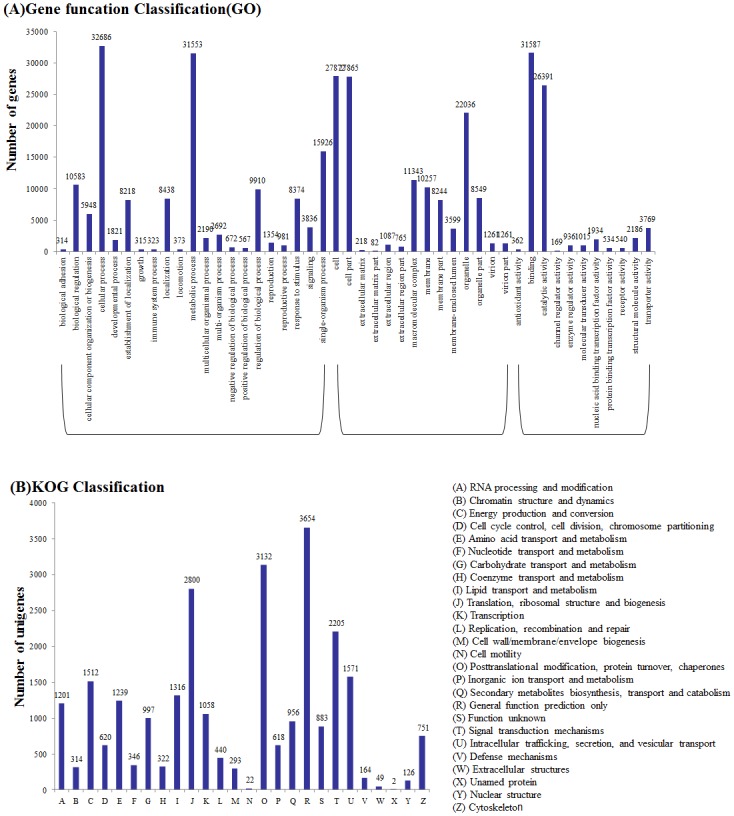
(A) Comparison of Gene Ontology (GO) classifications of common wild rice. Unigene numbers assigned to the same GO terms are indicated above the bars; the x-axis indicates the subcategories, and the y-axis indicates the number of genes in a category. (B) Histogram of the Eukaryotic Orthologous Groups (KOG) classification.
For KOG classifications, 23,905 sequences were assigned among the 65,856 NR annotated unigenes (Table 2). These sequences were assigned to 25 KOG categories, the cluster for general function prediction was the largest group (3654, 15.29%), others followed by posttranslational modification (3132, 13.10%), translation (2800, 11.71%), signal transduction (2205, 9.22%), intracellular trafficking and secretion (1571, 6.57%), and energy production and conversion (1512, 6.33%) (Fig 2B).
Functional Classification by KEGG Pathway
Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis with a cutoff of E<1E-5 was used to further understanding of the transcriptome data. Of the 119,332 unigenes, 18,286 (13.96%) with significant matches from the database were assigned to 5 main categories, including 254 KEGG pathways (Table 2, S4 Table). As shown in Fig 3, among the 5 main categories, metabolism was the biggest category (9120, 49.87%), followed by genetic information (5170, 28.27%), organismal systems (3720, 20.34%), cellular processes (2567, 14.04%) and environmental information processing (1739, 9.51%) (Fig 3).
Fig 3. Pathway assignment based on Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis.
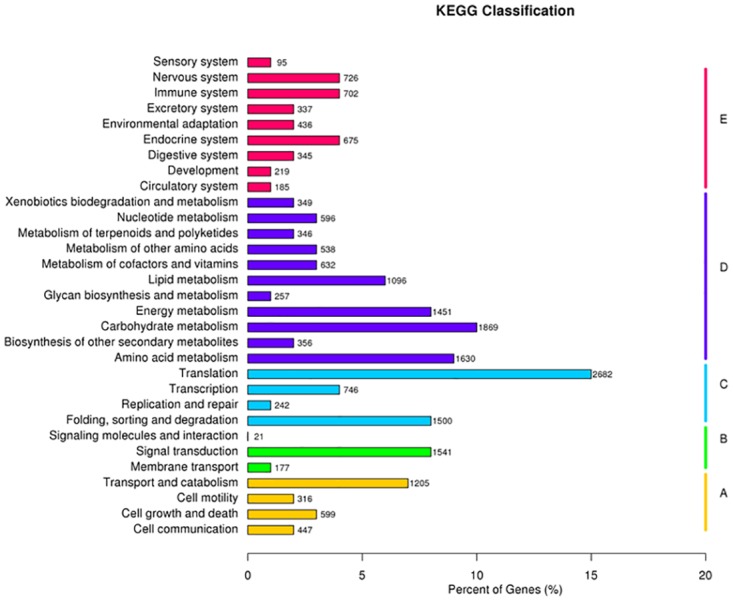
Classification based on the (A) cellular processes categories, (B) environmental information processing categories, (C) genetic information processing categories, (D) metabolism categories, (E) the organismal systems categories.
