Abstract
BACKGROUND
The prediction of clinical behavior, response to therapy, and outcome of infiltrative glioma is challenging. On the basis of previous studies of tumor biology, we defined five glioma molecular groups with the use of three alterations: mutations in the TERT promoter, mutations in IDH, and codeletion of chromosome arms 1p and 19q (1p/19q codeletion). We tested the hypothesis that within groups based on these features, tumors would have similar clinical variables, acquired somatic alterations, and germline variants.
METHODS
We scored tumors as negative or positive for each of these markers in 1087 gliomas and compared acquired alterations and patient characteristics among the five primary molecular groups. Using 11,590 controls, we assessed associations between these groups and known glioma germline variants.
RESULTS
Among 615 grade II or III gliomas, 29% had all three alterations (i.e., were triplepositive), 5% had TERT and IDH mutations, 45% had only IDH mutations, 7% were triple-negative, and 10% had only TERT mutations; 5% had other combinations. Among 472 grade IV gliomas, less than 1% were triple-positive, 2% had TERT and IDH mutations, 7% had only IDH mutations, 17% were triple-negative, and 74% had only TERT mutations. The mean age at diagnosis was lowest (37 years) among patients who had gliomas with only IDH mutations and was highest (59 years) among patients who had gliomas with only TERT mutations. The molecular groups were independently associated with overall survival among patients with grade II or III gliomas but not among patients with grade IV gliomas. The molecular groups were associated with specific germline variants.
CONCLUSIONS
Gliomas were classified into five principal groups on the basis of three tumor markers. The groups had different ages at onset, overall survival, and associations with germline variants, which implies that they are characterized by distinct mechanisms of pathogenesis.
The past 25 years of research into glioma biology have led to the discovery of hundreds of molecular alterations in grade II, III, and IV gliomas (grade II and III gliomas are sometimes described as lower-grade gliomas, and grade IV gliomas are commonly described as glioblastoma multiforme).1–3 Among these molecular alterations, three are particularly noteworthy, because they occur early during glioma formation, are prevalent in glioma, or are strongly associated with overall survival. The first alteration to be identified was the codeletion of chromosome arms 1p and 19q (1p/19q codeletion), which is associated with the oligodendroglial histologic type and with sensitivity to chemotherapy with alkylating agents.4–6 The second was mutation in either IDH1 or IDH2 (these genes are very similar to one another and hereafter are collectively referred to as IDH), which was not restricted to a specific histopathological type of glioma but instead was associated with a distinctive tumor-cell metabolism.7 The third was mutation in the promoter of TERT, which encodes telomerase. Curiously, mutation in the TERT promoter, which results in enhanced telomerase activity and lengthened telomeres, is seen in both the most aggressive human glioma (grade IV astrocytoma) and the least aggressive diffuse human glioma (grade II oligodendroglioma). This suggests that telomere maintenance may be a necessary precondition for the formation of brain cancer.8,9 In addition, certain germline polymorphisms are associated with specific histologic types of glioma10–12 and particularly with the presence or absence of tumor-specific mutations, such as those affecting IDH.13 For example, one single-nucleotide polymorphism (SNP) on chromosome 8q24 (rs55705857 in dbSNP) is associated with a risk of IDH-mutated glioma14 that is increased by up to a factor of 6 and is associated with progression-free survival among patients who have been treated with procarbazine, lomustine, and vincristine.15 Thus, using this knowledge of tumor biology, we defined molecular groups16,17 that are based on the presence or absence of TERT promoter mutations, IDH mutations, and 1p/19q codeletion and sought to determine whether (and in some cases, confirm that) these groups have specific clinical features, are enriched for specific acquired somatic alterations, and have associations with germline variants.14
Methods
Study Participants
Written informed consent was obtained from all participants in all studies. In this study, we included patients who had infiltrative glioma of histologic grade II, III, or IV. Grade I tumors (pilocytic astrocytoma) are clinically and pathologically distinct and thus were not included.
The Mayo Clinic glioma case–control series, as well as the cases and controls from the University of California, San Francisco (UCSF), Adult Glioma Study, have been described previously.10,13 A total of 317 cases and 789 controls from the Mayo Clinic series were used as the discovery set in this study, and 351 cases and as many as 4504 controls (depending on the SNP being analyzed) from the UCSF Adult Glioma Study were used as the first replication set.
A total of 153 grade IV gliomas and 266 grade II or grade III gliomas from the Cancer Genome Atlas (TCGA) could be assigned to one of the five molecular groups and thus were used as the second replication set in this study (Table S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org).
