Abstract
Temporal sequences of transcription factors (tTFs) in Drosophila neural progenitors generate neuronal diversity. Mattar et al. (2015) identify Casz1/Castor as a late temporal identity factor in mouse retinal progenitors that is regulated by the early factor Ikzf1/Hunchback, thus generalizing the notion of tTFs.
The nervous system consists of diverse neurons organized into complex circuits. A goal of developmental neurobiology is to understand the molecular mechanisms that guide neurogenesis. It is well established that spatial positioning of neural progenitors contributes to the production of unique neural fates. In addition, studies of neurogenesis in different species reveal that specific neurons are born in an invariant order (temporal patterning; for review, see: Cepko, 2014). For example, neurons in the vertebrate cortex are sequentially born in an inside-out fashion to populate the six cortical layers. Other examples of temporal patterning have been described in vertebrates and invertebrates. This suggests that birth-order is a second axis of information, which, coupled with spatial position, confers specific cell fates.
How are neurons born sequentially? An interesting model first described in the Drosophila embryonic ventral nerve cord (VNC) is that neural progenitors, termed neuroblasts, sequentially express a series of ‘temporal Transcription Factors’ (tTF) as they age. Once provided with spatial patterning cues, each neuroblast progresses through the tTF sequence to produce lineage-specific neuronal types in an invariant order (Brody & Odenwald, 2000; Isshiki et al., 2001).
In the fly VNC, Hunchback, Krüppel, Pdm, Castor and Grainyhead are sequentially expressed in neuroblasts as they age (Brody & Odenwald, 2000; Pearson & Doe, 2003). During each tTF time window, neuroblasts generate specific subsets of VNC neurons. In the developing fly optic lobes, two related tTF sequences have been identified in neuroblasts: Homothorax, Klumpfuss, Eyeless, Sloppy-paired, Dichaete, Tailless in the center of the outer proliferation center (Li et al., 2013), and Distalless, Eyeless, Sloppy-paired, Dichaete in the tips of the outer proliferation center (Bertet et al., 2014). Intermediate neural progenitors (INPs), which are also present in the subventricular zone of the adult mammalian brain (Doetsch et al., 1999), expand Drosophila neuroblasts lineages by progressing through a different tTF cascade (Dichaete, Grainyhead, Eyeless) that is overlaid onto the temporal progression of parental neuroblasts (Bayraktar & Doe, 2013). These studies suggest that different tTF sequences are used by multiple neural progenitors in a context-dependent manner to intrinsically determine age (Figure 1B). The parallels shared between Drosophila and vertebrate neural progenitors, particularly the sequential birth of neuronal types, hint that the molecular mechanisms may be similar. However, temporal patterning of neuronal progenitors by tTFs has not been described in vertebrates.
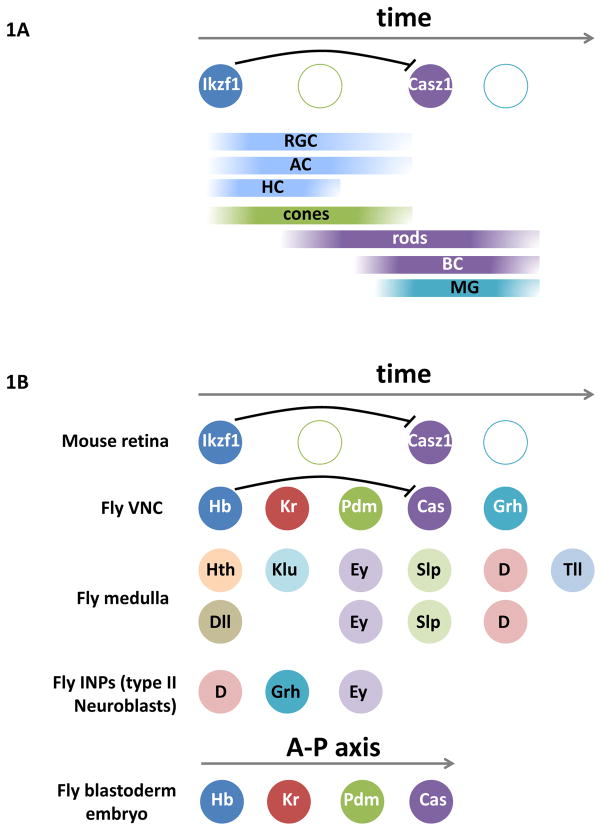
Figure 1. Temporal patterning in mouse and fly.
