Abstract
Facioscapulohumeral dystrophy (FSHD) is one of the most prevalent muscular dystrophies. The majority of FSHD cases are linked to a decreased copy number of D4Z4 macrosatellite repeats on chromosome 4q (FSHD1). Less than 5% of FSHD cases have no repeat contraction (FSHD2), most of which are associated with mutations of SMCHD1. FSHD is associated with the transcriptional derepression of DUX4 encoded within the D4Z4 repeat, and SMCHD1 contributes to its regulation. We previously found that the loss of heterochromatin mark (i.e., histone H3 lysine 9 trimethylation (H3K9me3)) at D4Z4 is a hallmark of both FSHD1 and FSHD2. However, whether this loss contributes to DUX4 expression was unknown. Furthermore, additional D4Z4 homologs exist on multiple chromosomes, but they are largely uncharacterized and their relationship to 4q/10q D4Z4 was undetermined. We found that the suppression of H3K9me3 results in displacement of SMCHD1 at D4Z4 and increases DUX4 expression in myoblasts. The DUX4 open reading frame (ORF) is disrupted in D4Z4 homologs and their heterochromatin is unchanged in FSHD. The results indicate the significance of D4Z4 heterochromatin in DUX4 gene regulation and reveal the genetic and epigenetic distinction between 4q/10q D4Z4 and the non-4q/10q homologs, highlighting the special role of the 4q/10q D4Z4 chromatin and the DUX4 ORF in FSHD.
Keywords: FSHD1, FSHD2, DUX4, H3K9me3, SMCHD1, D4Z4, heterochromatin
Introduction
Facioscapulohumeral dystrophy (FSHD) is an autosomal dominant muscular dystrophy characterized by progressive wasting of facial, shoulder, and upper arm musculature [van der Maarel and Frants, 2005]. The majority of FSHD cases (>95%) are caused by monoallelic partial deletion of D4Z4 repeat sequences at the subtelomeric region of chromosome (chr) 4q (4qter D4Z4) (FSHD1; MIM# 158900) [van der Maarel and Frants, 2005; van der Maarel et al., 2011]. D4Z4 is a 3.3 kb macrosatellite repeat that contains an open reading frame (ORF) for the double-homeobox transcription factor DUX4 retrogene (MIM# 606009) [Gabriëls et al., 1999; Snider et al., 2010; Geng et al., 2012]. There are only 1–10 D4Z4 repeats in the contracted allele in FSHD1, in contrast to 11–150 copies in the intact allele. In the more rare form of FSHD (<5% of cases), there is no D4Z4 repeat contraction (FSHD2) [van Overveld et al., 2003; de Greef et al., 2010]. A recent study found that the SMCHD1 gene (MIM# 614982) is mutated in >80% of FSHD2 cases (MIM# 158901) [Lemmers et al., 2012].
FSHD occurs only in individuals with a 4qA haplotype with specific single-nucleotide polymorphisms in the chromosomal region distal to the last D4Z4 repeat (creating a noncanonical polyadenylation signal for the DUX4 transcript) [Lemmers et al., 2002, 2004, 2007, 2010a] with some exceptions [Scionti et al., 2012]. While multiple transcripts encoding different parts of the DUX4 protein have been identified [Snider et al., 2009], expression of the full-length DUX4 transcript (DUX4fl) is most closely associated with FSHD [Lemmers et al., 2010a; Snider et al., 2010]. Overexpression of DUX4fl caused differentiation defects in human myoblasts and mouse C2C12 muscle cells, and FSHD-like phenotypes in zebrafish [Bosnakovski et al., 2008; Vanderplanck et al., 2011; Mitsuhashi et al., 2013]. Furthermore, though the DUX4fl expression can occasionally be observed in unaffected individuals at very low levels (suggesting the presence of additional disease modifier genes), activation of a subset of the DUX4fl target genes has been observed in patient cells in multiple studies, supporting the significance of DUX4fl in FSHD [Geng et al., 2012; Jones et al., 2012; Rahimov et al., 2012; Broucqsault et al., 2013; Ferreboeuf et al., 2014].
The chromatin environment plays a significant role in gene regulation in normal development and disease [Feinberg, 2007]. Epigenetic alteration of D4Z4 chromatin was found to be a common link between FSHD1 and FSHD2 [van Overveld et al., 2003; Zeng et al., 2009]. D4Z4 repeats contain transcriptionally repressive heterochromatin harboring DNA hypermethylation and histone H3 lysine 9 trimethylation (H3K9me3) together with H3K27me3 [van Overveld et al., 2003; Zeng et al., 2009]. Using chromatin immunoprecipitation (ChIP) analysis, we found a specific loss of H3K9me3 at the D4Z4 repeat sequences in both FSHD1 and FSHD2 patient proliferating cell cultures [Zeng et al., 2009]. Importantly, this change is highly specific for FSHD; no significant change of H3K9me3 was observed in other muscular dystrophies, some of which share similar clinical phenotypes [Zeng et al., 2009]. This change is seen not only in affected muscle cells, but also in patient fibroblasts from skin biopsies and lymphoblasts from blood samples [Zeng et al., 2009]. This indicates that the loss of H3K9me3 is not an epiphenomenon of dystrophic muscle. Although D4Z4 DNA was also shown to be hypomethylated in FSHD, we showed that the H3K9me3 loss is not a downstream consequence of DNA hypomethylation since H3K9me3 is intact in the phenotypically unrelated immunodeficiency-centromeric instability-facial anomalies syndrome, in which D4Z4 is severely DNA hypomethylated [Zeng et al., 2009] due to mutations in the DNA methyltransferase 3B (DNMT3B) gene [Hansen et al., 1999; Xu et al., 1999]. Nevertheless, the loss of DNA methylation and H3K9me3 indicates the perturbation of heterochromatin structure at D4Z4 in FSHD, strongly suggesting that FSHD is an epigenetic abnormality disease associated with the impairment of heterochromatin at D4Z4. We also found that the heterochromatin-binding protein HP1γ and the higher order chromatin organizer cohesin are corecruited to D4Z4 in an H3K9me3-dependent and cell type specific manner, and are lost in FSHD as a consequence of the loss of H3K9me3 [Zeng et al., 2009]. FSHD-specific loss of heterochromatin in this region is thought to contribute to the derepression of DUX4 [Lemmers et al., 2010a; Snider et al., 2010; Geng et al., 2012]. However, this hypothesis has not been explicitly tested.
Our previous study indicated that loss of H3K9me3 occurs not only at 4q D4Z4, but also at 10q D4Z4 in FSHD1 and FSHD2 [Zeng et al., 2009]. DNA hypomethylation was observed at both 4q and 10q D4Z4 in FSHD2, though it appears to be restricted to the contracted 4q allele in FSHD1 [van Overveld et al., 2003; de Greef et al., 2007, 2009]. The extent of heterochromatin change in other regions of the FSHD genome has not been determined. There are additional D4Z4-like sequences present in several other chromosomes [Hewitt et al., 1994; Winokur et al., 1994; Lyle et al., 1995; Tam et al., 2004]. These D4Z4 homologs are largely uncharacterized, and it is unknown whether they also encode functional DUX4 genes and undergo similar chromatin changes as do 4q and 10q D4Z4. Understanding the extent of genetic and epigenetic similarity or difference of these homologs is essential to elucidate the disease mechanism, particularly in FSHD2, in which heterochromatin status may be globally altered, and also to properly distinguish the 4q/10q D4Z4-specific events. In this study, we report the effect of the inhibition of H3K9me3 on DUX4fl expression and characterization of the non-4q/10q D4Z4 homologs from seven chromosomes. We found that decreased H3K9me3 results in the reduced SMCHD1 binding to D4Z4 and derepression of DUX4fl expression, demonstrating the significance of the loss of the H3K9me3 heterochromatin at D4Z4 in gene regulation. While the DUX4 ORF is highly conserved in 4q/10q D4Z4 repeats, it is frequently disrupted by high degrees of nucleotide polymorphism in the D4Z4 homologs. Furthermore, while H3K9me3/HP1γ/cohesin heterochromatin marks and DNA methylation are also present at these D4Z4 homologs in normal cells, they remain unchanged in both FSHD1 and FSHD2 patient cells. The results reveal that the preservation of the DUX4 ORF and the unique FSHD-associated epigenetic alterations that contributes to DUX4 expression are restricted to 4q and 10q D4Z4, indicating their distinct role in FSHD pathogenesis.