Differentially Expressed Gene (DEG) Analysis among the Three Libraries
To find the drought-induced genes in root, DEG analysis of the three transcriptome libraries was performed. First, comparing the CR and CL libraries showed that 3617 genes were up-regulated, and 4171 genes were down-regulated (Fig 4A, S5 Table). Among all DEGs, 535 were expressed in roots but not expressed in shoots (S5 Table).
Fig 4. Three digital gene expression (DGE) libraries of the common wild rice.
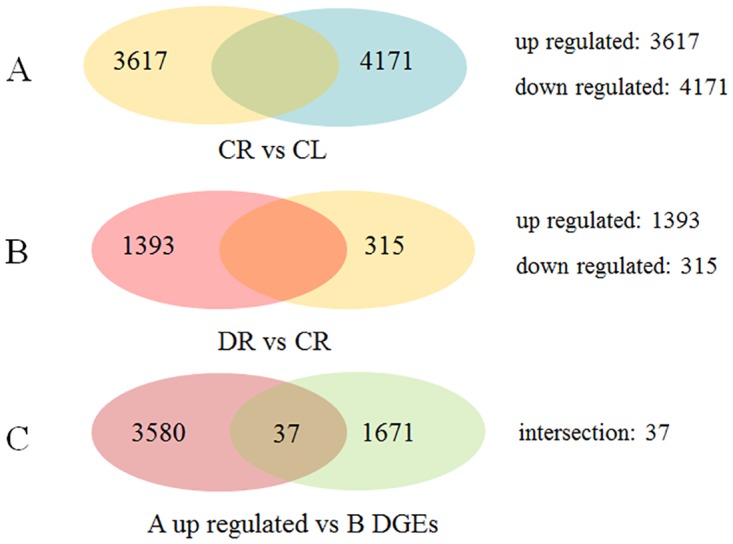
(A) DEGs obtained by comparing the CR and CL libraries. (B) DEGs obtained by comparing the CR and DR libraries. (C) DEGs identified from the up-regulated genes in A compared with DEGs in B.
According to the analysis of the differentially expressed genes between the roots (CR) and shoots (CL) shown in Fig 4A, Gene Ontology (GO) analysis on DEGs was performed based on the NR annotation. Among the 7788 DEGs, 4262 were assigned to 60 GO terms (S2A Fig), and of these genes, 2062 of the 3617 up-regulated genes and 2200 of the 4171 down-regulated genes were assigned to at least one of the GO terms(molecular function, cellular component, or biological process). Some GO terms, such as carbohydrate metabolic process, response to oxidative stress were enriched in both sets of transcripts from the two tissues. The results showed that there must be some same biological process for root activities and leaf photosynthesis. However, some significant differences were found between the two sets of enriched GO terms. In particular, GO terms related to photosynthesis and organelles were highly enriched in DEGs for roots and shoots, respectively, which also showed the expression of specific genes in different tissues. For the biological process category, 11 GO terms were found to be involved in responses to various stimuli (Table 4); response to stimulus was the most common (1364, 22.40%), followed by response to stress (757, 12.43%), response to chemical stimulus (408, 6.70%), response to abiotic stimulus (326, 5.35%), response to oxidative stress (156, 2.56%) and response to temperature stimulus (109, 1.79%) (Table 4).
Table 4. GO terms for the response to abiotic stimuli related to differentially expressed genes.
| GO Term | Name Ontology | CR vs CL (DEG_ item) | Percentage (%) | P | |
|---|---|---|---|---|---|
| Up | Down | ||||
| GO:0006979 | response to oxidative stress | 90 | 66 | 2.56 | 1.2E-31 |
| GO:0009628 | response to abiotic stimulus | 133 | 193 | 5.35 | 3.38E-28 |
| GO:0006950 | response to stress | 411 | 346 | 12.43 | 2.94E-20 |
| GO:0042221 | response to chemical stimulus | 197 | 211 | 6.70 | 7.45E-20 |
| GO:0050896 | response to stimulus | 696 | 668 | 22.40 | 8.44E-20 |
| GO:0009416 | response to light stimulus | 38 | 98 | 2.23 | 2.61E-19 |
| GO:0009314 | response to radiation | 40 | 99 | 2.28 | 3.67E-19 |
| GO:0010035 | response to inorganic substance | 87 | 101 | 3.09 | 4.95E-15 |
| GO:0009266 | response to temperature stimulus | 41 | 68 | 1.79 | 1.06E-12 |
| GO:0009409 | response to cold | 30 | 50 | 1.31 | 1.68E-11 |
| GO:0010038 | response to metal ion | 66 | 62 | 2.10 | 1.08E-10 |
To demonstrate the degree of enrichment of the response-related genes, the 11 GO terms were analyzed by the TopGO analysis. This analysis found that degree of response to abiotic stimuli was enriched the most, followed by the response to oxidative stress (S2B Fig). Among the up-regulated genes in the CR library, the strongest up-regulation occurred for the responses to stress, response to abiotic and response to oxidative stress (S2 Fig). These results showed that gene expression responses were different in different tissues; that is, genes in common wild rice show specificity of expression. Meanwhile, the DR and CR libraries were also compared, which showed the up-regulation of 1393 genes and the down-regulation of 315 genes in the DR library compared with the CR library (Fig 4B, S6 Table).
