Abstract
Breast cancer is one of the most common causes of cancer death in women; therefore, the study of molecular aspects of breast cancer for finding new biomarkers is important. Recent studies have shown that WW domain-binding protein 2 (WBP2) is important for the oncogenic property of breast cancer. WWP2 N-terminal-like (WBP2NL) is a testis-specific signaling protein that induces meiotic resumption and oocyte activation events. Our previous study revealed that WBP2NL gene expression is elevated in actively dividing cells and it might be associated with cellular proliferation and tumorigenic process. However, the clinical relevance and importance of WBP2NL gene in cancer has not been understood yet. Therefore, we were interested in analyzing the expression of WBP2NL gene in human breast cancer tissues and breast cancer cell lines, for the first time. We used reverse transcription-polymerase chain reaction (RT-PCR) and semi-nested RT-PCR to evaluate the expression of WBP2NL in malignant breast cancer and adjacent noncancerous tissue (ANCT) samples, as well as MCF-7 and MDA-MB-231 cell lines. The WBP2NL gene was expressed in 45 out of 50 (90%) breast cancer tissues and overexpressed in the MDA-MB-231 cell line. We suggest that WBP2NL may play roles in breast cancer activation maybe through binding to a group I WW domain protein. The elevated expression of WBP2NL gene in breast cancer and MDA-MB-231 cell line leads us to suggest that WBP2NL might be considered as a novel prognostic factor for early diagnosis of breast cancer.
Keywords: breast cancer, WBP2NL, gene expression, cancer/testis genes
Introduction
Breast cancer is the most frequent cause of cancer death in women worldwide. Approximately, 1 out of 10 women develops breast cancer during her lifetime.1 Genetic tests that evaluate the biology of cancer are usually used to understand more about breast cancer, particularly to help predict the cancer recurrence risk, and thereby to help choose appropriate treatment. Therefore, study on the molecular aspects of breast cancer to find the genes associated with breast cancer risk and new biomarkers for early diagnosis of breast cancer is very important.
Sertoli cells in the testis make a blood–testis barrier that provides an immune-privileged site for germ cell’s antigens. Cancer/testis (CT) genes are a family of genes that are normally expressed during spermatogenesis, usually in spermatocytes, and also in various human cancers, but absent in normal somatic cells.2,3 Most CT genes induce cell proliferation and differentiation in normal testis, and have roles in meiosis.3 The aberrant expression of CT genes in cancerous cells might cause abnormality in chromosome segregation and cell divisions.3 Because of the blood–testis barrier and the immune-privileged status of testicular tissue, and also because of the probable roles of CT genes in oncogenesis, these immunogenic proteins are being strongly considered as potential targets for cancer vaccines and immunotherapy.4,5
Recent studies have demonstrated that WW-binding protein 2 (WBP2) activates the transcription of both estrogen and progesterone receptors (ERα/PR), and thereby is involved in the regulation of ERα/PR target genes and breast cancer.6–8 It has been revealed that WW domain-mediated interaction with WBP2 is important for the oncogenic properties of the PDZ-binding motif (TAZ) in breast cancer.9–11
WWP2 N-terminal like (WBP2NL), also known as post-acrosomal sheath WW domain-binding protein (PAWP), is a testis-specific protein and has no expression in other tissues. WBP2NL is a 32-kDa signaling molecule localized in the post-acrosomal sheath (PAS) of spermatozoa.12,13 In humans, the WBP2NL gene is located on chromosome 22q13.2 and consists of six exons extending over 29/69 kb. The N-terminal part of WBP2NL has homology with WW domain binding protein-2, whereas the C-terminal part contains a PPXY motif that allows it to interact with group I WW domain proteins such as TAZ and the yes-associated protein (YAP). The WBP2NL protein enters the oocyte during fertilization, and induces meiotic resumption and oocyte activation events.14–16
A previous study by Aarabi et al has shown that WBP2NL contributes to sperm-induced calcium release and thereby initiates oocyte activation events.17 Our previous study revealed that WBP2NL is expressed in embryonic stem cells and actively dividing cancerous cell lines, and we suggested that it might be associated with cellular proliferation and tumorigenic processes.18 However, the mechanisms of WBP2NL in cell proliferation and its clinical relevance have not been understood yet. In the present study, we examined the expression of WBP2NL in normal and breast cancer tissues as well as in breast cancer cell lines in order to get more insights into its potential role in cellular proliferation and carcinogenesis, for the first time.