Materials and Methods
Cells and DNA Mapping Panel
The NIGMS human/rodent somatic cell hybrid mapping panel #2, version 3 was from Coriell Cell Repositories (Camden, NJ), in which human chrs 1, 16, 17, 20, and 21 hybrids are from mice, whereas the others are from Chinese hamsters. Mouse somatic cell hybrids containing human chrs 4, 10, 13, 14, 15, or 21 (catalog number GM11687, 11688, 11689, 10479, 11715, or 08854, respectively, from Coriell Cell Repositories) were grown in DMEM medium supplemented with 10% FBS, penicillin/streptomycin, and 2 mM L-glutamine. Normal, FSHD1 and FSHD2 myoblasts were grown in F-10 medium (Invitrogen, Carlsbad, CA) supplemented with 20% FBS, 10 ng/ml bFGF (Invitrogen), and 20 ng/ml dexamethasone sodium phosphate (Sigma–Aldrich, St. Louis, MO) except FSHD1 myoblasts (GM17731 and GM17940 from Coriell Cell Repositories) were grown in F-10 medium supplemented with 15% FBS. KD3 human myoblasts immortalized by the expression of telomerase, cyclin D1, and mutated cyclin-dependent kinase 4 are grown as previously described [Shiomi et al., 2011]. Control (NFGr) and FSHD2 (KII-II) fibroblasts were grown in DMEM/F-12 (1:1) supplemented with 10% FBS, 2 mM GlutaMAX-I (Invitrogen-Gibco, Carlsbad, CA), 10 mM HEPES buffer (Invitrogen-Gibco), and 1 mM sodium pyruvate (Invitrogen-Gibco). Control and patient cells used for ChIP analysis are shown in Supp. Table S1.
Antibodies
Antigen affinity-purified rabbit polyclonal antibodies specific for Rad21 and the preimmune IgG control were published previously [Gregson et al., 2001]. Antibodies against HP1γ, H3K9me3, and H3K27me3 were purchased from EMD/Millipore (Billerica, MA). Antibody against 5-methylcytidine (metC) was from Eurogentec North America Inc. (San Diego, CA). SMCHD1 antibody was from Abcam (Cambridge, MA) (ab31865).
Chaetocin, Auranofin, and H2O2 Treatment
KD3 myoblasts that reached approximately 80% confluency were treated with 0.4 mM chaetocin (Sigma-Aldrich C9492) and harvested after 24 h. Cells were also treated with the thioredoxin reductase (TrxR) inhibitor auranofin (Sigma-Aldrich A6733) (1 μM) for 24 h or 0.5 mM H2O2 for 3 h. The effects of different treatments on D4Z4 H3K9me3 and DUX4fl expression were analyzed by ChIP-PCR and RT-nested set PCR as described below.
Lentiviral shRNA Transduction
Nontarget shRNA control (Sigma-Aldrich: SHC002) and the lentiviral shRNA against human SUV39H1 (TRCN0000157251, Sigma-Aldrich) were transfected into 293T cells along with lentiviral packaging plasmids, using Lipofectamine 2000 (Invitrogen). Supernatants were collected 36 and 60 h posttransfection, passed through a 0.45 μm nitrocellulose filter, and applied on KD3 cells in the presence of polybrene (1 μg/mL). The next day, cells were selected with puromycin for 48 h (2 μg/ml, Sigma). Forty-eight hours after infection, cells were transferred to 10 or 15 cm dishes and maintained 2 days before harvesting for experimental purposes.
DUX4 Nested RT-PCR
Total RNA was extracted using the Qiagen RNeasy Plus kit. Two to five micrograms of RNA was used for double-stranded cDNA synthesis according to manufacturer’s protocol (Life Technologies, Grand Island, NY) with the exception of using enzymes purchased from New England Biolabs, Inc. (Ipswich, MA). cDNA was purified by Qiagen PCR purification kit and eluted in 40 μl EB buffer. The nested PCR was done using the primer sets (182–183 and 1A–184) previously published [Snider et al., 2010]. The PCR cycling protocol is as follows: 95°C 2 min, 95°C 30 sec, 62°C 30 sec, 72°C 1 min 40 sec (repeat 34 times), then 72°C 10 min. PCR enhancer system (Invitrogen 11495-017) was used for the nested PCR. The PCR products were loaded on agarose gel and the observed bands were cut out for sequencing to confirm the DUX4 identity. At least three independent experiments were performed and the representative experiment was shown.
ChIP Analysis
The ChIP analysis was performed based on the protocol from the Upstate ChIP assay kit with some modification as previous described [Zeng et al., 2009]. Briefly, we cross-linked the cells with 1% formaldehyde and used 1 × 106 cells for one histone ChIP and 3 × 106 cells for the other ChIP assays. Protein A beads were preincubated with 1 mg/ml BSA and 0.2 mg/ml ssDNA for 20 min at 4°C. Typically, 4 μg IgG was used per assay. The mixtures of antibody and nuclear extracts precleared with protein A beads were incubated at 4°C overnight followed by precipitation with protein A beads. After washing, immunoprecipitated materials were eluted with 0.1 M NaHCO3 and 1% SDS, and cross-links were reversed at 65°C for 4–6 h. The metC ChIP assay was performed according to the previously published protocol [Weber et al., 2005]. Primer sequences are listed in the Supp. Table S2 or were published previously [Zeng et al., 2009]. The endpoint gel analysis of the ChIP-PCR products was carried out using the Luminescent Image Analyzer LAS-4000 (FujiFilm Medical Systems U.S.A., Stamford, CT). Real-time Q-PCR was performed using the iCycler iQ real-time PCR detection system (Bio-Rad Laboratories, Inc., Hercules, CA) with iQ SYBR Green Supermix (Bio-Rad). The ChIP-PCR signal was normalized by the subtraction of the preimmune IgG ChIP-PCR signal, which was further divided by input genomic PCR (for normalization of different D4Z4 or D4Z4 homolog repeat numbers in different cells) minus PCR with no template as previously described [Zeng et al., 2009]. Alternatively, ChIP was normalized with pan-histone H3 antibody ChIP (Abcam ab1791) (Fig. 1D and Supp. Fig. S4) as recently described [Thijssen et al., 2013]. Both normalization methods yielded comparable results.
Figure 1.
Inhibition of H3K9me3 results in DUX4fl upregulation. A: The effect of chaetocin on H3K9me3 at D4Z4 and DUX4 expression in KD3 human myoblasts. Left: H3K9me3 ChIP-PCR analysis of D4Z4 and rDNA regions in DMSO and chaetocin-treated cells. Right: RT-PCR analysis of DUX4fl expression. Nested set RT-PCR analysis of DUX4fl expression as described [Snider et al., 2010] in cells treated with DMSO only or chaetocin as indicated at the top. The PCR products were sequenced to confirm their identity. GAPDH RT-PCR serves as a control. B: Comparison of the effects of chaetocin and auranofin treatments on D4Z4 H3K9me3 and DUX4 expression. Left: H3K9me3 ChIP-PCR analysis of the D4Z4 region in auranofin- and chaetocin-treated cells. Right: DUX4fl and GAPDH RT-PCR analyses as in (A). Untreated control, H2O2, DMSO-treated, auranofin, or cheatocin-treated cells were compared. C: The effect of SUV39H1 depletion on D4Z4 H3K9me3 and DUX4 expression. Lentiviral shRNA against SUV39H1 was used for depletion. Left: RT-qPCR analysis of SUV39H1 depletion in control and SUV39H1 shRNA-treated cells. Middle: ChIP-PCR analysis of H3K9me3 at D4Z4 in control and SUV39H1 shRNA-treated cells. Right: Top, DUX4fl and GAPDH RT-PCR as in (A); Bottom, Western blot analysis of control and SUV39H1-depleted cells with antibodies specific for H3K9me3 and H2A. D: ChIP-PCR analysis of SMCHD1 binding at D4Z4 in chaetocin and SUV39H1 shRNA-treated cells. Y-axis: (SMCHD1 ChIP–preimmune IgG)/input, which was further normalized by histone H3 ChIP. Right: Western blot analysis of cheatocin (top) or SUV39H1 shRNA (bottom) treated cells compared to DMSO (control) or control shRNA-treated cells, respectively, using antibodies indicated. Histone H2A (top) and actin (bottom) served as a loading control.
Genomic- and ChIP-PCR Cloning and Sequencing
The Q-PCR forward (Q-PCR-F) primer and the 4q-Hox reverse (4q-Hox-R) primer [Zeng et al., 2009] were used to amplify a subdomain of the D4Z4 homologs from the mapping panel and somatic cell hybrid genomic DNA with an annealing temperature of 58°C. The PCR enzyme was cloned using Pfu DNA polymerase (Stratagene, La Jolla, CA) and the blunt-end PCR product was ligated into the pCRBlunt vector (Invitrogen). The bacterial colonies transformed with the clones were picked, grown in 96-well plates, and subjected to single-pass plasmid sequencing (Agencourt, Beverly, MA). The total number of clones sequenced for each chromosome was 20 for chr 3, 50 for chr 4, 40 for chr 10, 37 for chr 13, 41 for chr 14, 28 for chr 15, 22 for chr 21, 16 for chr 22, and 18 for chr Y, respectively. Cloning and sequencing analyses of 4q/10q and non-4q/10q D4Z4 repeats were performed using either genomic or H3K9me3 ChIP DNA from multiple human cells. We sequenced 32 clones from four independent cell samples (individuals) for Q-PCR-F + 4q-Hox-R, 41 clones from five individual cell samples for Q-PCR-F + Q-PCR-reverse, 44 clones from five individual cell samples for Q-PCR-F + chr 3-reverse, 50 clones from five individual cell samples for chr 15-forward + 4q-Hox-R, and 136 clones for Q-PCR-F + chr 22-reverse (87 were from genomic DNA and 49 from H3K9me3 ChIP DNA) from 14 different individual cell samples (Supp. Table S3).