(A) Retinal cell types are born sequentially. Retinal progenitor cells first express Ikzf1 to specify early-born cell types (blue) before switching to express Casz1 to identify later-born retinal cell types (purple). Although the birth of cones (green) overlaps with other early-born cell types, Ikzf1 does not specify cone identity. In addition, Casz1 does not specify last-born Müller Glia (aqua), suggesting an additional temporal identity window. (B) Known temporal sequences in fly and mouse. Mattar et al. identified a regulatory interaction between Ikzf1/Hb and Casz1/Cas in mouse RPCs that is similar in Drosophila VNC neuroblasts.
The only indication of temporal patterning in vertebrates comes from the observation that a mouse homolog of Hunchback, Ikzf1, is expressed in early retinal progenitor cells (RPCs; Elliott et al., 2008). RPCs produce all neuronal retinal cells, as well as glia: input sensory neurons (cone and rod photoreceptors), interneurons (horizontal, bipolar, and amacrine cells), output neurons (retinal ganglion cells), and Müller glial cells. The first cells to be born are retinal ganglion cells, then horizontal cells, cones and amacrine cells. Rods are produced in a second wave of neurogenesis, while bipolar and Müller glial cells are the last cell types to be born. The different cells are produced within specific time windows that overlap extensively (Young, 1985; Cepko, 2014). Ikzf1 is necessary and sufficient for the generation of all early-born retinal cell types apart from cones (Figure 1A; Elliott et al., 2008). However, one gene is far from a temporal series and no other reports of tTF genes in neural precursors have been published since.
Mattar et al. (2015) studied the expression pattern of Casz1, the ortholog of Castor, during mouse retinal development. They discovered that Casz1 is expressed in RPCs at mid-retinogenesis (Figure 1A). Conditional deletion of Casz1 in RPCs increases early-born cell types as well as Müller glia, the latest cell type produced by RPCs. Furthermore, retroviral transfection of Casz1 in early RPCs reduces early-born neurons and late-born Müller glia, while concurrently increasing mid-phase bipolar cells and rods. In both cases, no effect on clone size is observed. These results suggest that Casz1 suppresses early and late cell fates, and promotes the production of rods and bipolar cells, without affecting proliferation or cell death. Interestingly, a division of labor exists in the production of mid-phase neurons between the two isoforms of Casz1, although their expression pattern seems identical; Casz1v1 increases the number of bipolar cells, while Casz1v2 produces extra rods. These results are consistent with the hypothesis that Casz1 is a temporal identity factor defining the mid-stage of retinal progenitor cells (Figure 1A).
Hunchback and Castor, the fly homologs of Ikzf1 and Casz1, participate in a tTF sequence during Drosophila embryonic VNC development. Hunchback represses Castor expression in young neuroblasts and promotes early-born cell fates (Tran et al., 2010). Similarly, Mattar et al. show that Ikzf1 lies upstream and acts as a repressor of Casz1. However, Ikzf1 repression of Casz1 is achieved indirectly, as Ikzf1 does not bind Casz1 cis-regulatory elements, perhaps indicating the existence of an intermediate tTF between Ikzf1 and Casz1 (Figure 1B). The fact that the Ikzf1-Casz1 regulatory logic appears to be conserved in the mouse retina and fly VNC raises an exciting question of evolutionary significance: Is this an example of deep homology, or is it a mechanism reached independently in the two cases driven by chance and necessity?