Materials and Methods
Patient and normal tissue samples
The study included 50 breast cancer tissues and 30 adjacent noncancerous tissue (ANCT) samples obtained from Imam Khomeini Hospital affiliated to Tehran University of Medical Sciences (Table 1). All patients had been diagnosed on the basis of clinical and laboratory findings (Table 2). Normal testis tissue, as positive control for WBP2NL gene expression, was obtained from a prostate cancer patient after orchiectomy. The study complied with the good clinical practice according to the principles of the Dec laration of Helsinki. Written informed consent was obtained from all subjects participating in the study.
Table 1.
Major pathological features and characteristics of patients.
| SUBJECT CHARACTERISTICS | NO. OF SUBJECTS (n = 50) |
|---|---|
| Age (years) | 35–65 (mean: 50) |
| Histology | |
| Ductal | 46 (92%) |
| Others | 4 (8%) |
| Grade | |
| 1 | 10 (20%) |
| 2–3 | 40 (80%) |
| HER2/neu | |
| Negative | 37 (74%) |
| Positive | 13 (26%) |
| ER/PR | |
| Negative | 12 (25%) |
| Positive | 38 (75%) |
Table 2.
Classification of pathological features and WBP2NL expression from breast cancer patients.
| NO. | PATIENT CODE | AGE | CANCER HISTOLOGY | CANCER GRADE | HER2/neu EXPRESSION | ER EXPRESSION | PR EXPRESSION | WBP2NL OVER EXPRESSION (FIRST PCR) | NESTED PCR |
|---|---|---|---|---|---|---|---|---|---|
| 1 | T201 | 47 | ISDC | II | − | +++ | + | − | + |
| 2 | T202 | 57 | IDC | II | +++ | − | − | + | + |
| 3 | T203 | 62 | IDC | III | ++ | + | + | ++ | + |
| 4 | T204 | 44 | ISDC | I | − | + | + | − | + |
| 5 | T205 | 57 | IDC | II | ++ | − | − | − | + |
| 6 | T206 | 48 | ISDC | II | − | + | − | − | − |
| 7 | T207 | 35 | IDC | III | ++ | ++ | + | + | + |
| 8 | T208 | 39 | IDC | II | − | − | + | − | + |
| 9 | T209 | 52 | ISDC | II | − | + | ++ | − | + |
| 10 | T210 | 41 | IDC | III | +++ | − | − | +++ | + |
| 11 | T211 | 46 | ISDC | I | − | + | ++ | − | − |
| 12 | T212 | 63 | ISDC | II | − | + | + | − | + |
| 13 | T213 | 51 | IDC | II | − | − | − | ++ | + |
| 14 | T214 | 49 | ISDC | I | − | +++ | − | − | + |
| 15 | T215 | 55 | ISDC | II | − | + | ++ | − | + |
| 16 | T216 | 44 | ISDC | II | − | − | − | − | + |
| 17 | T217 | 61 | ISDC | I | − | ++ | + | − | + |
| 18 | T218 | 42 | IDC | III | +++ | − | − | +++ | + |
| 19 | T219 | 47 | ISDC | II | − | + | +++ | − | + |
| 20 | T220 | 35 | IMC | II | + | + | + | − | + |
| 21 | T221 | 58 | IDC | II | +++ | − | − | − | + |