Results
Inhibition of H3K9me3 Results in DUX4fl Expression and the Loss of SMCHD1 Binding
We previously determined that SUV39H1 histone methyltransferase (HMTase) is responsible for H3K9me3 at D4Z4 in HeLa cells [Zeng et al., 2009]. Based on these results, we treated immortalized human KD3 myoblasts with chaetocin, a SUV39 HMTase inhibitor (Fig. 1). We confirmed that H3K9me3 is present at D4Z4 in this cell line, and found that the chaetocin treatment indeed decreased H3K9me3 at D4Z4 (Fig. 1A, left). Consistent with this, chaetocin treatment resulted in transcriptional derepression of DUX4fl (Fig. 1A, right). The identity of the DUX4fl-specific PCR product was confirmed by sequencing. Chaetocin was also shown to inhibit TrxR [Tibodeau et al., 2009]. Upregulation of HMOX1 is a marker for TrxR inhibition [Mostert et al., 2003; Trigona et al., 2006]. We found that HMOX1 is indeed upregulated in cells treated with chaetocin similar to cells treated with auranofin, a TrxR inhibitor, indicating that chaetocin under our condition also inhibits TrxR (Supp. Fig. S1A). Unlike chaetocin, however, treating cells with auranofin failed to affect H3K9me3 at D4Z4 or DUX4fl expression, indicating that the observed chaetocin effect on H3K9me3 and DUX4fl expression at D4Z4 is not due to TrxR inhibition (Fig. 1B). Since TrxR inhibition leads to oxidative damage induction [Tibodeau et al., 2009], and oxidative stress has been shown to be associated with FSHD [Turki et al., 2012], we also treated cells with H2O2. This failed to exhibit any effect on DUX4fl expression, indicating that the chaetocin treatment’s effect on DUX4 expression is not the result of oxidative stress (Fig. 1B, right). Chaetocin was also shown to affect G9a HMTase [Iwasa et al., 2010]. Under our treatment condition, we found that H3K9me3 at the c-Myc region, which was shown to be mediated by G9a in HeLa cells [Duan et al., 2005], was also suppressed, suggesting that G9a is also inhibited (Supp. Fig. S1B). Thus, although G9a depletion had no effect on H3K9me3 at D4Z4 in HeLa cells [Zeng et al., 2009], we cannot exclude the possibility that G9a may contribute to D4Z4 heterochromatin organization in the context of myoblasts. Nevertheless, we observed H3K9me3 reduction at D4Z4 in SUV39H1 shRNA-transduced cells, strongly supporting the significant role of SUV39H1 in H3K9me3 at D4Z4 in myoblasts (Fig. 1C). Importantly, SUV39H1 depletion led to DUX4fl induction (Fig. 1C, right). Taken together, the results indicate that the inhibition of H3K9me3 at D4Z4 under these experimental conditions contributes to the derepression of DUX4.
SMCHD1 was found to bind to D4Z4 and depletion of SM-CHD1 results in transcriptional derepression of DUX4fl [Lemmers et al., 2012]. Since SMCHD1 associates with the H3K9me3 domains of the inactive X chromosomes to modulate chromatin compaction [Nozawa et al., 2013], we tested whether H3K9me3 also dictates SMCHD1 association at D4Z4. Both chaetocin treatment and SUV39H1 shRNA depletion resulted in the significant loss of SMCHD1 binding to D4Z4 (Fig. 1D). Neither treatment affected the SMCHD1 protein level (Fig. 1D, right). Thus, the results indicate that one important downstream effector of H3K9me3 is SMCHD1 and suggest that the loss of H3K9me3 at D4Z4 in FSHD contributes to decreased SMCHD1 binding to D4Z4 leading to DUX4fl expression.
Sequencing of Subregions of D4Z4 Homologs on Different Chromosomes
The study of D4Z4 chromatin has been difficult due to the sequence similarity found in the D4Z4-like homologs. Having established the significance of H3K9me3 reduction at D4Z4, we decided to characterize non-4q/10q D4Z4 repeats to see if they also undergo similar epigenetic changes and contribute to DUX4 expression. We examined these repeats on individual chromosomes separately using a DNA mapping panel and somatic cell hybrids carrying a single human chromosome. We previously designed two pairs of PCR primers (4q-Hox and Q-PCR) that selectively amplify 4q and 10q D4Z4 but not other D4Z4 homologs, enabling specific analysis of 4q and 10q D4Z4 in human cells (Fig. 2) [Zeng et al., 2009]. When Q-PCR-F and 4q-Hox-R primers were used against genomic DNA from a DNA mapping panel, PCR products were also obtained from seven other chromosomes (chrs 3, 13, 14, 15, 21, 22, and Y) (Fig. 2). Cloning and sequencing of these PCR products revealed that they all encompass the overlapping region of D4Z4, corresponding to the 5′-untranslated region (5′-UTR) and 5′ one-third of the DUX4 ORF (Fig. 3A). These chromosomes were previously reported to contain D4Z4 homologs, with the exception of chr 3 [Lyle et al., 1995]. Thus, we have identified a novel D4Z4 homolog on chr 3. Interestingly, a D4Z4 homolog previously shown to be on chr 1 [Lyle et al., 1995] was not amplified by the selected primers, suggesting that the corresponding region is highly divergent in the chr 1 homolog or that the presence of this homolog on chr 1 is variable.
Figure 2.

Amplification of the D4Z4 homolog subdomain from chromosomes other than 4q and 10q. A: The combination of the Q-PCR forward primer and 4q-Hox reverse primer was used to amplify the D4Z4 homologs on chromosomes 3, 13, 14, 15, 21, 22, and Y from the mapping panel (top panel). The PCR results of the 4q-Hox primer pair and the Q-PCR primer pair, both of which only amplify 4q and 10q D4Z4, are also shown (two middle panels). PCR amplification of mouse β-minor globin or hamster ribosomal DNA repeats serves as an input control (bottom panel). B: A schematic diagram of the regions covered by the PCR products as indicated by black bars. The location of the DUX4 ORF is also shown to demonstrate the relative locations of these PCR products.
Figure 3.
The sequence results of the D4Z4 homologs on other chromosomes. A: A heat map of the nucleotide variation across sequenced clones. Gray indicates no change, whereas red and green indicate a shift to A/T and C/G, respectively. Black indicates insertion and deletion (indel). The nucleotide numbers are based on the 4q D4Z4 repeat sequence with the accession number AF117653 in the NCBI database. B: The occurrence frequency of the D4Z4 homologs with different numbers of nucleotide variants from each individual chromosome. The frequency is indicated by the diameter of the dots used in the plot. C: The expected and actual frequency of C/G to A/T alteration, A/T to C/G alteration, and C-G or A-T interchanges on the other D4Z4 homologs in comparison to 4q D4Z4. The expected frequency is calculated based on the nucleotide composition of the sequenced 4q D4Z4 subdomain and the probability of alteration to the other three nucleotides is considered as random. D: The percentage of the same sequence present in D4Z4 homologs from different chromosomes. For example, the percentage of “chr 15 versus chr 13” is 17.9%, which means that 17.9% of sequenced clones in chr 15 have sequence(s) that is also present in chr 13.