Transcription factors (TFs) is a group of protein molecule that specifically bind to the 5'-end upstream sequence of targeted gene to ensure genes’ expression under specific temporal and spatial condition. On the other hand, transcription factors are associated with the regulation of plant drought stress responses. For example, DREB transcription factor is a dehydration-responsive elements binding protein which has important regulatory roles during drought, salt and cold stress responses in plants.
TFs analysis was performed based on the Swiss-Prot annotation. The present study found that the differentially expressed TF genes of belonged to a diverse range of TF families (Table 5). Numerous transcription factors showed differential expression between the CR and CL libraries, including 40 MYB genes, 31 AP2/ERF genes, 4 HSF genes, 5 NAC genes, and 21 WRKY genes, etc. The comparison between the DR and CR libraries showed that 4 MYB genes, 19 Znf genes and 2 DREB gene were up-regulated in roots after drought stress.
Table 5. Distribution of differentially expressed transcription factor families.
| TF family | DR vs CR | CR vs CL | ||
|---|---|---|---|---|
| Up-regulated | Down-regulated | Up-regulated | Down-regulated | |
| MYB | 4 | 3 | 19 | 21 |
| AP2/ERF | 1 | 0 | 17 | 14 |
| Znf (WRKY) | 0 | 0 | 15 | 6 |
| Znf (RING-finger) | 2 | 2 | 20 | 16 |
| Znf | 19 | 5 | 104 | 103 |
| NAC | 0 | 2 | 4 | 1 |
| bZIP | 7 | 1 | 21 | 15 |
| HSF | 0 | 0 | 2 | 2 |
| Aux/IAA | 1 | 0 | 3 | 3 |
| CBF | 1 | 0 | 18 | 1 |
| DREB | 2 | 0 | 1 | 3 |
Each transcription factor contains known DNA-binding domains defined by the Pfam database.
Finally, the 3617 genes showing higher expression in the CR than CL library were compared with the 1708 genes showing differential expression between the DR and CR libraries. Thirty-seven genes were identified in both comparisons (Fig 4C, Table 6). That mean these genes were drought-induced and expressed specifically in root tissue. Of these 37 genes, 17 could not be matched to any genes in the database, that mean maybe these 17 genes were might be specific to the response to drought stress in common wild rice. Besides different types of transcriptional factors, some other types of genes have been discovered in this study, such as peroxidases. In this study 76 peroxidases related genes were found (S5 Table). Among the 76 genes, 48 genes were up-regulated in roots and 28 genes were up-regulated in leaves, meanwhile, 12 and 2 genes were found not expressing in leaves and roots, respectively. And 6 peroxidases genes were found in the DEGs between DR and CR (S6 Table).
Table 6. The thirty-seven differentially expressed genes identified from the three libraries.
| Gene_id | PFAM description | BP Description | NR E-value |
|---|---|---|---|
| comp106888_c0 | — | — | — |
| comp107776_c0 | — | — | — |
| comp32290_c1 | — | — | — |
| comp33316_c0 | Alphaherpesvirus glycoprotein E | — | 6.2E-11 |
| comp44002_c0 | Ribosomal protein L4/L1 family | ribosome biogenesis | 3.7E-145 |
| comp48589_c0 | — | — | — |
| comp53838_c0 | — | — | 4.32E-11 |
| comp56774_c0 | ADP-ribosylation factor family | intracellular protein transport | 2.3E-110 |
| comp57306_c0 | Myb-like DNA-binding domain | — | 4.46E-34 |
| comp60508_c0 | Brevenin/esculentin/gaegurin/rugosin family | defense response | — |
| comp62152_c1 | Tap, RNA-binding | protein phosphorylation | 9.48E-92 |
| comp63362_c0 | — | — | — |
| comp65617_c0 | — | — | — |
| comp66822_c0 | PRONE (Plant-specific Rop nucleotide exchanger) | root hair cell development | 0 |
| comp70721_c2 | Peroxidase | response to oxidative stress | 2.6E-99 |
| comp73162_c3 | Cecropin family | — | 5.53E-18 |
| comp73227_c1 | — | — | 0 |
| comp74521_c0 | Tubulin C-terminal domain | protein polymerization | 0 |
| comp75305_c0 | Myb-like DNA-binding domain | — | 3.43E-75 |
| comp75490_c0 | — | — | — |
| comp75573_c1 | Membrane transport protein | oxidation-reduction process | 1.4E-115 |
| comp75601_c0 | — | — | — |
| comp76407_c5 | EamA-like transporter family | — | 3E-119 |
| comp76878_c0 | Type III restriction enzyme, res subunit | pathogenesis | 0 |
| comp77894_c0 | — | — | 3.6E-109 |
| comp78054_c0 | — | — | — |
| comp78880_c0 | Peroxidase | response to oxidative stress | 1.49E-41 |
| comp79092_c0 | Calponin homology (CH) domain | — | 2.5E-146 |
| comp81484_c0 | Glycosyltransferase family 28 N-terminal domain | carbohydrate metabolic process | 1.19E-43 |
| comp83193_c0 | Cytochrome b559, alpha (gene psbE) and beta (gene psbF) subunits | photosynthesis | — |
| comp83816_c0 | — | — | — |
| comp84209_c0 | Fz domain | — | — |
| comp84868_c0 | — | — | — |
| comp84983_c0 | Sarcoglycan alpha | — | — |
| comp87399_c0 | — | — | — |
| comp88336_c0 | — | — | — |
| comp97810_c0 | — | — | 1.4E-63 |
—, no hits in the specific database
Verification of the Expression of Selected Differentially Regulated Genes by Quantitative Real-Time PCR
To confirm the gene expression data, 18 up or down-regulated unigenes were randomly chosen from the three libraries for qRT-PCR analysis. Among the 18 genes, 10 genes belonged to the 37 selected DEGs. For most of the 18 genes, their expression patterns in the real-time PCR analysis were similar to those predicted except comp77129_c2 and comp39308_c0 by the transcriptome sequencing data, for example the comp75573_c1 and comp74521_c0 genes were not expressed in shoots (Fig 5). The expression level of six known drought response genes significantly increased after drought stress. These results were consistent with that of previous researches.