| 22 | T222 | 53 | IDC | II | ++ | − | − | ++ | + |
| 23 | T223 | 50 | ISDC | II | − | ++ | + | − | + |
| 24 | T224 | 48 | IMC | II | − | + | ++ | − | + |
| 25 | T225 | 37 | ISDC | II | − | − | − | − | + |
| 26 | T226 | 47 | IDC | II | ++ | + | − | +++ | + |
| 27 | T227 | 60 | ISDC | II | − | ++ | + | + | + |
| 28 | T228 | 45 | ISDC | I | − | + | ++ | − | + |
| 29 | T229 | 63 | IDC | II | − | + | − | − | + |
| 30 | T230 | 55 | IDC | II | − | ++ | + | +++ | + |
| 31 | T231 | 38 | ISDC | I | − | ++ | ++ | − | − |
| 32 | T232 | 65 | ISDC | II | − | ++ | + | − | + |
| 33 | T233 | 44 | IDC | II | − | − | − | ++ | + |
| 34 | T234 | 51 | ISLC | II | − | + | ++ | + | + |
| 35 | T235 | 58 | IDC | III | +++ | − | − | + | + |
| 36 | T236 | 43 | IDC | II | − | − | ++ | − | + |
| 37 | T237 | 41 | ISDC | I | − | + | +++ | − | − |
| 38 | T238 | 57 | ISDC | I | − | ++ | + | − | + |
| 39 | T239 | 50 | IDC | II | − | + | − | +++ | + |
| 40 | T240 | 49 | IMC | II | − | + | − | − | + |
| 41 | T241 | 52 | ISDC | II | − | + | ++ | − | + |
| 42 | T242 | 37 | ISDC | I | − | ++ | + | − | + |
| 43 | T243 | 48 | IDC | II | − | − | − | − | + |
| 44 | T244 | 62 | IDC | III | +++ | − | + | ++ | + |
| 45 | T245 | 44 | IDC | II | + | + | + | − | + |
| 46 | T246 | 57 | ISDC | I | − | + | ++ | − | − |
| 47 | T247 | 48 | ISDC | II | − | − | ++ | − | + |
| 48 | T248 | 35 | ISLC | II | − | + | − | − | + |
| 49 | T249 | 39 | IMC | II | − | ++ | + | − | + |
| 50 | T250 | 56 | ISDC | II | − | + | ++ | − | + |
Cell culture
The human breast cancer cell lines MCF-7 and MDA-MB-231 were obtained from Pasteur Institute of Iran and cultured according to the manufacturer’s instruction with slight modification. Cells were cultured in RPMI medium with 10% fetal bovine serum (FBS) at 37°C and 5% CO2. After 2 days, the cells were harvested and about 2 × 10⁶ cells were separated for RNA extraction, cDNA synthesis, and reverse-transcription polymerase chain reaction (RT-PCR) experiments.
RNA extraction and cDNA synthesis
Total RNA was extracted from tumor and normal samples and also breast cancer cell lines by TriPure isolating reagent (Roche) according to the manufacturer’s instructions with slight modifications. RNA was dissolved in DEPS-treated water and its concentration was determined by a spectrophotometer (NanoDrop 2000). About 2 μg of total RNA from various samples were subjected for cDNA synthesis using the M-MLV reverse transcription kit (Invitrogen) and random primer (Pharmacia, Sweden).