Non-4q/10q D4Z4 Homologs are Highly Divergent
We sequenced a total of 280 Q-PCR-F + 4q-Hox-R PCR products mentioned above for nine different chromosomes in a DNA mapping panel as well as additional somatic hybrids (see the Methods section for details). We found that the PCR products from 4q and 10q D4Z4 are highly conserved with very little nucleotide variability (Fig. 3A and B, and Supp. Fig. S2). The two major haplotypes of 4q D4Z4 (4qA and 4qB) were shown to differ by one nucleotide (nt) (a “T” instead of “C” at nt 7551 in a portion of 4qB) in this region [Lemmers et al., 2001]. In addition, 10q D4Z4 differs from 4qA D4Z4 by two nucleotides (nts 7515 and 7551, the latter is the same as in 4qB) [Lemmers et al., 2002]. No other heterogeneity was found in 4q and 10q D4Z4-derived clones, consistent with previous reports [Lemmers et al., 2002]. Interestingly, however, cloned PCR products from other homologs exhibited significant nucleotide variability both within and between chromosomes, and none were identical to the 4q or 10q D4Z4 sequences (Fig. 3A and Supp. Fig. S2). Nucleotide changes are scattered relatively evenly throughout the region examined, and there is no apparent mutation hotspot (Fig. 3A). The homologs on chrs 13, 14, 15, 21, and 22 contain multiple sequences with different patterns and degrees of polymorphism, suggesting that the majority of non-4q/10q D4Z4 homologs are susceptible to frequent mutations (Fig. 3B). It should be noted that all PCR clones from chrs 3 and Y each displayed one sequence containing a unique pattern of nucleotide changes (Fig. 3A and B). It is unlikely that this uniformity is due to the existence of only one copy of the D4Z4 homolog on these chromosomes based on the robust PCR signal from chr 3 comparable to that of chr 4 (Fig. 2A). Also, multiple D4Z4 homologs exist on chr Y according to the HG19 build of the human genome reference from the UCSC Genome Browser. Nevertheless, D4Z4 homologs on chrs 3 and Y are divergent from each other and from 4q/10q D4Z4. Furthermore, sequence comparison of the recently discovered single-copy DUXO gene on chr 3, which encodes DUX4c homolog [Sharon et al., 2012], revealed that the D4Z4 homolog on chr 3 amplified by our primer set is distinct from the DUXO gene (Supp. Fig. S3). Taken together, non-4q/10q D4Z4 sequences are highly divergent and variable compared to 4q/10q D4Z4.
D4Z4 Homologs are Enriched for C/G to A/T Conversions and Repeat Exchanges
Approximately 70% of the variation in D4Z4 homologs in comparison to the 4q D4Z4 sequence is a “C” or “G” alteration to “A” or “T” with another 15% corresponding to “A” or “T” alteration to “C” or “G.” The remaining 15% variation is C–G (C to G or G to C) or A–T (A to T or T to A) interchanges. These alterations result in about 80% of nucleotide polymorphisms being A or T (Fig. 3A and Supp. Fig. S2). The 4q D4Z4 repeat is a GC-rich sequence (~73% GC), and the sequenced region also contains 68% GC. If the alterations occurred randomly, however, the C/G to A/T alteration ratio should be 46% (Fig. 3C). Thus, there appears to be a bias for A or T conversion in this region in D4Z4 homologs compared to 4q D4Z4.
Despite previous evidence for repeat exchange between 4q and 10q D4Z4 repeats [Cacurri et al., 1998; Zhang et al., 2001; Lemmers et al., 2010a, b], we failed to detect any 4q sequences in 10q clones (n = 48) or 10q patterns in 4q clones (n = 50). Similarly, no 4q or 10q D4Z4 sequences were found in D4Z4 homologs on other chromosomes (n = 182). In contrast, the same nucleotide polymorphism was found in a subset (<20%) of clones from homologs on chrs 13, 15, and 21 (Fig. 3D). In particular, the same pattern of polymorphisms was observed in more than 10% of clones derived from chrs 13 and 15 D4Z4 homologs. Thus, the results suggest that repeat exchanges had occurred at much higher frequency among non-4q/10q D4Z4 homologs than between these homologs and 4q/10q D4Z4.
The DUX4 ORF is Frequently Disrupted in Non-4q/10q D4Z4 Homologs
The observed hypervariability of the non-4q/10q D4Z4 homologs resulted in alterations of the corresponding amino acid sequence of the putative DUX4 ORF within the sequenced region. Importantly, in 90% of the D4Z4 homolog clones, the nucleotide polymorphisms introduced nonsense codons (Fig. 4). This is in contrast to clones from 4q and 10q D4Z4, in which the DUX4 ORF is consistently open. The 4qB haplotype- and 10q-specific nucleotide polymorphisms do not change the DUX4-encoded amino acid sequence. Analysis of additional somatic cell hybrids revealed the enrichment of different patterns of mutations, also resulting in DUX4 ORF interruption, on chrs 14, 15, and 21 (Fig. 4). Taken together, these results reveal that 4q and 10q D4Z4 repeats are unusually conserved compared to non-4q/10q D4Z4 homologs on other chromosomes.
Figure 4.
The DUX4 ORF is closed in D4Z4 homologs. A: Schematic diagrams of ORFs. The location of the stop codon is marked by a vertical white dashed line and the read-through ratio of the reading frames is demonstrated with different color codes. For the D4Z4 homologs with the same stop codon in all the sequenced clones, the stop codon is indicated by a vertical solid white line with an asterisk. Two different somatic cell hybrid DNA sources were used for the analysis of chrs 13, 14, 15, and 21, and the results are shown separately. The 4q and 10q DUX4 ORFs continue beyond the sequenced area. B: Amino acid sequence comparison of the PCR-amplified N-terminal regions of 4q/10q DUX4 and corresponding regions derived from chrs 3, 15, and Y. Only one sequence each was found in chrs 3 and Y, while a sequence representing approximately 25% of chr 15–2 from (A) is shown.
Development and Analysis of D4Z4 Homolog-Specific PCR Primers
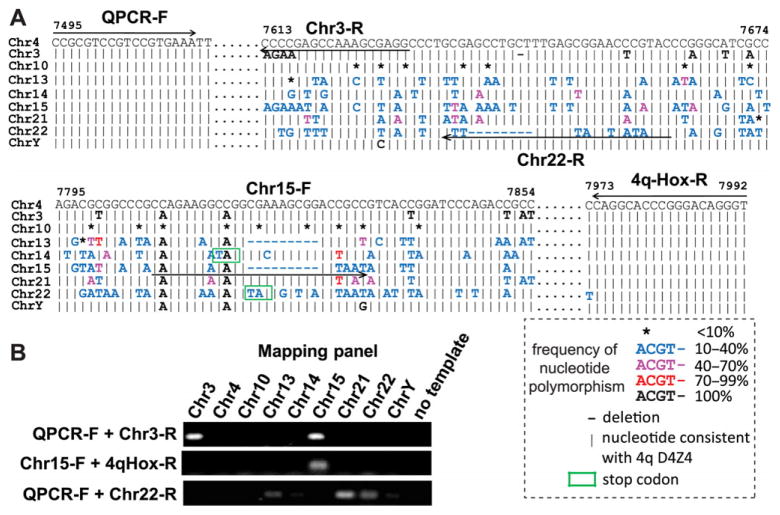
Sequence analysis of the D4Z4 homologs from the DNA mapping panel and somatic cell hybrids described above enabled us to design PCR primers specific for their unique regions (Fig. 5A). The specificity of these primer pairs was confirmed using a DNA mapping panel (Fig. 5B). The primer pair Q-PCR-F + chr 3-R amplified D4Z4 homologs from chrs 3 and 15, but not from others (Fig. 5B). Cross-hybridization of the chr 3-R PCR primer with D4Z4 homologs on chr 15 is expected because the same nucleotide variation in the primer-binding region was found in 14.3% of the chr 15-derived clones (Fig. 5A). The primer pair chr 15-F + 4q-Hox-R specifically amplified the D4Z4 homologs from chr 15, although a similar deletion corresponding to the chr 15-F primer-binding site was also found in a subset (8%) of clones from the chr 13-derived D4Z4 homologs (Fig. 5A and B). The primer pair Q-PCR-F + chr 22-R amplified the D4Z4 homologs not only from chr 22, but also from chrs 13 and 21, and more weakly from chrs 14 and Y (Fig. 5B). The chr 22-specific primer corresponds to a unique deletion found in the chr 22 homolog (Fig. 5A). A similar sequence was also found in a subpopulation (15.4%) of clones from chr 21. Although our initial sequencing analyses of the PCR products of the Q-PCR-F + 4q-Hox-R primers did not identify a sequence identical to chr 22 in the chrs 13-, 14-, and Y-derived clones, the presence of the Q-PCR-F +chr 22-R PCR products indicates that a subpopulation of the repeats on these chromosomes contain sufficient homology that allows chr 22-specific primer binding and amplification. The fact that the PCR signals are relatively weak for chrs 14 and Y suggests that they may represent relatively minor copies, which may explain why we did not see this sequence with our limited sequencing. In addition, it is possible that the 4q-HOX-R primer may not be able to bind to some of these repeats with the chr 22-specific primer binding site because of the sequence diversity within the 4q-HOX-R primer binding region. Importantly, there was no amplification of 4q and 10q D4Z4 using these primer pairs, indicating that they selectively amplify non-4q/10q D4Z4 homologs (Fig. 5B and Supp. Table S3).
Figure 5.
Designing primers specifically covering other D4Z4 homologs. A: PCR primers used to amplify D4Z4 homologs on chrs 3, 15, and 22 are indicated by the arrows. B: The specificity of the primer pairs was assessed on the mapping panel. Neither 4q nor 10q D4Z4 is amplified with these primer pairs. The positive signals are not restricted to chrs 3 or 22 because some of the non-4q/10q D4Z4 homologs contain the same sequence covered by the primers.