Data from the Mayo Genome Consortia phase 1 controls were used as the control data for SNP association analyses of TCGA glioma cases.18 The Mayo Genome Consortia phase 1 samples included 6297 controls across three studies.
Statistical Methods
We compared age at diagnosis across the five molecular groups, using contrast estimates from an analysis of variance model. Both unadjusted and adjusted Kaplan–Meier curves were used to estimate overall survival for each of the five molecular groups stratified according to grade. A stratified (according to grade) Cox proportional hazards model was used to determine whether the five molecular groups were associated with overall patient survival after adjustment for sex, age at diagnosis, tumor histologic type, and tumor grade. For each molecular group, an additive logistic-regression model was used to assess the association between each SNP and disease status, with genotype coded as 0, 1, or 2 copies of the minor allele. Detailed information regarding the statistical analysis, as well as additional information about the three case–control studies, clinical characteristics, pathologic features, tumor markers, and genotyping, is available in the Supplementary Appendix.
Results
Molecular Groups
The overall prevalence of TERT promoter mutations, IDH mutations, and 1p/19q codeletion was similar across the three data sets (the Mayo Clinic, the UCSF Adult Glioma Study, and TCGA) (Fig. S1 in the Supplementary Appendix). Of the eight possible combinations based on the presence or absence of the three tumor markers, five (each of which represented 4% or more of the full set) could be used to classify most of the 1087 glioma cases (Fig. 1, and Table S2A in the Supplementary Appendix): triple-positive (mutations in both TERT and IDH plus 1p/19q codeletion), mutations in both TERT and IDH, mutation in IDH only, triple-negative, and mutation in TERT only. The prevalences of these five groups were similar across the three data sets (Fig. S2 in the Supplementary Appendix). Among the 615 grade II or grade III gliomas in the combined data set, 29% were triple-positive, 5% had mutations in both TERT and IDH, 45% had mutations in IDH only, 7% were triple-negative, 10% had mutations in TERT only, and 5% had other combinations. Among the 472 grade IV gliomas in the combined data set, less than 1% were triple-positive, 2% had mutations in both TERT and IDH, 7% had mutations in IDH only, 17% were triple-negative, and 74% had mutations in TERT only. Only 28 (2.6%) of the gliomas could not be assigned to one of these five groups: 21 of these (75%) were oligodendrogliomas or mixed oligoastrocytomas with IDH mutations and 1p/19q codeletion (but no TERT mutation).
Figure 1. Prevalence of the Glioma Molecular Groups in the Combined Sample.
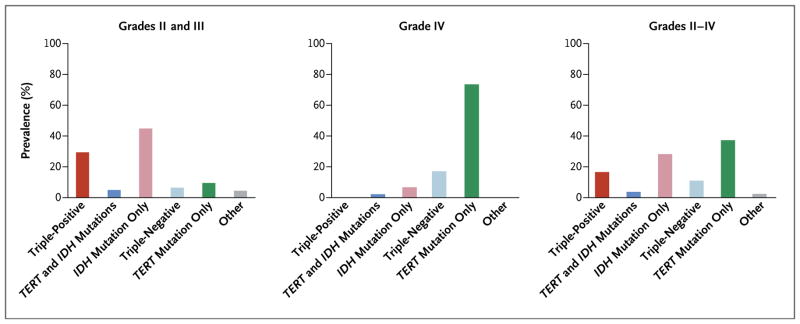
The prevalence of the molecular groups among gliomas of grade II or III (astrocytomas, mixed oligoastrocytomas, and oligodendrogliomas), grade IV (glioblastoma multiforme), and grades II through IV combined is shown.
The five molecular groups were distributed across all histologic types and grades of glioma (Fig. S3 and S4 in Supplementary Appendix). However, specific histologic types were found more frequently within certain groups (Table S2A in the Supplementary Appendix). For example, in the combined data set, all but three triple-positive gliomas were oligodendrogliomas or mixed oligoastrocytomas. The group with TERT and IDH mutations and the group with only IDH mutations contained a large proportion of grade II or III astrocytomas and mixed oligoastrocytomas. Although 85% of gliomas with only TERT mutations were grade IV gliomas, this group also contained 32 of 119 (27%) grade III astrocytomas and 7 of 74 (10%) grade II astrocytomas. Finally, 81 of 121 (67%) triple-negative gliomas were grade IV gliomas. Approximately 80% of triple-positive gliomas and gliomas with only IDH mutations occurred in the frontal lobe (see Table S2B and the Results section in the Supplementary Appendix).