Temporal sequences of transcription factors represent a very powerful and elegant mechanism for generating a wide variety of different neurons using a small set of neural progenitors. The existence of a temporal sequence requires the presence of three types of molecules with diverse functions. First, a molecular clock that either counts time intrinsically (in time units or cell divisions) or receives extrinsic signals that promote the progression of the sequence. The second type is temporal factors, like Ikzf1 and Casz1; these tTFs are controlled by the clock and orchestrate the neuronal output of progenitor cells. Finally, downstream effector genes integrate spatial and temporal information. These genes can encode transcription factors, signaling molecules, membrane receptors, etc. and function to control and maintain the differentiation of neurons born throughout development. Transcriptome analysis of neuroblasts and their progeny at multiple developmental stages will identify many of these factors.
This type of tTF circuit could be modular and perhaps reutilized during development of different neuronal tissues. The use of the Hunchback/Ikzf1-Castor/Casz1 regulatory module during fly VNC and mouse retina development could result from two different reasons:
This regulatory module might predate the existence of these temporal sequences and may have been adopted as a whole by dividing neural progenitors in flies and mice to distinguish different age states. Potential participation of other members of the Drosophila temporal sequence in the RPC temporal sequence would support this hypothesis. For example, in Drosophila VNC neuroblasts, the tTF sequence recapitulates the spatial expression of the same genes along the anteroposterior axis of the fly cellular blastoderm embryo (Figure 1B; Isshiki et al., 2001). This hints towards the presence of a common gene regulatory network that operates in both space and time.
Alternatively, the Hunchback-Castor and Ikzf1-Casz1 regulatory modules might have been assembled independently. There is a restricted number of transcription factors that can be used to orchestrate specific neuronal identity and morphological characters, which could explain why the same transcription factors are used at multiple stages of neurogenesis (e.g. during neuron specification and morphological differentiation). Casz1 is one example; it is expressed in mid-late RPCs to specify cell fate, but also in post-mitotic cones and a subset of amacrine cells born during the Ikzf1+/Casz1− early time window. Similarly, in flies, some tTFs participate in multiple temporal sequences and in different gene regulatory networks. For example, Dichaete functions in a tTF sequence with Sloppy-paired and Tailless in medulla neuroblasts, while it interacts with Grainyhead in INPs of type II neuroblasts in the Drosophila central brain (Figure 1B). This suggests that single tTFs, rather than whole networks, are recruited to temporal sequences. Finally, the ontogeny of retinal neurons might recapitulate their phylogeny (i.e., the birth order of retinal neurons reflects the order in which they appeared during evolution; Cepko, 2014). If this is true, the Ikzf1-expressing RPCs predate the Casz1-expressing RPCs and the regulatory module is probably the product of convergent evolution.
Another important point raised by this study is that, in vertebrates, tTFs bias cell fate decisions rather than specifying rigidly one cell fate. This is in contrast to Drosophila neuroblasts, in which precise cell fates are specified during each tTF windows. During rat retinal development, which is similar in many aspects to mice, RPCs proceed through successive divisions with a stochastic binary outcome, generating either two RPCs, or a RPC and a differentiating daughter cell, or two differentiating cells, each with different probabilities (Gomes et al., 2011). As RPCs age, the bias of this stochasticity is proposed to change in response to temporal factors. A similar process operates during zebrafish retinal development (He et al., 2012), arguing for a conserved mechanism amongst vertebrates. This stochasticity in the generation of diverse neuronal types may offer a level of plasticity that could be subject to regulation by extrinsic factors (e.g., light intensity, wavelength, polarization, etc.) or by evolution, making the animal more adaptable to diverse environments.
In mouse, Ikzf1 and Casz1 represent excellent candidates to influence the stochastic bias as RPCs age. To corroborate this hypothesis, a complete reconstruction of the molecular circuitry regulating the production of diverse neuronal types in the vertebrate retina needs to be achieved. Unraveling the primary players of this process will illuminate several interesting observations made by Mattar et al. For example, overexpression of Ikzf1-VP16, a fusion of Ikzf1 with the activation domain of VP16 that transforms the Ikzf1 repressor into an activator that upregulates the expression of Casz1, results in production of more Müller glia, contrary to the overexpression of Casz1 itself, which produces rods and bipolar cells at the expense of Müller glia and early-born retinal types. This suggests that additional targets of Ikzf1 participate in cell fate determination. Finally, the differential effects of the two Casz1 isoforms in the production of bipolar cells and rod photoreceptors argues for a complex circuitry that segments temporal windows into more elaborate stages.