Epigenetic Analysis of D4Z4 Homologs Using Specific PCR Primers
With 4q/10q and non-4q/10q D4Z4-specific primers, we examined the heterochromatin status of the D4Z4 homologs. A significant decrease of H3K9me3, but not H3K27me3, was observed at 4q/10q D4Z4 in six independent FSHD1 myoblast samples and one FSHD2 myoblast sample compared to four control myoblasts consistent with our previous results [Zeng et al., 2009] (Fig. 6A). A similar decrease of H3K9me3, but not H3K27me3, was observed in FSHD2 fibroblasts [Zeng et al., 2009]. A parallel loss of cohesin and HP1γ binding was also confirmed (Fig. 6B and C). When we used the three sets of homolog-specific PCR primers described above (Fig. 5), the same heterochromatin marks (H3K9me3 and H3K27me3 as well as cohesin and HP1γ binding) were found in control cells, indicating a similar chromatin organization in non-4q/10q D4Z4 homologs as in 4q/10q D4Z4 (Fig. 6). However, no significant decrease in H3K9me3, cohesion, and HP1γ binding was observed at the non-4q/10q homologs in FSHD1 and FSHD2 cells. Although some of the non-4q/10q ChIP signals appeared to be somewhat weaker, the results are variable with some samples even higher than the control. No consistent decrease in H3K9me3 was observed at non-4q/10q homologs in three additional FSHD2 myoblast samples tested (Supp. Fig. S4). D4Z4 DNA is also hypermethylated in normal somatic cells and hypomethylated only at the contracted 4q D4Z4 allele in FSHD1 and at both the 4q and 10q alleles in FSHD2 [van Overveld et al., 2003; de Greef et al., 2007]. Consistent with this, anti-metC ChIP confirmed the decreased DNA methylation at D4Z4 in both FSHD1 and FSHD2 cells, with FSHD2 more severe than FSHD1 (Fig. 6A and C). However, there was no significant decrease in metC in the non-4q/10q D4Z4 homologs. Taken together, the results indicate that heterochromatin marks (including H3K9me3, HP1γ, and cohesin) as well as DNA methylation do not significantly change in non-4q/10q D4Z4 homologs in both FSHD1 and FSHD2 cells, demonstrating that the loss of heterochromatin is a restricted event associated only with 4q and 10q D4Z4.
Figure 6.
Cohesin/HP1γ binding, H3K9me3, and DNA methylation are intact on non-4q/10q D4Z4 homologs in FSHD1 and FSHD2 cells. A: ChIP-qPCR analysis using antibodies specific for H3K9me3, H3K27me3, and methylcytidine (metC) using 4q/10q-specific (4q/10q-Q-PCR) and non-4q/10q-specific (Q-PCR-F + chr 3-R, chr 15-F + 4q-Hox-R, Q-PCR-F + chr 22-R) PCR primers as indicated at the top. Four normal control (white bars), six FSHD1 (grey bars), and one FSHD2 (black bar) myoblast samples were analyzed. The quantitative real-time PCR was normalized with input DNA and preimmune controls as indicated. P-values for significant differences between normal and FSHD samples are indicated. B: ChIP-qPCR analysis of cohesin and HP1γ in control and FSHD1 myoblasts. C: ChIP-qPCR analysis of normal control and FSHD2 fibroblasts using antibodies specific for H3K9me3, H3K27me3, cohesin, HP1γ, and met-C as indicated.
Sequence Analysis of Non-4q/10q D4Z4 Homologs in Multiple Human Cells
Based on the above results, we characterized the sequences of the endogenous non-4q/10q homologs in human cells (Fig. 7). H3K9me3 ChIP or genomic DNA from primary human myoblasts, fibroblasts, and lymphoblasts as well as from the HeLa cervical cancer cell line, all representing different individuals, were used for PCR. The PCR products were cloned and approximately 6–10 clones from each cell sample were sequenced. Interestingly, the 4q and 10q D4Z4 sequences were preferentially amplified from H3K9me3 ChIP DNA using the Q-PCR-F and 4q-Hox-R primers (Fig. 7, top). This primer pair, however, also efficiently amplified non-4q/10q homologs when individual chromosomes were separately available in the DNA mapping panel (Fig. 2). Therefore, the amplification bias may reflect copy number differences in the context of the whole human genome (Fig. 7, top). The Q-PCR-F and Q-PCR-R primers that specifically amplify 4q/10q D4Z4 [Zeng et al., 2009] indeed amplified the identical D4Z4 sequence except for the two SNPs associated with 10q and 4qB. In contrast, similar to the results from the mapping panel and somatic cell hybrids, significant sequence variability was found in the PCR products obtained with the non-4q/10q D4Z4 primers in human cells (Fig. 7 and Supp. Table S3). Approximately 90% of these sequences matched the sequence variations identified in the mapping panel and in the somatic cell hybrids (Figs. 3A and 7, and Supp. Table S3). Chr 15-specific PCR products from five different cell samples (five different individuals) exhibited one identical sequence (Fig. 7, right), in contrast to the highly variable sequences of the Q-PCR-F/chr 22-R PCR products both within and between different cell samples representing 14 different individuals (Fig. 7, bottom). The Q-PCR-F + chr 22-R PCR products from both H3K9me3 ChIP and genomic DNA were highly variable compared to 4q/10q D4Z4 (Fig. 7, bottom). This is consistent with amplification of D4Z4 homologs from multiple chromosomes by this primer pair (Fig. 5B). Over 70% of the sequences of the Q-PCR-F/chr 22-R PCR products lack the start codon for the DUX4 ORF. Taken together, these results reveal the high conservation of 4q/10q D4Z4 sequences as opposed to non-4q/10q homologs in human cells and confirm that the H3K9me3 heterochromatin mark is indeed present at non-4q/10q homologs. No significant enrichment of a specific sequence(s) in either normal or FSHD cells suggests that H3K9me3 is widely distributed among homologs rather than restricted to a small subregion regardless of the disease status, consistent with the ChIP results (Fig. 6). Our results also demonstrate that these primers are useful for distinguishing 4q/10q from the non-4q/10q D4Z4 repeats in human cells.
Figure 7.
Sequencing analysis of non-4q/10q D4Z4 homologs in human cells. H3K9me3 ChIP and genomic DNA from the indicated human cell samples were used for PCR analysis using 4q/10q-specific (Q-PCR-F + Q-PCR-R) and non-4q/10q homolog-specific (Q-PCR-F + chr 3-R, chr 15-F + 4q-Hox-R, Q-PCR-F + chr 22-R) primers. Sequencing results using Q-PCR and 4q-Hox-R as used in Figure 3A are shown at the top. Nucleotide numbers are indicated. A heat map of the nucleotide variation was made similar to Figure 3A. Gray indicates no change, whereas red and green indicate a shift to A/T and C/G, respectively. Black indicates insertion and deletion (indel). The results are also summarized in the Supp. Table S3. The bracket for each cell line is proportional to the number of clones analyzed.
Discussion
In the current study, we demonstrated the significance of the H3K9me3 heterochromatin in DUX4 gene regulation and SMCHD1 assembly at D4Z4. We also performed genomic and epigenomic characterization of the DUX4 5′ region in D4Z4 homologs on multiple human chromosomes. Despite awareness of the presence of D4Z4-like repeats scattered on different chromosomes, the extent of their homology and epigenetic similarity with 4q and 10q D4Z4 had not been assessed. We discovered unexpected differences between 4q/10q D4Z4 and other D4Z4 homologs, highlighting the unique link between 4q/10q D4Z4 and FSHD.
H3K9me3 Affects DUX4fl Expression and SMCHD1 Binding at D4Z4
Although the loss of H3K9me3 at D4Z4 observed in both FSHD1 and FSHD2 patients was postulated to contribute to disease-specific gene alterations [Zeng et al., 2009], this has not been experimentally proven. We previously demonstrated that SUV39H plays a critical role in mediating H3K9me3 at D4Z4 [Zeng et al., 2009]. Using immortalized myoblasts, we showed that DUX4fl expression is indeed increased when H3K9me3 is inhibited either by a chemical inhibitor or by shRNA against SUV39H1. This provides important evidence that H3K9me3 plays a role in DUX4fl repression, supporting our hypothesis that disruption of D4Z4 H3K9me3 heterochromatin affects gene expression in FSHD. While these immortalized cells retain differentiation capability in vivo and in vitro with expression of appropriate marker genes [Shiomi et al., 2011], further analysis is necessary to determine to what extent loss of H3K9me3 affects expression of DUX4 and other genes in the context of primary patient muscle cells and tissues. Furthermore, other H3K9 HMTases, such as G9a, may contribute to H3K9me3 regulation at D4Z4 in a myogenic context, though SUV39H1 depletion alone had a significant effect on DUX4fl expression in our study.