Fig 5. Confirmation of the transcriptomic profiles of selected genes by qRT-PCR.
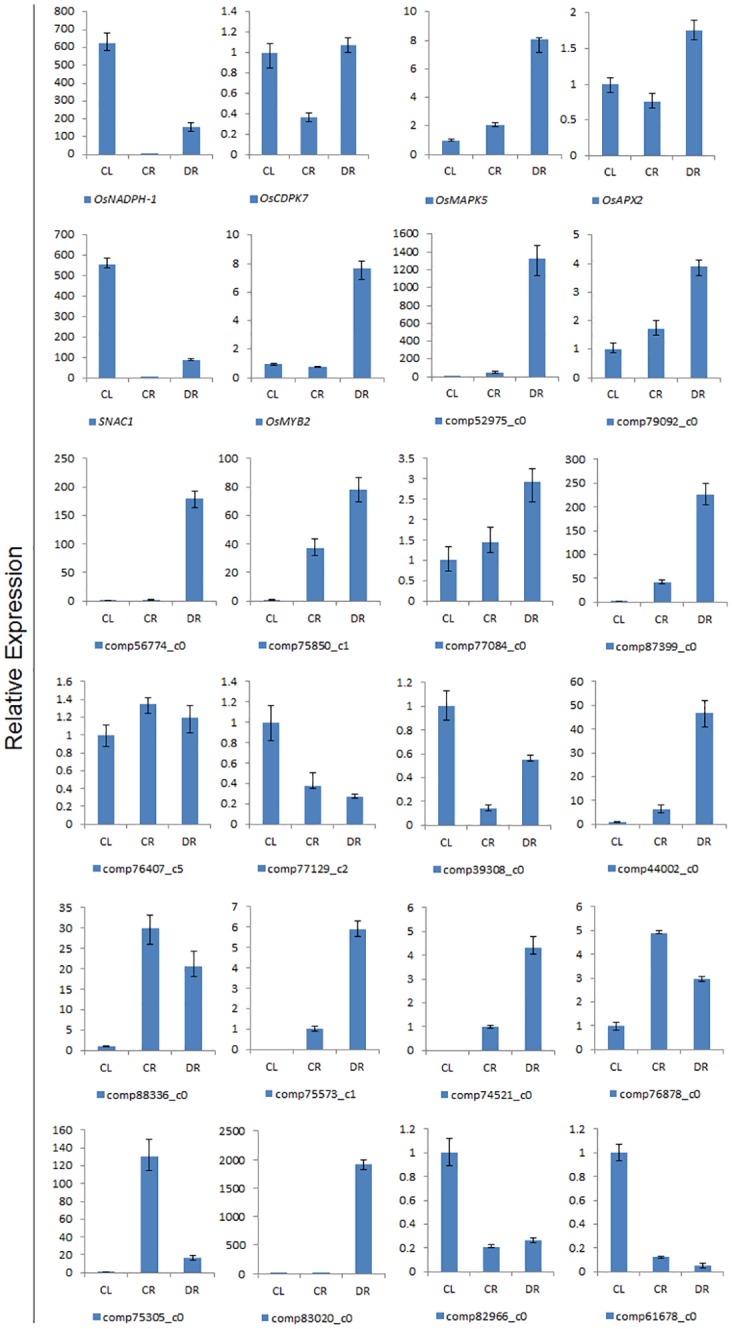
The x-axis shows the three samples; the expression level of CL was used as a control. The y-axis shows the relative quantitative expression level of each unigene.