This paper by Mattar et al. raises more questions than it answers, as is often the case for important findings. Namely, are other Drosophila VNC tTFs also acting in mouse RPCs? Have new members been recruited in the mouse temporal sequence, which differ from those in Drosophila? More importantly, are other systems, such as cerebral cortex progenitors, shaped by the Ikzf1-Casz1 regulatory logic? Interestingly, a recent study found that Ikzf1 specifies early-born fate in the cortex (Alsiö et al., 2013). Finally, multiple developmental systems in different animals must be interrogated for their potential use of novel temporal sequences.
Temporal patterning in mammals was discovered long ago but its mechanisms remain mysterious. This work took advantage of homologies with Drosophila to explain a small part of the huge neuronal diversity that exists in the brain, and in the retina. It is a beginning that illustrates potential mechanisms, which, combined with spatial patterning and plasticity, could one day explain how to make the thousands of neural types that make up our brain.
References
- Alsiö JM, Tarchini B, Cayouette M, Livesey FJ. Ikaros promotes early-born neuronal fates in the cerebral cortex. Proc Natl Acad Sci. 2013;110(8):E716–25. doi: 10.1073/pnas.1215707110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayraktar OA, Doe CQ. Combinatorial temporal patterning in progenitors expands neural diversity. Nature. 2013;498(7455):449–55. doi: 10.1038/nature12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertet C, Li X, Erclik T, Cavey M, Wells B, Desplan C. Temporal patterning of neuroblasts controls Notch-mediated cell survival through regulation of Hid or Reaper. Cell. 2014;158(5):1173–86. doi: 10.1016/j.cell.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody T, Odenwald WF. Programmed transformations in neuroblast gene expression during Drosophila CNS lineage development. Developmental Biology. 2000;226(1):34–44. doi: 10.1006/dbio.2000.9829. [DOI] [PubMed] [Google Scholar]
- Cepko C. Intrinsically different retinal progenitor cells produce specific types of progeny. Nature Reviews Neuroscience. 2014;15(9):615–627. doi: 10.1038/nrn3767. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular Zone Astrocytes Are Neural Stem Cells in the Adult Mammalian Brain. Cell. 1999;97(6):703–716. doi: 10.1016/S0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Elliott J, Jolicoeur C, Ramamurthy V, Cayouette M. Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron. 2008;60(1):26–39. doi: 10.1016/j.neuron.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Gomes FLaF, Zhang G, Carbonell F, Correa Ja, Harris Wa, Simons BD, Cayouette M. Reconstruction of rat retinal progenitor cell lineages in vitro reveals a surprising degree of stochasticity in cell fate decisions. Development. 2011;138(2):227–35. doi: 10.1242/dev.059683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Zhang G, Almeida AD, Cayouette M, Simons BD, Harris Wa. How variable clones build an invariant retina. Neuron. 2012;75(5):786–98. doi: 10.1016/j.neuron.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentiall express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106(4):511–521. doi: 10.1016/S0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Li X, Erclik T, Bertet C, Chen Z, Voutev R, Venkatesh S, Morante J, Celik A, Desplan C. Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature. 2013;498(7455):456–62. doi: 10.1038/nature12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattar P, Ericson J, Blackshaw S, Cayouette M. A conserved regulatory logic controls temporal identity in mouse neural progenitors. Neuron. 2015;xxx:xxx–xxx. doi: 10.1016/j.neuron.2014.12.052. xxx. xxx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ. Regulation of neuroblast competence in Drosophila. Nature. 2003;425(6958):624–28. doi: 10.1038/nature02024.1. [DOI] [PubMed] [Google Scholar]
- Tran KD, Miller MR, Doe CQ. Recombineering Hunchback identifies two conserved domains required to maintain neuroblast competence and specify early-born neuronal identity. Development. 2010;137(9):1421–30. doi: 10.1242/dev.048678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RW. Cell differentiation in the retina of the mouse. The Anatomical Record. 1985;212(2):199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]