RT-PCR and semi-nested PCR
A couple of specific primers of the housekeeping gene hypoxanthine-guanine phosphoribosyltransferase-1 (HPRT1) were used to ascertain the cDNA quality. PCR amplification of HPRT was performed in five minutes at 94°C as start denaturation, 30 cycles of denaturation at 94°C for 40 seconds, annealing at 59°C for 30 seconds, extension at 72°C for 30 seconds, and a 7-minute final extension at 72°C. A pair of specific primers was designed to amplify 567 base pair fragments of human WBP2NL. The total reaction volumes were 25 μL containing 1 μL cDNA, two specific primers, PCR set, and smart Taq polymerase (Invitrogen). The PCR amplification of WBP2NL was performed with 35 cycles of denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, extension at 72°C for 30 seconds, and a 7-minute final extension at 72°C following an initial denaturation of 4 minutes at 94°C. The primers were positioned in different exons of each gene to avoid false positive results because of probable genomic DNA contamination. If no expression of WBP2NL was detected in RT-PCR runs on agarose gel, semi-nested PCR was performed using 2 μL of the first PCR reactions. Negative control reaction of the first PCR was used for the semi-nested PCR to identify false positive results caused by genomic DNA contamination. In order to carry out semi-nested PCR, an internal forward primer was used to amplify 168 base pair fragments of WBP2NL. The semi-nested amplification of WBP2NL was performed with 35 cycles of denaturation at 94°C for 30 seconds, annealing at 57°C for 20 seconds, extension at 72°C for 30 cycles, and a 7-minute final extension at 72°C, following an initial denaturation of 4 minutes at 94°C. PCR products were subjected to electrophoresis in 1.5% aga-rose gel and then visualized under ultraviolet light after DNA staining with ethidium bromide. Amplification reaction carried out using following designed primers:
HPRT Forward: 5-ACCAGTCAACAGGGGACATAA-3′
Reverse: 5′-CTTCGTGGGGTCCTTTTCACC-3′
WBP2NL Forward: 5′-GCCATTGAATTTGCCCAGT TG-3′
Reverse1: 5′-TTAAGAATGGACCTGAGAAGAGG-3′
Reverse2: 5′-GGCTCCATAGACAATAACTGAAC-3′
Statistical analysis
Statistical analysis was carried out using the SPSS software (SPSS Inc.), and significance was defined as P < 0.05.
Results
Complementary DNA amplification obtained from breast tumor samples was carried out with specific primers for WBP2NL gene using RT-PCR. We carried out semi-nested RT-PCR method in order to analyze the semiquantitative expression level of WBP2NL.
We detected WBP2NL expression in 45 out of 50 (90%) breast cancer samples (Fig. 1). Fifteen (30%) cases expressed WBP2NL in the first round of RT-PCR, representing a high level of gene expression (Fig. 1A), and 30 (60%) cases expressed WBP2NL in the second amplification by semi-nested RT-PCR, representing a low level of WBP2NL expression in the samples (Fig. 1B). Negative results from semi-nested RT-PCR showed a nondetectable WBP2NL transcript in the breast cancer samples (Fig. 1B). No expression of WBP2NL was seen in ANCT samples (Fig. 2). The different levels of WBP2NL gene expression in normal and breast cancer tissues are summarized in Tables 2 and 3.
Figure 1.
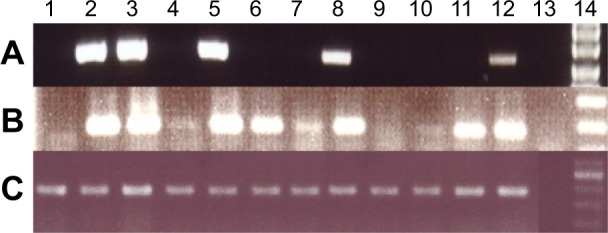
RT-PCR results from breast cancer samples, testis tissue, and breast cancer cell lines. (A) RT-PCR was performed to analyze of WBP2NL gene expression in breast cancer samples, testis tissue, and cell lines. Lane 1 and 2 are cDNA from MCF-7 and MDA-MB-231 cell line, respectively. Lane 3 is cDNA from an adult human testis as a positive control. Lanes 4–12 are cDNAs from breast cancer samples, Lane 13 is the negative control (water), and Lane 14 is 100 bp DNA marker. (B) Nested PCR was performed to analyze low expression of WBP2NL gene in breast cancer and cell lines. (C) All samples were controlled for presence of cDNA using the housekeeping gene HPRT.