Acquired Genetic Alterations
Table 1 summarizes the alterations that were consistently observed in the five glioma groups. A detailed summary of these alterations, including the prevalence of MGMT methylation, is provided in the Supplementary Appendix. Table S3 in the Supplementary Appendix provides a list of selected copy number and point mutations for which data were available in the three study cohorts. A comparison among the five molecular groups with regard to acquired mutations and TCGA glioblastoma RNA expression subtypes is provided in Figures S4 and S5 in the Supplementary Appendix.
Table 1.
Summary of Acquired and Germline Alterations in Adult Gliomas.*
| Alteration Type | Triple-Positive | TERT and IDH Mutation | IDH Mutation Only | Triple-Negative | TERT Mutation Only |
|---|---|---|---|---|---|
| Grouping alterations | IDH, 1p/19q, and TERT | TERT and IDH | IDH | — | TERT |
| Common acquired mutations | CIC, FUBP1, NOTCH1, and PIK3CA or PIK3R1 | TP53 and ATRX | TP53 and ATRX | EGFR, PTEN, and NF1 | EGFR, EGFRvIII, PTEN, NF1, RB1, and PIK3CA or PIK3R1 |
| Common acquired copy-number alterations | Loss of chromosome 4, hemizygous loss of CDKN2A/B; “genomically quiet” | Gain of chromosome 7, duplication of 8q24 (MYC), homozygous loss of CDKN2A/B, deletion of PTEN | Duplication of 7q, duplication of 8q24 (MYC), hemizygous loss of CDKN2A/B, deletion of 19q (without deletion of 1p) | Similar to TERT mutation only (at lower prevalence) | Loss of chromosome 4, gain of chromosome 7, gain of chromosome 19, amplification of EGFR, homozygous loss of CDKN2A/B, deletion of PTEN, other amplifications |
| TCGA glioblastoma expression subtype | Proneural | Mesenchymal, neural, proneural | Proneural | Classical, mesenchymal, neural, proneural | Classical, mesenchymal |
| Germline SNPs | 8q24 | 8q24, TP53 3′ untranslated region | 8q24, PHLDB1 | CDKN2A/B, RTEL1, TERT | CDKN2A/B, RTEL1, TERT, TERC |
SNP denotes single-nucleotide polymorphism, and TCGA the Cancer Genome Atlas.
Clinical Significance of Glioma Molecular Groups
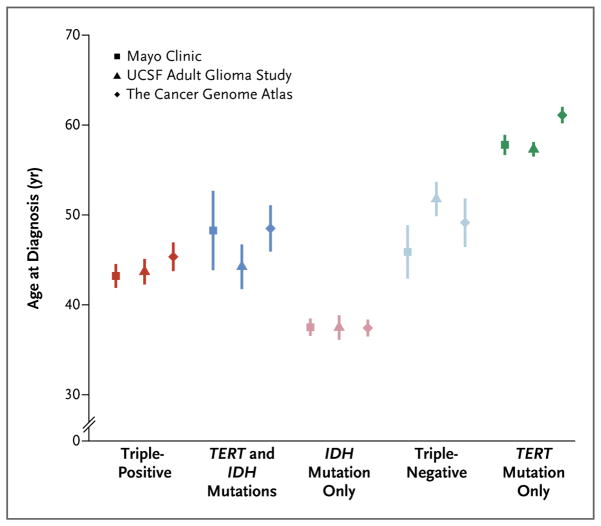
The age at diagnosis varied significantly among the molecular groups (Fig. 2, and Tables S2B and S2C in the Supplementary Appendix). The mean age at diagnosis across all three data sets was 44 years in the triple-positive group, 46 years in the group with TERT and IDH mutations, 37 years in the group with only IDH mutations, 50 years in the triple-negative group, and 59 in the group with only TERT mutations. Patients who had gliomas with only TERT mutations were significantly older than those with triple-negative gliomas (P<0.001) (Table S2C in the Supplementary Appendix), even though nearly all patients in both groups had grade IV tumors. In addition, patients who had gliomas with only IDH mutations were significantly younger than those with triple-positive gliomas, gliomas with TERT and IDH mutations, or triple-negative gliomas (P<0.001).
Figure 2. Mean Age at Diagnosis for Each Glioma Molecular Group.
Data from three glioma case series were used: the Mayo Clinic; the University of California, San Francisco (UCSF), Adult Glioma Study; and the Cancer Genome Atlas. Vertical lines represent standard deviations.