Next, we tested the expression patterns of four of the 37 differentially expression genes to testing the expression pattern in response to drought stress (15% PEG-6000) in root tissues. The transcription sequencing results showed that two of these four genes, comp83816_c0 and comp60508_c0, gene were up regulated by drought, and the other two genes, comp78054_c0 and comp88336_c0, were down-regulated by drought. In the qRT-PCR experiment, the transcription levels of the comp83816_c0 and comp60508_c0 genes gradually increased in response to drought stress at 1 to 2 days and then decreased at 3 days. The transcript level of the comp88336_c0 gene gradually decreased in response to drought stress on days 1 to 3, and the level of comp88336_c0 quickly decreased in response to drought stress (Fig 6).
Fig 6. Expression characters of four selected unigenes in the roots of PEG-treated plants at 0 h, 24 h, 48 h and 72 h.
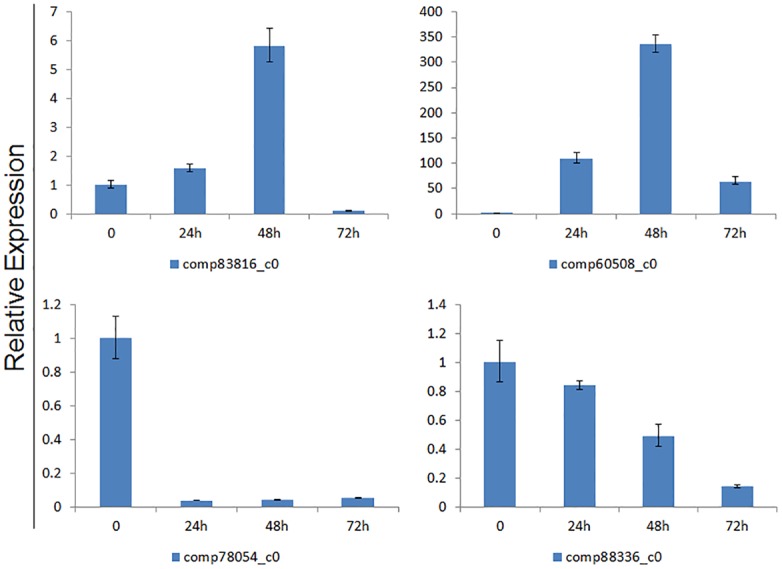
The relative expression value was calculated using the 2-ΔΔCt method and 0 h as a control sample.
Microsatellite Distribution Analysis and EST-SSR Marker Development
In order to analyze the microsatellites distribution in the common wild rice genome, the 119,332 unigenes generated in this study were used to mine potential microsatellites that were defined as mono- to hexanucleotide motifs with a minimum of three repetitions. A total of 26,574 potential simple sequence repeats (SSRs) were identified in 20,586 unigenes. Of these 20,586 unigenes, 15,973 and 4613 unigenes contained one or more than one SSR, respectively (Table 7). The number of potential EST-SSRs per unigene varied from 1 to 9, with an average of 1.29. For the motif types, the Tri-nucleotide type was the most one. Using the SSR-containing sequences, 12,324 SSR sites were designed EST-SSR primers with the Primer Premier 3.0 software. Information about these EST-SSR primers is shown in S7 Table. In the 12,324 SSR sites, about 2220 unigenes which had SSR sites were differentially express between CR and CL, while only 145 DEGs found between DR and CR.
Table 7. Summary of the EST-SSRs that were identified in the transcriptome data.
| Search item | Numbers |
|---|---|
| Total number of examined unigenes | 119,332 |
| Total size of examined sequences (bp) | 85,352,510 |
| Total number of identified EST-SSRs | 26,574 |
| Number of EST-SSRs containing sequences | 20,586 |
| Number of sequences containing more than one SSR | 4,613 |
| Mono-nucleotide | 9,889 |
| Di-nucleotide | 3,871 |
| Tri-nucleotide | 12,378 |
| Tetra-nucleotide | 392 |
| Penta-nucleotide | 29 |
| Hexa-nucleotide | 15 |
Discussion
As one of the important component in wild rice, common wild rice is a very useful resource which has many useful genes could be used for cultivated rice improvement. Many genes associated with important agronomic traits, such as domestication, disease resistance, and insect resistance, have been identified [30, 33, 34, 44–46]. In previous studies researchers used common wild rice and cultivated rice to do hybridization and got many inbred lines to increase the drought resistance [35]. While until now there is little information about the drought-related genes resources in common wild rice. In the current study, we used next-generation sequencing technologies to analyze the transcriptomic changes that occur in common wild rice during drought stress to identify the genes that are induced in roots under this condition. As expected, we identified many known genes, including transcription factors, and some possible new genes to be involved in the drought-response pathway in common wild rice.