Figure 2.
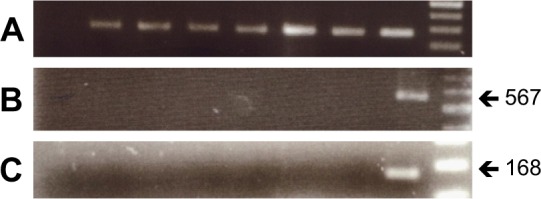
RT-PCR results from ACNT samples. (A) Presence of cDNA was checked in all samples using the housekeeping gene HPRT. (B) Lanes 2–7 are results of RT-PCR from some ACNT samples which showed no WBP2NL gene expression. (C) Nested PCR was carried out, which showed no expression of WBP2NL gene in ACNT samples (Lanes 2–7). Lane 1 was a negative control (water) and Lane 8 was RT-PCR from an adult human testis as a positive control. Lane 9 is a 100-bp DNA marker.
Table 3.
WBP2NL gene expression in all samples after two rounds amplification (first PCR and semi-nested PCR).
| WBP2NL GENE EXPRESSION | SAMPLE TYPE | |||
|---|---|---|---|---|
| ANCT | BC | MDA-MB-231 | MCF-7 | |
| Positive in the first PCR (high expression) | 0 (0%) | 15 (30%) | 2 (100%) | 0 (0%) |
| Positive in the semi-nested PCR out of negatives in the first PCR (low expression) | 0 (0%) | 30 (60%) | 2 (100%) | 0 (0%) |
| Negative in the semi-nested PCR of negatives in the first PCR (no expression) | 20 (100%) | 5 (10%) | 0 (0%) | 2 (100%) |
| Total positive expression | 0 (0%) | 45 (90%) | 2 (100%) | 0 (0%) |
| Total samples | 20 | 50 | 2 | 2 |
Analysis of the WBP2NL gene expression levels was examined in women of two different age groups with breast cancer: young (35–50 years) and elderly (51–65 years), but no significant association was detected between age and the gene expression.
For more information, we evaluated the correlation between the histological feature and grade status of tumor samples and WBP2NL expression. A significant difference was detected in WBP2NL gene expression levels and the type of infiltrating ductal carcinoma (P < 0.01). We also analyzed the correlation between WBP2NL gene expression levels and additional factors including ER/PR and HER2/neu expression of tumor samples. A significant difference was detected in WBP2NL gene expression levels and HER2/neu expression (P < 0.05).
Fifteen peripheral blood samples from breast cancer patients were examined for the expression of WBP2NL, but the transcript was not found in the peripheral blood samples from patients using RT-PCR (data not shown).
We also evaluated WBP2NL expression in the breast cancer cell lines MCF-7 and MDA-MB-231 (Table 3). The latter cell line showed expression of WBP2NL in the first round of RT-PCR, whereas the expression of WBP2NL was negative in RT-PCR and semi-nested RT-PCR (Fig. 1).
Discussion
Breast cancer is the most frequent cause of cancer death in women worldwide.1 Nowadays, progressive studies have been developed to find the genes involved in the disease as well as new biomarkers for early diagnosis and treatment of the patients. In this respect, CT genes are among the best candidates for new therapeutics for breast cancer.