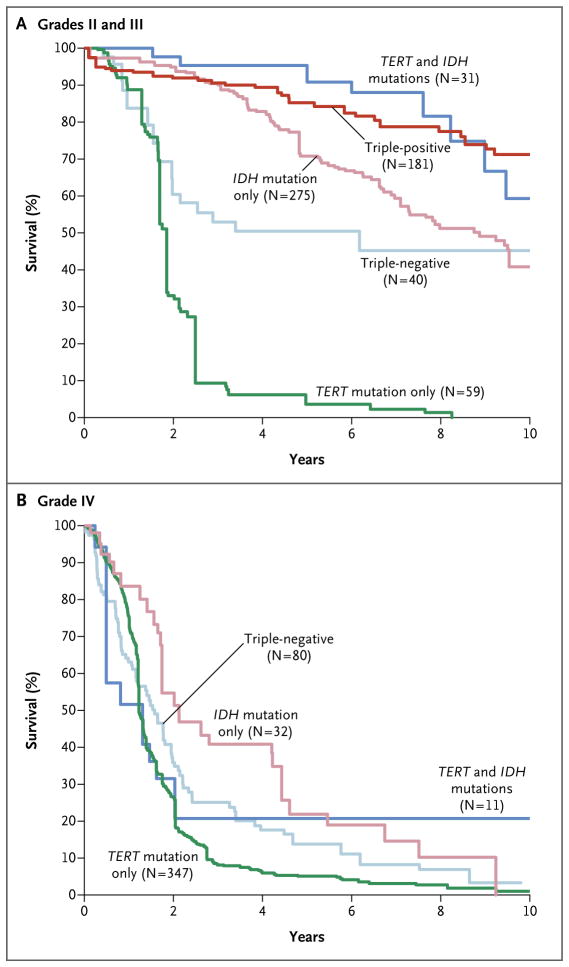
An analysis with the use of a multivariate Cox model showed a significant interaction between glioma grade and molecular group (P = 0.006). Thus, all Cox models were evaluated separately for grade II and III gliomas and for grade IV gliomas. Among patients with grade II or III gliomas, the age at diagnosis, grade, and molecular group were independently associated with overall survival (Table 2). After adjustment for the age at diagnosis and for grade, patients who had gliomas with only TERT mutations had poorer overall survival than did patients who had gliomas that were triple-negative, gliomas with TERT and IDH mutations, gliomas with only IDH mutations, or triple-positive gliomas (Fig. 3A, and Table S4 in the Supplementary Appendix). Patients with triple-negative gliomas had poorer overall survival after adjustment for age and grade than did patients who had gliomas with TERT and IDH mutations or triple-positive gliomas. Among patients with grade IV gliomas, molecular group was associated with overall survival in the univariate model but not in the multivariate model (Table 2 and Fig. 3B). Site-specific unadjusted Kaplan– Meier curves showed consistency across the data sets (Fig. S6 in the Supplementary Appendix).
Table 2.
Univariate and Multivariate Cox Proportional-Hazards Models for Grades II and III and Grade IV Gliomas.
| Variable | No. of Patients | Hazard Ratio (95% CI) | |
|---|---|---|---|
| Univariate | Multivariate | ||
| Grade II or III gliomas | |||
| Age at diagnosis* | 1.05 (1.03–1.06)† | 1.03 (1.02–1.05)† | |
| Sex | |||
| Female | 227 | Reference | Reference |
| Male | 359 | 1.14 (0.82–1.60) | 0.90 (0.63–1.28) |
| Histologic type | |||
| Oligodendroglioma | 205 | Reference | Reference |
| Oligoastrocytoma | 190 | 1.48 (0.96–2.30) | 1.21 (0.74–1.99) |
| Astrocytoma | 191 | 2.90 (1.94–4.34)† | 1.42 (0.84–2.39) |
| Grade | |||
| II | 311 | Reference | Reference |
| III | 275 | 2.02 (1.45–2.82)† | 1.49 (1.03–2.15)† |
| Molecular group | |||
| Triple-positive | 181 | Reference | Reference |
| TERT and IDH mutations | 31 | 1.64 (0.73–3.69) | 1.31 (0.55–3.12) |
| IDH mutation only | 275 | 2.12 (1.33–3.37)† | 2.08 (1.22–3.57)† |
| Triple-negative | 40 | 4.05 (2.13–7.71)† | 3.74 (1.91–7.36)† |
| TERT mutation only | 59 | 21.92 (12.75–37.70)† | 11.74 (6.15–22.41)† |
| Grade IV gliomas | |||
| Age at diagnosis* | 1.03 (1.02–1.04)† | 1.03 (1.02–1.04)† | |
| Sex | |||
| Female | 169 | Reference | Reference |
| Male | 301 | 1.0 (0.81–1.23) | 1.03 (0.83–1.27) |
| Molecular group | |||
| IDH mutation only | 32 | Reference | Reference |
| TERT and IDH mutations | 11 | 0.97 (0.42–2.26) | 0.81 (0.35–1.88) |
| Triple-negative | 80 | 1.70 (1.07–2.70)† | 1.29 (0.80–2.07) |
| TERT mutation only | 347 | 2.12 (1.40–3.21)† | 1.28 (0.82–2.00) |
The hazard ratio is for each 1-yr increase in age.