SMCHD1, an epigenetic gene silencer mutated in >80% of FSHD2 patients and severe cases of FSHD1, binds to D4Z4 and its depletion results in expression of DUX4fl [Lemmers et al., 2012; Sacconi et al., 2013]. How SMCHD1 is recruited to D4Z4 chromatin, however, was unclear. We found that suppression of H3K9me3 affects SMCHD1 association with D4Z4, indicating that SMCHD1 is a component of D4Z4 H3K9me3 heterochromatin. This raises the possibility that SMCHD1 binding to D4Z4 is already impacted by the loss of H3K9me3 in FSHD even in those FSHD1 and FSHD2 cases in which the SMCHD1 gene is intact, and this may be further worsened in the cases with the haploinsufficiency mutations of SM-CHD1. It will be important to test whether SMCHD1 binding to D4Z4 is indeed reduced in primary patient myoblasts. Because SM-CHD1 was found to mediate DNA methylation [Ashe et al., 2008; Blewitt et al., 2008; Gendrel et al., 2012], it is possible that SM-CHD1 contributes to DNA hypermethylation observed at D4Z4, which is lost in FSHD [van Overveld et al., 2003]. Thus, we propose that H3K9me3 contributes to the recruitment of SMCHD1, which in turn mediates DNA methylation at D4Z4 (Fig. 8A). Although SMCHD1 associates with the inactive X chromosome through interaction with XIST RNA in the chromatin domains enriched for H3K27me3 (though H3K27me3 itself is not required), it also associates with the H3K9me3 domains via the HP1-binding protein HBiX1 [Nozawa et al., 2013]. Thus, it would be interesting to further examine the relationship between SMCHD1 and HP1γ/cohesin that assembles at D4Z4 in an H3K9me3-dependent manner [Zeng et al., 2009]. Taken together, our results suggest that diminished binding of downstream effectors of H3K9me3 may be a key event leading to pathogenic alteration of DUX4 gene expression (Fig. 8B) and possibly of additional target genes in FSHD.
Figure 8.

Schematic models of H3K9me3 heterochromatin at D4Z4. A: The heterochromatin domain of D4Z4 is marked by DNA hypermethylation, H3K9me3, and K27me3. HP1γ and cohesin are recruited to this region in a mutually dependent manner, which requires H3K9me3 mediated primarily by SUV39H. SMCHD1 is also recruited to this domain in an H3K9me3-dependent manner, and may contribute to DNA methylation. Possible interaction between SMCHD1 and HP1γ is shown with a question mark. B: The H3K9me3 heterochromatin assembled at D4Z4 is specifically lost in FSHD, contributing to DUX4fl expression.
Frequent Repeat Sharing among D4Z4 Homologs, but Not with 4q/10q D4Z4
Although homologous repeat exchange between 4q and 10q D4Z4 during hominoid evolution has been reported [Lemmers et al., 2010b], we failed to detect any 4q sequence patterns in 10q D4Z4 clones, or vice versa, which may require sequencing of additional DNA samples. In contrast, however, we were able to detect patterns of nucleotide polymorphisms that are shared among non-4q/10q D4Z4 homologs on different chromosomes distinct from 4q/10q D4Z4. This suggests possible evolutionary interactions and repeat exchanges between D4Z4 homologs. We failed to detect any 4q or 10q D4Z4 sequence in the clones from non-4q/10q D4Z4 homologs or vice versa, suggesting that the involvement of 4q/10q D4Z4 repeats in repeat exchange with other D4Z4 homologs is less frequent, consistent with recent observations [Lemmers et al., 2010b]. It should be noted that while 4q and 10q D4Z4 clusters are both in the subtelomeric regions, other D4Z4 homologs are mostly located near centromeres [Lyle et al., 1995]. Furthermore, while 4q/10q D4Z4 repeats are direct repeats, homologs on acrocentric chrs 13, 14, 15, 21, and 22 were shown to be interspersed with ribosomal DNA and β-satellite repeats [Winokur et al., 1996]. Although they are likely to be derived from genomic duplication events, it is possible that differences in chromosomal location and repeat structure organization might have restricted the frequency of repeat exchange and interfered with “concerted evolution” [Liao, 1999] between 4q/10q D4Z4 and non-4q/10q homologs.
Hypervariability of D4Z4 Homologs within the DUX4 Coding Region
Sequence comparison between 4q/10q D4Z4 and other homologs revealed striking differences in the frequency of nucleotide polymorphisms. This was observed not only in the somatic cell hybrids, but also in multiple human cell samples. Many polymorphisms found in non-4q/10q D4Z4 homologs resulted in stop codons in the DUX4 ORF. The biased C/G to A/T changes in non-4q/10q D4Z4 homologs when compared to 4q/10q D4Z4 sequence may be related to the high frequency of methyl-C to T mutations [Rideout et al., 1990; Mugal and Ellegren, 2011; Xia et al., 2012]. The reason why 4q/10q D4Z4, in particular the DUX4 ORF, is free from similar mutational changes is currently unclear. Chromosomal location (subtelomeres vs. peri-centromeres) or repeat organization (see above) may contribute to this difference. However, it is likely that there is specific selection pressure to preserve the primary sequence and/or the coding capacity of the DUX4 ORF in 4q and 10q D4Z4. The DUX4 gene is conserved in primates and is relatively abundantly expressed in human testis [Clapp et al., 2007; Snider et al., 2010], suggestive of its conserved role in development. While D4Z4 transgenic mice failed to exhibit any dystrophic phenotype [Krom et al., 2013], DUX4fl and its target gene expression in muscles were nevertheless shown to highly correlate with FSHD in humans [Dixit et al., 2007; Lemmers et al., 2010a; Snider et al., 2010; Geng et al., 2012; Jones et al., 2012; Rahimov et al., 2012; Broucqsault et al., 2013; Ferreboeuf et al., 2014]. Taken together, the results strongly suggest the biological significance of the preservation of the DUX4 ORF, and support the notion that its misregulation contributes significantly to FSHD.
The Loss of H3K9me3/HP1γ/Cohesin and DNA Methylation at 4q and 10q D4Z4 is Not Transmitted to Other D4Z4 Homologs in FSHD
Using specific PCR primers, we were able for the first time to analyze the epigenetic marks on the non-4q/10q D4Z4 homologs. Although heterochromatin marks similar to those associated with 4q/10q D4Z4 were found on these homologs in control myoblasts/fibroblasts, they are retained in both FSHD1 and FSHD2 cells, suggesting regulatory differences between the heterochromatin of 4q/10q D4Z4 and that of the non-4q/10q D4Z4 homologs in FSHD. In particular, this establishes that even FSHD2, which does not carry any genetic alteration at the 4q D4Z4 locus, exhibits a region-restricted epigenetic change, further emphasizing the specificity and significance of D4Z4 chromatin change in FSHD. Our results indicate that the heterochromatin loss can contribute to derepression of DUX4 transcription. It is attractive to speculate that this derepression is meaningful, and is therefore associated with the disease, because the DUX4 ORF on 4q/10q is open. Although we found that the DUX4 ORF is mostly interrupted in non-4q/10q D4Z4 homologs due to mutations, the 4q D4Z4 region was shown to express the DUX4 C-terminal short transcript [Snider et al., 2010] and additional noncoding RNA transcripts [Snider et al., 2009], which may still be expressed from the non-4q/10q D4Z4 homologs. The lack of any significant epigenetic change suggests, however, that the expression of these transcripts, if any, from the non-4q/10q D4Z4 homologs is most likely unaffected in FSHD.
Conclusions
Our results indicate that the decrease in H3K9me3, the epigenetic change that signifies FSHD, results in derepression of DUX4fl in myoblasts. Our study has revealed a striking difference between 4q/10q D4Z4 and the non-4q/10q D4Z4 homologs on other chromosomes. The 4q and 10q D4Z4 repeats are highly uniform and conserved, at least in the region encompassing the N-terminus of the DUX4 ORF, and are epigenetically coregulated in FSHD [Zeng et al., 2009]. In contrast, the corresponding regions in the non-4q/10q homologs are highly heterogeneous with frequent interruptions of the DUX4 ORF and do not undergo epigenetic alteration in FSHD. Thus, our results reveal the unique characteristics of 4q and 10q D4Z4 among the related repeats, linking the epigenetic alteration and DUX4 expression in FSHD. The homolog-specific nucleotide polymorphism information reported here should also facilitate further D4Z4 repeat analysis in the FSHD field.
Supplementary Material
Acknowledgments
Contract grant sponsors: National Institutes of Health (NS069539 and AR058548); Muscular Dystrophy Association (MDA4026); FSH Society (FSHS-DHY-001, FSHS-DHY-002 and FSHS-22011-05); National Library of Medicine (NIH T15LM07443); Eunice Kennedy Shriver National Institute of Child Health and Human Development (T32-HD060555); National Cancer Institute (NIH T32 CA009054).