The CT genes are a group of tumor antigens that are normally expressed during spermatogenesis, usually in spermatocytes, but absent in normal somatic cells.2,3 This special feature makes CT antigens a promising tumor-specific marker and very good candidate for cancer vaccines and immunotherapy approaches.4,5,19 Previous studies have shown the expression of some CT antigens in breast cancer tumors.20–23
The testis-specific protein WBP2NL is a signaling molecule expressed in spermatocytes and localized in the PAS of elongated spermatocytes.12,13 The WBP2NL protein enters in the oocyte during fertilization, and induces meiotic resumption and oocyte activation events.14–16 The N-terminal part of WBP2NL has homology with WBP2, whereas its C-terminal contains a functional proline-rich (PPXY) motif that allows it to interact with group I WW domain proteins such as TAZ and YAP.24–26 Both transcription coactivators YAP and TAZ are the key downstream effectors of the mammalian Hippo pathway,27 which plays important roles in cell proliferation and tumorigenesis processes.28–30 Recent studies have revealed that the interaction of WBP2 via PPxY-containing C-terminal region with WW domain in TAZ is important for the oncogenic properties of TAZ.9–11 It has been reported that TAZ is also overexpressed in other human cancers and promotes malignant cell growth.31–34 Some recent studies have suggested WBP2 as a target of cancer therapy, and proposed that either inhibition of WBP2 expression or interfering in its relation with YAP and TAZ could suppress tumor cell proliferation.35,36
Previous study by Aarabi et al revealed that WBP2NL contributes to sperm-induced calcium release and thereby initiates oocyte activation events.17 Our recent study has shown that WBP2NL is elevated in the embryonic stem cells and actively dividing cancerous cell lines.18 The WBP2NL structural considerations, together with previously published data, leads us to suggest that WBP2NL may have a potential role in the proliferation and activation of breast cancer cells maybe through binding to a group I WW domain protein.
Conclusion
In conclusion, elevated expression of the WBP2NL gene in breast cancer cells and MDA-MB-231 cell line leads us to suggest that WBP2NL might be classified as a CT antigen that is implicated in tumorigenesis. Therefore, it might be considered as a novel prognostic marker for early diagnosis of breast cancer.
Acknowledgments
We would like to thank Department of Medical Genetic, Faculty of Medicine, Tehran University of Medical Sciences for technical and financial support. We appreciate Mia Feldman (University of Pittsburgh) for her careful proofreading of the manuscript and helpful comments and suggestions.
Footnotes
ACADEMIC EDITOR: Barbara Guinn, Editor in Chief
FUNDING: This paper was supported by the Department of Medical Genetics, Faculty of Medicine, Tehran University of Medical Sciences, Tehran, Iran. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflict of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: SN, MD, MA, MHM. Analyzed the data: SN, MD, MBM, MA. Wrote the first draft of the manuscript: SN, MD, MBM. Contributed to the writing of the manu script: SN, MD, MBM, MA, MHM. Agreed with the manuscript, results, and conclusions: SN, MD, MBM, GK, MA, MHM. Jointly developed the structure and arguments for the paper: SN, MD, MBM, GK, MA, MHM. Made critical revisions and approved the final version: SN, MD, MBM, GK, MA, MHM. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Key TJ, Verkasalo PK, Banks E. Epidemiology of breast cancer. Lancet Oncol. 2001;2(3):133–140. doi: 10.1016/S1470-2045(00)00254-0. [DOI] [PubMed] [Google Scholar]
- 2.Scanlan MJ, Simpson AJ, Old LJ. The cancer/testis genes: review, standardization, and commentary. Cancer Immun. 2004;4:1. [PubMed] [Google Scholar]