The hazard ratio was significant (P<0.05).
Figure 3. Adjusted Kaplan–Meier Estimates of Overall Survival in the Glioma Molecular Groups.
Overall survival estimates were adjusted for sex and age at diagnosis (on the basis of the 2010 U.S. white population) with the use of the reweighted (direct adjustment) method. Because there was only one triple-positive case among patients with grade IV gliomas, this group was not included in Panel B.
Association between Glioma Molecular Groups and Germline Variants
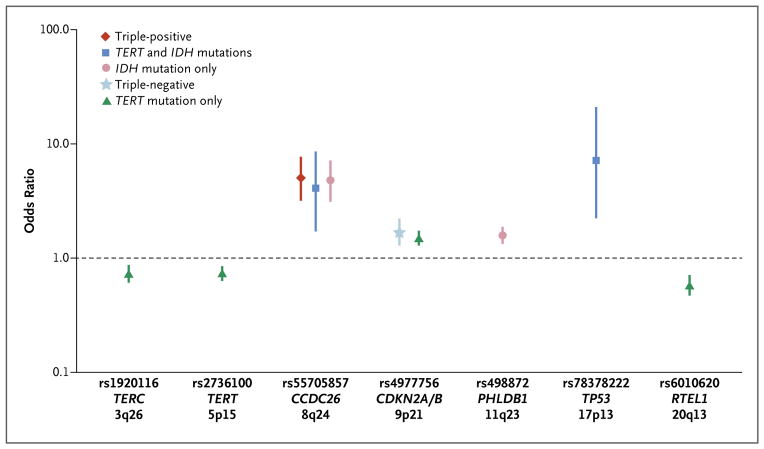
We evaluated the associations between the five glioma groups and nine regions that had previously been shown in genomewide association studies to be associated with glioma risk: TERC (3q26), TERT (5p15), EGFR (7p12, containing two independent regions), CCDC26 (8q24), CDKN2A or CDKN2B (hereafter referred to as CDKN2A/B) (9p21), PHLDB1 (11q23), TP53 (17p13), and RTEL1 (20q13) (Fig. 4, and Fig. S7 and Table S5 in the Supplementary Appendix).10–14 Most notably, the CCDC26 SNP (rs55705857) was associated with an increased risk of the development of any glioma with IDH mutations. The PHLDB1 SNP (rs498872) was associated with gliomas with only IDH mutations. The TERC (rs1920116), TERT (rs2736100), and RTEL1 (rs6010620) SNPs were protective against gliomas with only TERT mutations. The CDKN2A/B SNP (rs4977756) was associated with an increased risk of both triple-negative gliomas and gliomas with only TERT mutations. The TP53 SNP (rs78378222) was associated with an increased risk of gliomas that have TERT and IDH mutations. The CCDC26 and TERT SNPs were also associated with an increased risk of proneural gliomas (Table S6 and the Results section in the Supplementary Appendix).
Figure 4. Associations between Glioma Molecular Groups and Glioma-Related SNPs.
Shown are the odds ratios for the five molecular groups of glioma for each of the single-nucleotide polymorphisms (SNPs) that have been found in case–control studies to be associated with glioma. Only SNPs with a P value of 0.0011 or lower (Bonferroni-corrected P value for testing nine regions in each of the five molecular groups) are shown. Vertical lines indicate the 95% confidence intervals associated with the odds ratios. For this figure, the cases and controls from Mayo Clinic, UCSF Adult Glioma Study, the Cancer Genome Atlas, and the Mayo Genome Consortia were combined. See Figure S7 in Supplementary Appendix for an illustration of results for the nine regions for all five glioma groups. See Table S6 in Supplementary Appendix for a complete summary of the data for all 22 SNPs evaluated.