More than 33 million raw reads were generated by Illumina sequencing. With de novo assembly, the raw reads were further assembled into 119,332 unigenes, which was much higher than the number of genes in found in cultivated Japonica rice. One possible reason for this difference in gene number is the high heterozygosity of common wild rice [47,48]. The N50 length of the unigenes was 1147 bp, and the average length was 715 bp; these results were comparable to other recently published plant transcriptomic analyses, such as that for Reaumuria soongorica (N50 = 1109 bp, average length = 677 bp) [49]. More than half of the unigenes identified in this study (65856, 55.18%) were successfully assigned to annotated genes by BLAST against public databases. Among the unigenes, a BLAST against the nt database showed that 62,572 (52.40%) had significant homology to functional genes encoding specific proteins in Oryza sativa Japonica. As expected, most of the genes (64.7%) were matched to cultivated rice, indicating that the genome of cultivated rice is highly similar to that of common wild rice. However, other than the matched genes, no genes could be matched to any database, indicating that common wild rice contains some new and nonconserved genes. After unigene assembly, the DEGs were screened using comparative analyses of the CL, CR and DR libraries. When comparing the CL and CR libraries, 366 DEGs were found to be only expressed in leaves, and 535 were only expressed in roots, indicating that these genes were tissue-specific. This result was consistent with results in previous studies. Some tissue specific gene, such as root-specific gene OsEXPA8 and cell-wall invertase genes of rice at flowering have been identified [50,51]. Furthermore, the gene expression patterns of different organs of rice shoots under the drought and high-salinity stress have also been revealed [52]. Many of the tissue-specific genes identified in this study were associated with abiotic stresses, including water, cold, and heat stress (S8 Table), suggesting that common wild rice tissue has a unique strategy for managing environmental stress.
Previous researches have shown that gene expression altering when plants exposed to drought stress. To discover more drought-related genes that are specifically expressed in roots, we enlarged the threshold value to P-value<0.1. After performing DEG analysis, 37 possible drought-related and tissue-specific genes were identified, including some known transcriptional factors, such as the MYB genes [53].
After analyzing the DEG information, we selected six drought related genes, including one perpxidases gene to do real-time PCR to check the expression character during the drought treatment (Fig 6). The differentially expressed genes between CR and CL showed that this type of gene has the character of tissue specific in plants, and the differentially expressed genes between DR and CR showed that the increasing expression level of perpxidases genes could enhance the peroxisome level in cells to protect the cells in plants.
Among the 37 DEGs regulated by drought stress in common wild rice, over 50% had no homologs in the NCBI database, indicating that some of these genes may represent new drought-related transcripts that have not been reported in plants and that these genes may be useful resources for the genetic improvement of crops.
Roots and leaves are important tissues which could percept the drought stress information. In this study, we didn’t analyze the sequence data from drought treated leaves as we want to extract the important information from roots, and the gene information from drought leaves will be further analyzed in the future.
Supporting Information
The lengths of the interval assembly transcripts/unigenes are plotted on the horizontal axis, and the lengths of each assembled transcript are plotted on the vertical axis.
(TIF)
(A) Gene Ontology (GO) categorization of the DEGs. (B) DAG graph of the topGO analysis for the DEGs. (C) DAG graph of the topGO analysis for the DEGs that are up-regulated by drought. Each cycle or square represents one GO term. The more dark colour showed that the gene enrichment density was higher.
(TIF)
(XLSX)
(XLS)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the National Special Program of Transgenic Research (Nos. 2014ZX08011-001).
References
- 1. Minh-Thu PT, Hwang DJ, Jeon JS, Nahm BH, Kim YK (2013) Transcriptome analysis of leaf and root of rice seedling to acute dehydration. Rice 6: 38 10.1186/1939-8433-6-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Friedel S, Usadel B, von Wiren N, Sreenivasulu N (2012) Reverse engineering: a key component of systems biology to unravel global abiotic stress cross-talk. Front Plant Sci 3: 294 10.3389/fpls.2012.00294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58: 221–227. [DOI] [PubMed] [Google Scholar]
- 4. Hadiarto T, Tran LS (2011) Progress studies of drought-responsive genes in rice. Plant Cell Rep 30: 297–310. 10.1007/s00299-010-0956-z [DOI] [PubMed] [Google Scholar]
- 5. Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K (2011) Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol 11: 163 10.1186/1471-2229-11-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Santos AP, Serra T, Figueiredo DD, Barros P, Lourenco T, Chander S, et al. (2011) Transcription regulation of abiotic stress responses in rice: a combined action of transcription factors and epigenetic mechanisms. OMICS 15: 839–857. 10.1089/omi.2011.0095 [DOI] [PubMed] [Google Scholar]