- 3.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5(8):615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 4.Caballero OL, Chen YT. Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Sci. 2009;100(11):2014–2021. doi: 10.1111/j.1349-7006.2009.01303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siliņa K, Zayakin P, Kalniņa Z, et al. Sperm-associated antigens as targets for cancer immunotherapy: expression pattern and humoral immune response in cancer patients. J Immunother. 2011;34(1):28–44. doi: 10.1097/CJI.0b013e3181fb64fa. [DOI] [PubMed] [Google Scholar]
- 6.Dhananjayan SC, Ramamoorthy S, Khan OY, et al. WW domain binding protein-2, an E6-associated protein interacting protein, acts as a coactivator of estrogen and progesterone receptors. 2006;20:2343–2354. doi: 10.1210/me.2005-0533. [DOI] [PubMed] [Google Scholar]
- 7.Lim SK, Orhant-Prioux M, Toy W, Tan KY, Lim YP. Tyrosine phosphorylation of transcriptional coactivator WW-domain binding protein 2 regulates estrogen receptor α function in breast cancer via the Wnt pathway. FASEB J. 2011;25(9):3004–3018. doi: 10.1096/fj.10-169136. [DOI] [PubMed] [Google Scholar]
- 8.Buffa L, Saeed AM, Nawaz Z. Molecular mechanism of WW-domain binding protein-2 coactivation function in estrogen receptor signaling. IUBMB Life. 2013;65(1):76–84. doi: 10.1002/iub.1105. [DOI] [PubMed] [Google Scholar]
- 9.Chan SW, Lim CJ, Guo K, et al. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 2008;68:2592–2598. doi: 10.1158/0008-5472.CAN-07-2696. [DOI] [PubMed] [Google Scholar]
- 10.Chan SW, Lim CJ, Huang C, et al. WW domain-mediated interaction with Wbp2 is important for the oncogenic property of TAZ. Oncogene. 2011;30:600–610. doi: 10.1038/onc.2010.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao D, Zhi X, Zhou Z, Chen C. TAZ antagonizes the WWP1-mediated KLF5 degradation and promotes breast cell proliferation and tumorigenesis. Carcinogenesis. 2012;33(1):59–67. doi: 10.1093/carcin/bgr242. [DOI] [PubMed] [Google Scholar]
- 12.Wu AT, Sutovsky P, Xu W, Vander Spoel AC, Platt FM, Oko R. The postacrosomal assembly of sperm head protein, PAWP, is independent of acrosome formation and dependent on microtubular manchette transport. Dev Biol. 2007;312(2):471–483. doi: 10.1016/j.ydbio.2007.08.051. [DOI] [PubMed] [Google Scholar]
- 13.Kennedy CE, Krieger KB, Sutovsky M, et al. Protein expression pattern of PAWP in bull spermatozoa is associated with sperm quality and fertility following artificial insemination. Mol Reprod Dev. 2014;81(5):436–449. doi: 10.1002/mrd.22309. [DOI] [PubMed] [Google Scholar]
- 14.Sutovsky P, Manandhar G, Wu A, Oko R. Interactions of sperm perinuclear theca with the oocyte: implications for oocyte activation, anti-polyspermy defense, and assisted reproduction. Microsc Res Tech. 2003;61(4):362–378. doi: 10.1002/jemt.10350. [DOI] [PubMed] [Google Scholar]
- 15.Wu AT, Sutovsky P, Manandhar G, et al. PAWP, a sperm specific ww-domain binding protein, promotes meiotic resumption and pronuclear development during fertilization. J Biol Chem. 2007;282:12164–12175. doi: 10.1074/jbc.M609132200. [DOI] [PubMed] [Google Scholar]
- 16.Oko R, Sutovsky P. Biogenesis of sperm perinuclear theca and its role in sperm functional competence and fertilization. J Reprod Immunol. 2009;83(1–2):2–7. doi: 10.1016/j.jri.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 17.Aarabi M, Balakier H, Bashar S, Moskovtsev SI, Sutovsky P, Librach CL, Oko R. Sperm-derived WW domain-binding protein, PAWP, elicits calcium oscillations and oocyte activation in humans and mice. FASEB J. 2014;2014;10:4434–4440. doi: 10.1096/fj.14-256495. [DOI] [PubMed] [Google Scholar]