Discussion
Despite extensive efforts to characterize the molecular biologic characteristics of glioma,1,2,16,17,19,20 in practice there remains no simple way to reliably categorize patients with glioma in clinically and etiologically similar groups. We defined a priori groups that were based on the presence or absence of TERT promoter mutations, IDH mutations, and 1p/19q codeletion. With the resulting classification of patients in one of five groups, we extend previous observations that have been based on histologic type, grade, IDH mutation, and 1p/19q codeletion.3,15 We found consistent associations between the molecular groups and age at diagnosis, survival, patterns of acquired alterations, and germline variants across the three data sets. This consistency is remarkable, given the different methods of case ascertainment that were used in the three different series that we analyzed (cases in the UCSF Adult Glioma Study were population-based and clinic-based, cases from the Mayo Clinic were referral-based, and cases from the TCGA project were selected primarily on the basis of tumor size). The Cancer Genome Atlas Research Network3 independently identified similar groups, using unsupervised clustering analyses of DNA mutation, RNA expression, DNA copy number, and DNA methylation data.
Triple-positive gliomas are most strongly associated with the oligodendroglial histologic type, the minor allele of the SNP rs55705857 (8q24), and better overall survival. If it is assumed that nearly all gliomas with 1p/19q codeletion have mutations in TERT and IDH, recent data from the Radiation Therapy Oncology Group suggest that such tumors will have greater benefit from adjuvant chemotherapy and radiation.15 These tumors are “genomically quiet” relative to those in the other groups (i.e., they have few additional acquired alterations other than the 1p/19q codeletion, TERT promoter mutation, and IDH mutation). Approximately half of these tumors do not have CIC and FUBP1 mutations, but whether CIC and FUBP1 are altered by other mechanisms or whether different tumor suppressor genes on 1p and 19q acquire mutations remains to be determined. This group and the group with only TERT mutations have a high prevalence of loss of chromosome 4 and acquired PIK3CA or PIK3R1 mutations; however, the age at diagnosis, overall survival, and germline variant associations differ between these two groups. Experimental models of triple- positive gliomas should enhance our understanding of the dynamic relationships among germline variants, chromatin structure, IDH mutation, telomere maintenance, and chemosensitivity.
Among adults with gliomas, those with tumors that have only IDH mutations have the earliest mean age at onset (37 years) and an intermediate prognosis; they also account for half of all patients who have gliomas with an oligodendroglial component and two thirds of those who have grade II or III astrocytomas. These tumors almost always acquire mutations in TP53 and ATRX. On the basis of their uniform mutational profile, gliomas with only IDH mutations become an important group for experimental modeling and novel therapeutic discovery.
Gliomas with only TERT mutations are primarily grade IV gliomas; however, 39 of 406 (9.6%) in our study were grade II or III. Grade II or III gliomas in this group have an aggressive course and are associated with poor survival, which suggests the need for early adjuvant therapies and meticulous follow-up. Additional prognostic information may be gained by stratifying these gliomas according to epidermal growth factor receptor (EGFR) status.21 The relationship between germline variants in telomere components (TERC, TERT, and RTEL1) and the acquisition of TERT promoter mutations is noteworthy and deserves further study.22
Gliomas that harbor mutations in both TERT and IDH without an accompanying 1p/19q codeletion are relatively rare, accounting for 4% of all gliomas in the combined data set. The germline TP53 SNP rs78378222 is associated with gliomas with TERT and IDH mutations but not with the other four groups. Some tumors also acquire ATRX alterations and thus show that ATRX mutation and TERT mutation are not always mutually exclusive. Patients who have grade IV tumors with mutations in TERT and IDH have poor survival, similar to that in patients who have gliomas with TERT mutations only. Thus, the presence of an IDH mutation does not always indicate a favorable prognosis. When these tumors manifest as grade II or III, they are associated with an overall survival similar to that associated with triple-positive gliomas. Because triple-positive gliomas are defined by whole-arm 1p/19q codeletion mediated by a 1;19 translocation,4 it is possible that some gliomas with TERT and IDH mutations acquire functionally similar alterations that simulate deletion of 1p and 19q (or alterations within genes that reside on these chromosomes).
Triple-negative gliomas constitute 7% of grade II or III gliomas and 17% of grade IV gliomas. Although they typically develop in younger patients (approximately 10 years younger) than do gliomas with TERT mutations only, these two types of gliomas have a similar level of MGMT promoter methylation and similar associations with germline variants.
TERT promoter and ATRX gene alterations are common in all gliomas. Alterations of both genes are associated with enhanced telomere maintenance.9,23 Our data suggest that TERT promoter alterations and ATRX alterations are usually mutually exclusive, but some gliomas appear to have both, and some appear to have neither. In our study, triple-negative gliomas typically did not acquire ATRX alterations. Of the gliomas that could not be classified within our scheme, most had acquired the 1p/19q codeletion and IDH mutations (without TERT or ATRX mutations). If enhancement of telomere maintenance is required for glioma development, then it is reasonable to hypothesize that gliomas without alterations in TERT or ATRX acquire alterations that we did not capture in our analysis or alterations in other components of the enzymatic machinery that maintains telomeres, such as POT1.24 Some germline variants (such as those in TERT or TERC) may also be associated with critical telomere lengthening.22 In terms of survival, mutation in the TERT promoter is generally unfavorable in the absence of IDH mutation and favorable in the presence of IDH mutation and 1p/19q codeletion. TERT promoter mutation is associated with an older age of the patient at presentation, regardless of whether IDH mutation is present.