We thank Dr. Marina Mora in the Division of Neuromuscular Diseases at the C. Besta Neurological Institute for providing myoblasts through the biobank “Cells, tissues, and DNA from patients with neuromuscular diseases,” as part of the Telethon Genetic Network of Biobanks and the EuroBioBank network.
Footnotes
Disclosure statement: The authors declare no conflict of interest.
Additional Supporting Information may be found in the online version of this article.
References
- Ashe A, Morgan DK, Whitelaw NC, Bruxner TJ, Vickaryous NK, Cox LL, Butterfield NC, Wicking C, Blewitt ME, Wilkins SJ, Anderson GJ, Cox TC, et al. A genome-wide screen for modifiers of transgene variegation identifies genes with critical roles in development. Genome Biol. 2008;9:R182. doi: 10.1186/gb-2008-9-12-r182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blewitt ME, Gendrel AV, Pang Z, Sparrow DB, Whitelaw N, Craig JM, Apedaile A, Hilton DJ, Dunwoodie SL, Brockdorff N, Kay GF, Whitelaw E. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet. 2008;40:663–669. doi: 10.1038/ng.142. [DOI] [PubMed] [Google Scholar]
- Bosnakovski D, Xu Z, Gang EJ, Galindo CL, Liu M, Simsek T, Garner HR, Agha-Mohammadi S, Tassin A, Coppée F, Belayew A, Perlingeiro RR, et al. An iso-genetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 2008;27:2766–2779. doi: 10.1038/emboj.2008.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broucqsault N, Morere J, Gaillard MC, Dumonceaux J, Torrents J, Salort-Campana E, Maues De Paula A, Bartoli M, Fernandez C, Chesnais AL, Ferreboeuf M, Sarda L, et al. Dysregulation of 4q35- and muscle-specific genes in fetuses with a short D4Z4 array linked to facioscapulohumeral dystrophy. Hum Mol Genet. 2013;22:4206–4214. doi: 10.1093/hmg/ddt272. [DOI] [PubMed] [Google Scholar]
- Cacurri S, Piazzo N, Deidda G, Vigneti E, Galluzzi G, Colantoni L, Merico B, Ricci E, Felicetti L. Sequence homology between 4qter and 10qter loci facilitates the instability of subtelomeric KpnI repeat units implicated in facioscapulohumeral muscular dystrophy. Am J Hum Genet. 1998;63:181–190. doi: 10.1086/301906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapp J, Mitchell LM, Bolland DJ, Fantes J, Corcoran AE, Scotting PJ, Armour JA, Hewitt JE. Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2007;81:264–279. doi: 10.1086/519311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Greef JC, Lemmers RJ, Camaño P, Day JW, Sacconi S, Dunand M, van Engelen BG, Kiuru-Enari S, Padberg GW, Rosa AL, Desnuelle C, Spuler S, et al. Clinical features of facioscapulohumeral muscular dystrophy 2. Neurology. 2010;75:1548–1554. doi: 10.1212/WNL.0b013e3181f96175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Greef JC, Lemmers RJ, van Engelen BG, Sacconi S, Venance SL, Frants RR, Tawil R, van der Maarel SM. Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum Mol Genet. 2009;30:1449–1459. doi: 10.1002/humu.21091. [DOI] [PubMed] [Google Scholar]
- de Greef JC, Wohlgemuth M, Chan OA, Hansson KB, Smeets D, Frants RR, Weemaes CM, Padberg GW, van der Maarel SM. Hypomethylation is restricted to the D4Z4 repeat array in phenotypic FSHD. Neurology. 2007;69:1018–1026. doi: 10.1212/01.wnl.0000271391.44352.fe. [DOI] [PubMed] [Google Scholar]
- Dixit M, Ansseau E, Tassin A, Winokur S, Shi R, Qian H, Sauvage S, Mattéotti C, van Acker AM, Leo O, Figlewicz D, Barro M, et al. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci USA. 2007;104:18157–18162. doi: 10.1073/pnas.0708659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Zarebski A, Montoya-Durango D, Grimes HL, Horwitz M. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol Cell Biol. 2005;25:10338–10351. doi: 10.1128/MCB.25.23.10338-10351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- Ferreboeuf M, Mariot V, Bessières B, Vasiljevic A, Attié-Bitach T, Collardeau S, Morere J, Roche S, Magdinier F, Robin-Ducellier J, Rameau P, Whalen S, et al. DUX4 and DUX4 downstream target genes are expressed in fetal FSHD muscles. Hum Mol Genet. 2014;23:171–181. doi: 10.1093/hmg/ddt409. [DOI] [PubMed] [Google Scholar]
- Gabriëls J, Beckers MC, Ding H, De Vriese A, Plaisance S, van der Maarel SM, Padberg GW, Frants RR, Hewitt JE, Collen D, Belayew A. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236:25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Apedaile A, Coker H, Termanis A, Zvetkova I, Godwin J, Tang YA, Huntley D, Montana G, Taylor S, Giannoulatou E, Heard E, et al. Smchd1-dependent and -independent pathways determine developmental dynamics of CpG island methylation on the inactive X chromosome. Dev Cell. 2012;23:265–279. doi: 10.1016/j.devcel.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng LN, Yao Z, Snider L, Fong AP, Cech JN, Young JM, van der Maarel SM, Ruzzo WL, Gentleman RC, Tawil R, Tapscott SJ. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev Cell. 2012;22:38–51. doi: 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregson HC, Schmiesing JA, Kim J-S, Kobayashi T, Zhou S, Yokomori K. A potential role for human cohesin in mitotic spindle aster assembly. J Biol Chem. 2001;276:47575–47582. doi: 10.1074/jbc.M103364200. [DOI] [PubMed] [Google Scholar]
- Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, Gartler SM. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci USA. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt JE, Lyle R, Clark LN, Valleley EM, Wright TJ, Wijmenga C, van Deutekom JC, Francis F, Sharpe PT, Hofker M, Frants RR, Williamson R. Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum Mol Genet. 1994;3:1287–1295. doi: 10.1093/hmg/3.8.1287. [DOI] [PubMed] [Google Scholar]
- Iwasa E, Hamashima Y, Fujishiro S, Higuchi E, Ito A, Yoshida M, Sodeoka M. Total synthesis of (+)-chaetocin and its analogues: their histone methyltransferase G9a inhibitory activity. J Am Chem Soc. 2010;132:4078–4079. doi: 10.1021/ja101280p. [DOI] [PubMed] [Google Scholar]
- Jones TI, Chen JC, Rahimov F, Homma S, Arashiro P, Beermann ML, King OD, Miller JB, Kunkel LM, Emerson CPJ, Wagner KR, Jones PL. Facioscapulohumeral muscular dystrophy family studies of DUX4 expression: evidence for disease modifiers and a quantitative model of pathogenesis. Hum Mol Genet. 2012;21:4419–4430. doi: 10.1093/hmg/dds284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krom YD, Thijssen PE, Young JM, den Hamer B, Balog J, Yao Z, Maves L, Snider L, Knopp P, Zammit PS, Rijkers T, van Engelen BG, et al. Intrinsic epigenetic regulation of the D4Z4 macrosatellite repeat in a transgenic mouse model for FSHD. PLoS Genet. 2013;9:e1003415. doi: 10.1371/journal.pgen.1003415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, de Kievit P, Sandkuijl L, Padberg GW, van Ommen GJ, Frants RR, van der Maarel SM. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat Genet. 2002;32:235–236. doi: 10.1038/ng999. [DOI] [PubMed] [Google Scholar]
- Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camaño P, Dauwerse JG, Snider L, Straasheijm KR, van Ommen GJ, Padberg GW, Miller DG, Tapscott SJ, et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010a;329:1650–1653. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, van der Vliet PJ, van der Gaag KJ, Zuniga S, Frants RR, de Knijff P, van der Maarel SM. Worldwide population analysis of the 4q and 10q subtelomeres identifies only four discrete interchromosomal sequence transfers in human evolution. Am J Hum Genet. 2010b;86:364–377. doi: 10.1016/j.ajhg.2010.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, Wohlgemuth M, Frants RR, Padberg GW, Morava E, van der Maarel SM. Contractions of D4Z4 on 4qB subtelomeres do not cause facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2004;75:1124–1130. doi: 10.1086/426035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, Wohlgemuth M, van der Gaag KJ, van der Vliet PJ, van Teijlingen CM, de Knijff P, Padberg GW, Frants RR, van der Maarel SM. Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2007;81:884–894. doi: 10.1086/521986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJL, de Kievit P, van Geel M, van der Wielen MJ, Bakker E, Padberg GW, Frants RR, van der Maarel SM. Complete allele information in the diagnosis of facioscapulohumeral muscular dystrophy by triple DNA analysis. Ann Neurol. 2001;50:816–819. doi: 10.1002/ana.10057. [DOI] [PubMed] [Google Scholar]