- 18.Nourashrafeddin S, Aarabi M, Miryounesi M, et al. Expression analysis of PAWP during mouse embryonic stem cell-based spermatogenesis in vitro. In Vitro Cell Dev Biol Anim. 2014;50(5):475–481. doi: 10.1007/s11626-013-9722-1. [DOI] [PubMed] [Google Scholar]
- 19.Lim SH, Zhang Y, Zhang J. Cancer-testis antigens: the current status on antigen regulation and potential clinical use. Am J Blood Res. 2012;2(1):29–35. [PMC free article] [PubMed] [Google Scholar]
- 20.Curigliano G, Viale G, Ghioni M, et al. Cancer-testis antigen expression in triple-negative breast cancer. Ann Oncol. 2011;22(1):98–103. doi: 10.1093/annonc/mdq325. [DOI] [PubMed] [Google Scholar]
- 21.Dianatpour M, Mehdipour P, Nayernia K, et al. Expression of testis specific genes TSGA10, TEX101 and ODF3 in breast cancer. Iran Red Crescent Med J. 2012;14(11):722–726. doi: 10.5812/ircmj.3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chapman KB, Prendes MJ, Kidd JL, Sternberg H, West MD, Wagner J. Elevated expression of cancer/testis antigen FSIP1 in ER-positive breast tumors. Biomark Med. 2013;7(4):601–611. doi: 10.2217/bmm.13.58. [DOI] [PubMed] [Google Scholar]
- 23.Hou SY, Sang MX, Geng CZ, et al. Expressions of MAGE-A9 and MAGE-A11 in breast cancer and their expression mechanism. Arch Med Res. 2014;45(1):44–51. doi: 10.1016/j.arcmed.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Sudol M. Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene. 1994;9:2145–2152. [PubMed] [Google Scholar]
- 25.Kanai F, Marignani PA, Sarbassova D, et al. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin Z. Contribution of a sperm protein, PAWP, to the signal transduct pathway during vertebrate fertilization. 2008. http://hdl.handle.net/1974/992.
- 27.Hong W, Guan KL. The YAP and TAZ transcription co-activators: key downstream effectors of the mammalian hippo pathway. Semin Cell Dev Biol. 2012;23(7):785–793. doi: 10.1016/j.semcdb.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macias MJ, Wiesner S, Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002;513(1):30–37. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- 29.Varelas X, Sakuma R, Samavarchi-Tehrani P, et al. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol. 2008;10(7):837–848. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- 30.Wang K, Degerny C, Xu M, Yang XJ. YAP, TAZ, and Yorkie: a conserved family of signal responsive transcriptional coregulators in animal development and human disease. Biochem Cell Biol. 2009;87(1):77–91. doi: 10.1139/O08-114. [DOI] [PubMed] [Google Scholar]
- 31.de Cristofaro T, Di Palma T, Ferraro A, et al. TAZ/WWTR1 is overexpressed in papillary thyroid carcinoma. Eur J Cancer. 2011;47:926–933. doi: 10.1016/j.ejca.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 32.Wang L, Chen Z, Wang Y, et al. WWTR1 promotes cell proliferation and inhibits apoptosis through cyclin A and CTGF regulation in non-small cell lung cancer. Tumour Biol. 2014;35(1):463–468. doi: 10.1007/s13277-013-1064-9. [DOI] [PubMed] [Google Scholar]
- 33.Xie M, Zhang L, He CS, et al. Prognostic significance of TAZ expression in resected non-small cell lung cancer. J Thorac Oncol. 2012;7(5):799–807. doi: 10.1097/JTO.0b013e318248240b. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Z, Hao Y, Liu N, Raptis L, Tsao MS, Yang X. TAZ is a novel oncogene in non-small cell lung cancer. Oncogene. 2011;30(18):2181–2186. doi: 10.1038/onc.2010.606. [DOI] [PubMed] [Google Scholar]
- 35.Sudol M. Newcomers to the WW domain-mediated network of the hippo tumor suppressor pathway. Genes Cancer. 2010;1(11):1115–1118. doi: 10.1177/1947601911401911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Milton CC, Poon CL, Hong W, Harvey KF. Wbp2 cooperates with Yorkie to drive tissue growth downstream of the Salvador-Warts-Hippo pathway. Cell Death Differ. 2011;18(8):1346–1355. doi: 10.1038/cdd.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]