- 7. Busk PK, Pages M (1998) Regulation of abscisic acid-induced transcription. Plant Mol Biol 37: 425–435. [DOI] [PubMed] [Google Scholar]
- 8. Guo L, Wang ZY, Lin H, Cui WE, Chen J, Liu M, et al. (2006) Expression and functional analysis of the rice plasma-membrane intrinsic protein gene family. Cell Res 16: 277–286. [DOI] [PubMed] [Google Scholar]
- 9. Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, et al. (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51: 617–630. [DOI] [PubMed] [Google Scholar]
- 10. Lovisolo C, Perrone I, Hartung W, Schubert A (2008) An abscisic acid-related reduced transpiration promotes gradual embolism repair when grapevines are rehydrated after drought. New Phytol 180: 642–651. 10.1111/j.1469-8137.2008.02592.x [DOI] [PubMed] [Google Scholar]
- 11. Islam MA, Du H, Ning J, Ye H, Xiong L (2009) Characterization of Glossy1-homologous genes in rice involved in leaf wax accumulation and drought resistance. Plant Mol Biol 70: 443–456. 10.1007/s11103-009-9483-0 [DOI] [PubMed] [Google Scholar]
- 12. Yamada M, Morishita H, Urano K, Shiozaki N, Yamaguchi-Shinozaki K, Shinozaki K, et al. (2005) Effects of free proline accumulation in petunias under drought stress. J Exp Bot 56: 1975–1981. [DOI] [PubMed] [Google Scholar]
- 13. Kim H, Lee K, Hwang H, Bhatnagar N, Kim DY, Yoon IS, et al. (2014) Overexpression of PYL5 in rice enhances drought tolerance, inhibits growth, and modulates gene expression. J Exp Bot 65: 453–464. 10.1093/jxb/ert397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo C, Ge X, Ma H (2013) The rice OsDIL gene plays a role in drought tolerance at vegetative and reproductive stages. Plant Mol Biol 82: 239–253. 10.1007/s11103-013-0057-9 [DOI] [PubMed] [Google Scholar]
- 15. Liu AL, Zou J, Liu CF, Zhou XY, Zhang XW, Luo GY, et al. (2013) Over-expression of OsHsfA7 enhanced salt and drought tolerance in transgenic rice. BMB Rep 46: 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, et al. (2013) Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat Genet 45: 1097–1102. 10.1038/ng.2725 [DOI] [PubMed] [Google Scholar]
- 17. Jeong JS, Kim YS, Redillas MC, Jang G, Jung H, Bang SW, et al. (2013) OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol J 11: 101–114. 10.1111/pbi.12011 [DOI] [PubMed] [Google Scholar]
- 18. Zhou L, Ni E, Yang J, Zhou H, Liang H, Li J, et al. (2013) Rice OsGL1-6 is involved in leaf cuticular wax accumulation and drought resistance. PLoS One 8: e65139 10.1371/journal.pone.0065139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhai R, Feng Y, Wang H, Zhan X, Shen X, Wu W, et al. (2013) Transcriptome analysis of rice root heterosis by RNA-Seq. BMC Genomics 14: 19 10.1186/1471-2164-14-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Henry A, Gowda VRP, Torres RO, McNally KL, Serraj R (2011) Variation in root system architecture and drought response in rice (Oryza sativa): Phenotyping of the OryzaSNP panel in rainfed lowland fields. Field Crops Research 120: 205–214. [Google Scholar]
- 21. Rabello AR, Guimaraes CM, Rangel PH, Da SF, Seixas D, Souza E, et al. (2008) Identification of drought-responsive genes in roots of upland rice (Oryza sativa L). BMC Genomics 9: 485 10.1186/1471-2164-9-485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moumeni A, Satoh K, Kondoh H, Asano T, Hosaka A, Venuprasad R, et al. (2011) Comparative analysis of root transcriptome profiles of two pairs of drought-tolerant and susceptible rice near-isogenic lines under different drought stress. BMC Plant Biol 11: 174 10.1186/1471-2229-11-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ma TL, Wu WH, Wang Y (2012) Transcriptome analysis of rice root responses to potassium deficiency. BMC Plant Biol 12: 161 10.1186/1471-2229-12-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takehisa H, Sato Y, Antonio BA, Nagamura Y (2013) Global transcriptome profile of rice root in response to essential macronutrient deficiency. Plant Signal Behav 8: e24409 10.4161/psb.24409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dubey S, Shri M, Misra P, Lakhwani D, Bag SK, Asif MH, et al. (2014) Heavy metals induce oxidative stress and genome-wide modulation in transcriptome of rice root. Funct Integr Genomics. 14(2):401–417. 10.1007/s10142-014-0361-8 [DOI] [PubMed] [Google Scholar]
- 26. Takehisa H, Sato Y, Igarashi M, Abiko T, Antonio BA, Kamatsuki K, et al. (2012) Genome-wide transcriptome dissection of the rice root system: implications for developmental and physiological functions. Plant J 69: 126–140. 10.1111/j.1365-313X.2011.04777.x [DOI] [PubMed] [Google Scholar]
- 27. Mizuno H, Kawahara Y, Sakai H, Kanamori H, Wakimoto H, Yamagata H, et al. (2010) Massive parallel sequencing of mRNA in identification of unannotated salinity stress-inducible transcripts in rice (Oryza sativa L.). BMC Genomics 11: 683 10.1186/1471-2164-11-683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jo K, Kwon HB, Kim S (2014) Time-series RNA-seq analysis package (TRAP) and its application to the analysis of rice, Oryza sativa L. ssp. Japonica, upon drought stress. Methods 67: 364–372. 10.1016/j.ymeth.2014.02.001 [DOI] [PubMed] [Google Scholar]