Specific germline variants segregate with gliomas that have IDH mutations and 1p/19q codeletion.13,14,25–27 SNPs at chromosome locus 8q24, especially rs55705857, were highly associated with IDH mutation, which suggests that this region contains a germline alteration that facilitates the initiation or progression of gliomas with IDH mutations. Risk alleles of 8q24 SNPs are associated with gliomas with duplication of chromosome 8q24 (data not shown) that include MYC and specific SNPs (such as rs55705857) in this region. Perhaps this region contains several “driver” DNA elements that facilitate glioma development, similar to the way that duplication of chromosome 8q24 facilitate the development of some breast cancers.28 A possible mechanism of action may involve long noncoding RNAs encoded within the large 8q24 genomic region near MYC.29 Another possibility is that the SNP rs55705857 encodes a DNA element that confers a predisposition to IDH mutation; alternatively, in the presence of IDH mutation, the encoded element may activate specific transcriptional regulatory networks (e.g., those involving MYC).
A multivariate Cox-model analysis (Table 2) suggested that survival among patients with grade II or III glioma, stratified according to molecular group, was independent of age at diagnosis, histologic type, and grade. This did not appear to be true for patients with grade IV gliomas: age at diagnosis and molecular group were each associated with outcome in patients with grade IV gliomas, and these variables were not independent of one another. Although the data on overall survival were robust with respect to sample size and showed consistency among data sets, they must be considered preliminary. Confounders such as treatment and MGMT methylation status may have affected survival. Furthermore, the biologic characteristics of the molecular groups may reflect additional prognostic variables. For example, these groups may be associated with the intracerebral location and infiltrative characteristics of the tumor,17 which may influence resectability, radiation planning, and sequelae, and therefore, outcome. Further study of outcomes associated with the groups, stratified according to key clinical variables, is warranted.
Historically, histologic classification and grading of infiltrating gliomas have been used to determine prognosis and therapeutic interventions. However, histologic type and grade have poor reproducibility among pathologists, especially for tumors with oligodendroglial components. We show that classic observations relating glioma prognosis to age at diagnosis, histologic type, and grade can be captured within a conceptually simple framework that incorporates three molecular tests. Two tests (for IDH mutation and 1p/19q codeletion) are currently being performed in clinical laboratories that are certified according to the Clinical Laboratory Improvement Amendments and are components of the College of American Pathologists Proficiency Surveys.30 The third test (for TERT promoter alterations) is also relatively simple and could be incorporated into molecular pathology laboratories. These tests will have sensitivity and specificity that vary according to the laboratory that performs them, but misclassification should be minimal.
In summary, the presence or absence of TERT promoter mutations, IDH mutations, and 1p/19q codeletion can be used to define five principal groups of gliomas with characteristic distributions of age at diagnosis, clinical behavior, acquired genetic alterations, and associated germline variants. This framework could be further refined through the incorporation of alterations in ATRX, TP53, EGFR, or PTEN or other alterations that might be useful to consider in the diagnosis of glioma.3,31
Supplementary Material
Acknowledgments
Work at the Mayo Clinic was supported by National Institutes of Health (NIH; P50CA108961, P30 CA15083, and RC1NS068222Z), the Bernie and Edith Waterman Foundation, and the Ting Tsung and Wei Fong Chao Family Foundation. Work at the University of California, San Francisco (UCSF), was supported by the NIH (R01CA52689, P50CA097257, R25CA112355, R01CA139020, R01CA126831, R01CA163687, and UL1 RR024131) and by the National Center for Research Resources, National Center for Advancing Translational Sciences, National Brain Tumor Foundation, Stanley D. Lewis and Virginia S. Lewis Endowed Chair in Brain Tumor Research, and UCSF Robert Magnin Newman Endowed Chair in Neuro-oncology.