- Lemmers RJLF, Tawil R, Petek LM, Balog J, Block GJ, Santen GWE, Amell AM, van der Vliet PJ, Almomani R, Straasheijm KR, Krom YD, Klooster R, et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet. 2012;44:1370–1374. doi: 10.1038/ng.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D. Concerted evolution: molecular mechanism and biological implications. Am J Hum Genet. 1999;64:24–30. doi: 10.1086/302221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyle R, Wright TJ, Clark LN, Hewitt JE. The FSHD-associated repeat, D4Z4, is a member of a dispersed family of homeobox-containing repeats, subsets of which are clustered on the short arms of the acrocentric chromosomes. Genomics. 1995;28:389–397. doi: 10.1006/geno.1995.1166. [DOI] [PubMed] [Google Scholar]
- Mitsuhashi H, Mitsuhashi S, Lynn-Jones T, Kawahara G, Kunkel LM. Expression of DUX4 in zebrafish development recapitulates facioscapulohumeral muscular dystrophy. Hum Mol Genet. 2013;22:568–577. doi: 10.1093/hmg/dds467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostert V, Hill KE, Burk RF. Loss of activity of the selenoenzyme thioredoxin reductase causes induction of hepatic heme oxygenase-1. FEBS Lett. 2003;541:85–88. doi: 10.1016/s0014-5793(03)00309-0. [DOI] [PubMed] [Google Scholar]
- Mugal CF, Ellegren H. Substitution rate variation at human CpG sites correlates with non-CpG divergence, methylation level and GC content. Genome Biol. 2011;12:R58. doi: 10.1186/gb-2011-12-6-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa RS, Nagao K, Igami KT, Shibata S, Shirai N, Nozaki N, Sado T, Kimura H, Obuse C. Human inactive X chromosome is compacted through a PRC2-independent SMCHD1-HBiX1 pathway. Nat Struc Mol Biol. 2013;20:566–573. doi: 10.1038/nsmb.2532. [DOI] [PubMed] [Google Scholar]
- Rahimov F, King OD, Leung DG, Bibat GM, Emerson CPJ, Kunkel LM, Wagner KR. Transcriptional profiling in facioscapulohumeral muscular dystrophy to identify candidate biomarkers. Proc Natl Acad Sci USA. 2012;109:16234–16239. doi: 10.1073/pnas.1209508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout WMr, Coetzee GA, Olumi AF, Jones PA. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science. 1990;249:1288–1290. doi: 10.1126/science.1697983. [DOI] [PubMed] [Google Scholar]
- Sacconi S, Lemmers RJ, Balog J, van der Vliet PJ, Lahaut P, van Nieuwenhuizen MP, Straasheijm KR, Debipersad RD, Vos-Versteeg M, Salviati L, Casarin A, Pegoraro E, et al. The FSHD2 gene smchd1 is a modifier of disease severity in families affected by FSHD1. Am J Hum Genet. 2013;93:744–751. doi: 10.1016/j.ajhg.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scionti I, Greco F, Ricci G, Govi M, Arashiro P, Vercelli L, Berardinell iA, Angelini C, Antonini G, Cao M, Di Muzio A, Moggio M, et al. Large-scale population analysis challenges the current criteria for the molecular diagnosis of fascioscapulohumeral muscular dystrophy. Am J Hum Genet. 2012;90:628–635. doi: 10.1016/j.ajhg.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon N, Mor I, Zahavi E, Benvenisty N. DUXO, a novel double homeobox transcription factor, is a regulator of the gastrula organizer in human embryonic stem cells. Stem Cell Res. 2012;9:261–269. doi: 10.1016/j.scr.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Shiomi K, Kiyono T, Okamura K, Uezumi M, Goto Y, Yasumoto S, Shimizu S, Hashimoto N. CDK4 and cyclin D1 allow human myogenic cells to recapture growth property without compromising differentiation potential. Gene Ther. 2011;18:857–866. doi: 10.1038/gt.2011.44. [DOI] [PubMed] [Google Scholar]
- Snider L, Asawachaicharn A, Tyler AE, Geng LN, Petek LM, Maves L, Miller DG, Lemmers RJ, Winokur ST, Tawil R, van der Maarel SM, Filippova GN, et al. RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum Mol Genet. 2009;18:2414–2430. doi: 10.1093/hmg/ddp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider L, Geng LN, Lemmers RJ, Kyba M, Ware CB, Nelson AM, Tawil R, Filippova GN, van der Maarel SM, Tapscott SJ, Miller DG. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 2010;6:e1001181. doi: 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam R, Smith KP, Lawrence JB. The 4q subtelomere harboring the FSHD locus is specifically anchored with peripheral heterochromatin unlike most human telomeres. J Cell Biol. 2004;167:269–279. doi: 10.1083/jcb.200403128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thijssen PE, Tobi EW, Balog J, Schouten SG, Kremer D, Bouazzaoui FE, Henneman P, Putter H, Slagboom PE, Heijmans BT, van der Maarel SM. Chromatin remodeling of human subtelomeres and TERRA promoters upon cellular senescence: commonalities and differences between chromosomes. Epigenetics. 2013;8:512–521. doi: 10.4161/epi.24450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibodeau JD, Benson LM, Isham CR, Owen WG, Bible KC. The anticancer agent chaetocin is a competitive substrate and inhibitor of thioredoxin reductase. Antioxid Redox Signal. 2009;11:1097–1106. doi: 10.1089/ars.2008.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigona WL, Mullarky IK, Cao Y, Sordillo LM. Thioredoxin reductase regulates the induction of haem oxygenase-1 expression in aortic endothelial cells. Biochem J. 2006;394:207–216. doi: 10.1042/BJ20050712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turki A, Hayot M, Carnac G, Pillard F, Passerieux E, Bommart S, Raynaud de Mauverger E, Hugon G, Pincemail J, Pietri S, Lambert K, Belayew A, et al. Functional muscle impairment in facioscapulohumeral muscular dystrophy is correlated with oxidative stress and mitochondrial dysfunction. Free Radic Biol Med. 2012;53:1068–1079. doi: 10.1016/j.freeradbiomed.2012.06.041. [DOI] [PubMed] [Google Scholar]
- van der Maarel SM, Frants RR. The D4Z4 repeat-mediated pathogenesis of facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2005;76:375–386. doi: 10.1086/428361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maarel SM, Tawil R, Tapscott SJ. Facioscapulohumeral muscular dystrophy and DUX4: breaking the silence. Trends Mol Med. 2011;17:252–258. doi: 10.1016/j.molmed.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderplanck C, Ansseau E, Charron S, Stricwant N, Tassin A, Laoudj-Chenivesse D, Wilton SD, Coppée F, Belayew A. The FSHD atrophic myotube phenotype is caused by DUX4 expression. PLoS One. 2011;6:e26820. doi: 10.1371/journal.pone.0026820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overveld PG, Lemmers RJ, Sandkuijl LA, Enthoven L, Winokur ST, Bakels F, Padberg GW, van Ommen G-JB, Frants RR, van der Maarel SM. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet. 2003;35:315–317. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schübeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Winokur ST, Bengtsson U, Feddersen J, Mathews KD, Weiffenbach B, Bailey H, Markovich RP, Murray JC, Wasmuth JJ, Altherr MR. The DNA rearrangement associated with facioscapulohumeral muscular dystrophy involves a heterochromatin-associated repetitive element: implications for a role of chromatin structure in the pathogenesis of the disease. Chromosome Res. 1994;2:225–234. doi: 10.1007/BF01553323. [DOI] [PubMed] [Google Scholar]
- Winokur ST, Bengtsson U, Vargas JC, Wasmuth JJ, Altherr MR, Weiffenbach B, Jacobsen SJ. The evolutionary distribution and structural organization of the homeoboxs-containing repeat D4Z4 indicates a functional role for the ancestral copy in the FSHD region. Hum Mol Genet. 1996;5:1567–1575. doi: 10.1093/hmg/5.10.1567. [DOI] [PubMed] [Google Scholar]
- Xia J, Han L, Zhao Z. Investigating the relationship of DNA methylation with mutation rate and allele frequency in the human genome. BMC Genomics. 2012;13:S7. doi: 10.1186/1471-2164-13-S8-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GL, Bestor TH, Bourc’his D, Hsieh CL, Tommerup N, Bugge M, Hulten M, Qu X, Russo JJ, Viegas-Pequignot E. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- Zeng W, de Greef JC, Chen Y-Y, Chien R, Kong X, Gregson HC, Winokur ST, Pyle A, Robertson KD, Schmiesing JA, Kimonis VE, Balog J, et al. Specific loss of histone H3 lysine 9 trimethylation and HP1γ/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS Genet. 2009;5:e1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Forner J, Fournet S, Jeanpierre M. Improved characterization of FSHD mutations. Ann Genet. 2001;44:105–110. doi: 10.1016/s0003-3995(01)01075-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.