- 29. Tanksley SD, Mccouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild[J]. Science 277(5329): 1063–1066. [DOI] [PubMed] [Google Scholar]
- 30. Ishii T, Brar DS, Multani DS, Khush GS (1994) Molecular tagging of genes for brown planthopper resistance and earliness introgressed from Oryza australiensis into cultivated rice, O. sativa . Genome 37: 217–221. [DOI] [PubMed] [Google Scholar]
- 31. Liu FX, Sun CQ, Tan LB, Fu YC, Li DJ, Wang XK(2003) Identification and mapping of quantitative trait loci controlling cold-tolerance of Chinese common wild rice (O.rufipogon Griff.) at booting to flowering stages. Chinese Science Bulletin 48(19):2068–2071. [Google Scholar]
- 32. Li DJ, Sun CQ, Fu YC, Li C, Zhu ZF, Chen L, et al. (2002) Identification and mapping of genes for improving yield from Chinese common wild rice (O.rufipogon Griff.) using advanced backcross QTL analysis. Chinese Science Bulletin 18: 1533–1537. [Google Scholar]
- 33. Tan L, Li X, Liu F, Sun X, Li C, Zhu Z, et al. (2008) Control of a key transition from prostrate to erect growth in rice domestication. Nat Genet 40: 1360–1364. 10.1038/ng.197 [DOI] [PubMed] [Google Scholar]
- 34. Zhou Y, Lu D, Li C, Luo J, Zhu BF, Zhu J, et al. (2012) Genetic control of seed shattering in rice by the APETALA2 transcription factor SHATTERING ABORTION1 [C][W][OA] . Plant Cell 24: 1034–1048. 10.1105/tpc.111.094383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang X, Zhou S, Fu Y, Su Z, Wang X, Sun C (2006) Identification of a drought tolerant introgression line derived from Dongxiang common wild rice (O. rufipogon Griff.). Plant Mol Biol 62: 247–259. [DOI] [PubMed] [Google Scholar]
- 36. Yoshida S, Forno D, Cook J, Gomez K (1976) Laboratory manual for physiological studies of rice: Manila: International Rice Research Institute; 61–67 p. [Google Scholar]
- 37. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, et al. (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36: 3420–3435. 10.1093/nar/gkn176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, et al. (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34: W293–W297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 42. Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, et al. (2008) A large genome center's improvements to the Illumina sequencing system. Nat Methods 5: 1005–1010. 10.1038/nmeth.1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, et al. (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21 . Science 270: 1804–1806. [DOI] [PubMed] [Google Scholar]
- 45. Jeung JU, Kim BR, Cho YC, Han SS, Moon HP, Lee YT, et al. (2007) A novel gene, Pi40(t), linked to the DNA markers derived from NBS-LRR motifs confers broad spectrum of blast resistance in rice. Theor Appl Genet 115: 1163–1177. [DOI] [PubMed] [Google Scholar]
- 46. Xiao Jinhua, Grandillo Silvana, Sang Nag Ahn, McCouch SR (1996) Genes from wild rice improve yield. Nature: 223–224. [Google Scholar]
- 47. Zhang QJ, Zhu T, Xia EH, Shi C, Liu YL, Zhang Y, et al. (2014) Rapid diversification of five Oryza AA genomes associated with rice adaptation. Proc Natl Acad Sci U S A 10.1073/pnas.1418307111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang F, Yuan QH, Shi L, Qian Q, Liu WG, Kuang BG, et al. (2006) A large-scale field study of transgene flow from cultivated rice (Oryza sativa) to common wild rice (O. rufipogon) and barnyard grass (Echinochloa crusgalli). Plant Biotechnol J 4: 667–676. [DOI] [PubMed] [Google Scholar]
- 49. Shi Y, Yan X, Zhao P, Yin H, Zhao X, Xiao H, et al. (2013) Transcriptomic analysis of a tertiary relict plant, extreme xerophyte Reaumuria soongorica to identify genes related to drought adaptation. PLoS One 8: e63993 10.1371/journal.pone.0063993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ji XM, Raveendran M, Oane R, Ismail A, Lafitte R, Bruskiewich R, et al. (2005) Tissue-specific expression and drought responsiveness of cell-wall invertase genes of rice at flowering. Plant Mol Biol 59: 945–964. [DOI] [PubMed] [Google Scholar]
- 51. Ma N, Wang Y, Qiu S, Kang Z, Che S, Wang G, et al. (2013) Overexpression of OsEXPA8, a root-specific gene, improves rice growth and root system architecture by facilitating cell extension. PLoS One 8: e75997 10.1371/journal.pone.0075997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhou J, Wang X, Jiao Y, Qin Y, Liu X, He K, et al. (2007) Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle. Plant Mol Biol 63: 591–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Kim M, et al. (2010) Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol 153: 185–197. 10.1104/pp.110.154773 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The lengths of the interval assembly transcripts/unigenes are plotted on the horizontal axis, and the lengths of each assembled transcript are plotted on the vertical axis.
(TIF)
(A) Gene Ontology (GO) categorization of the DEGs. (B) DAG graph of the topGO analysis for the DEGs. (C) DAG graph of the topGO analysis for the DEGs that are up-regulated by drought. Each cycle or square represents one GO term. The more dark colour showed that the gene enrichment density was higher.
(TIF)
(XLSX)
(XLS)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.