We thank the study participants; clinicians and research staff at participating medical centers; Katherine Cornelius; staff at the Mayo Clinic Comprehensive Cancer Center Biospecimens and Processing and Genotyping Shared Resources; and staff at the UCSF Diller Cancer Center Genomics Core. The results published here are based in part on data generated by the Cancer Genome Atlas, managed by the National Cancer Institute and the National Human Genome Research Institute (http://cancergenome.nih.gov). Full acknowledgments are provided in the Supplementary Appendix.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 2.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Cancer Genome Atlas Research Network. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–98. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenkins RB, Blair H, Ballman KV, et al. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66:9852–61. doi: 10.1158/0008-5472.CAN-06-1796. [DOI] [PubMed] [Google Scholar]
- 5.Cairncross G, Wang M, Shaw E, et al. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol. 2013;31:337–43. doi: 10.1200/JCO.2012.43.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van den Bent MJ, Brandes AA, Taphoorn MJ, et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol. 2013;31:344–50. doi: 10.1200/JCO.2012.43.2229. [DOI] [PubMed] [Google Scholar]
- 7.Guo C, Pirozzi CJ, Lopez GY, Yan H. Isocitrate dehydrogenase mutations in gliomas: mechanisms, biomarkers and therapeutic target. Curr Opin Neurol. 2011;24:648–52. doi: 10.1097/WCO.0b013e32834cd415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Killela PJ, Reitman ZJ, Jiao Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–6. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vinagre J, Pinto V, Celestino R, et al. Telomerase promoter mutations in cancer: an emerging molecular biomarker? Virchows Arch. 2014;465:119–33. doi: 10.1007/s00428-014-1608-4. [DOI] [PubMed] [Google Scholar]
- 10.Wrensch M, Jenkins RB, Chang JS, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41:905–8. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shete S, Hosking FJ, Robertson LB, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stacey SN, Sulem P, Jonasdottir A, et al. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet. 2011;43:1098–103. doi: 10.1038/ng.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenkins RB, Wrensch MR, Johnson D, et al. Distinct germ line polymorphisms underlie glioma morphologic heterogeneity. Cancer Genet. 2011;204:13–8. doi: 10.1016/j.cancergencyto.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenkins RB, Xiao Y, Sicotte H, et al. A low-frequency variant at 8q24.21 is strongly associated with risk of oligodendroglial tumors and astrocytomas with IDH1 or IDH2 mutation. Nat Genet. 2012;44:1122–5. doi: 10.1038/ng.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cairncross JG, Wang M, Jenkins RB, et al. Benefit from procarbazine, lomustine, and vincristine in oligodendroglial tumors is associated with mutation of IDH. J Clin Oncol. 2014;32:783–90. doi: 10.1200/JCO.2013.49.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan X, Vengoechea J, Zheng S, et al. Molecular subtypes of glioblastoma are relevant to lower grade glioma. PLoS One. 2014;9(3):e91216. doi: 10.1371/journal.pone.0091216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorovets D, Kannan K, Shen R, et al. IDH mutation and neuroglial developmental features define clinically distinct subclasses of lower grade diffuse astrocytic glioma. Clin Cancer Res. 2012;18:2490–501. doi: 10.1158/1078-0432.CCR-11-2977. [DOI] [PubMed] [Google Scholar]
- 18.Bielinski SJ, Chai HS, Pathak J, et al. Mayo Genome Consortia: a genotype-phenotype resource for genome-wide association studies with an application to the analysis of circulating bilirubin levels. Mayo Clin Proc. 2011;86:606–14. doi: 10.4065/mcp.2011.0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–77. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–22. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labussière M, Boisselier B, Mokhtari K, et al. Combined analysis of TERT, EGFR, and IDH status defines distinct prognostic glioblastoma classes. Neurology. 2014;83:1200–6. doi: 10.1212/WNL.0000000000000814. [DOI] [PubMed] [Google Scholar]
- 22.Walsh KM, Codd V, Smirnov IV, et al. Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat Genet. 2014;46:731–5. doi: 10.1038/ng.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huse JT. Elucidating the oncogenic role of ATRX deficiency in glioma. Neuro Oncol. 2014;16(Suppl 3):iii45. [Google Scholar]
- 24.Bainbridge MN, Armstrong GN, Gramatges MM, et al. Germline mutations in shelterin complex genes are associated with familial glioma. J Natl Cancer Inst. 2014;107:384. doi: 10.1093/jnci/dju384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enciso-Mora V, Hosking FJ, Kinnersley B, et al. Deciphering the 8q24.21 association for glioma. Hum Mol Genet. 2013;22:2293–302. doi: 10.1093/hmg/ddt063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Stefano AL, Enciso-Mora V, Marie Y, et al. Association between glioma susceptibility loci and tumour pathology defines specific molecular etiologies. Neuro Oncol. 2013;15:542–7. doi: 10.1093/neuonc/nos284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice T, Zheng S, Decker PA, et al. Inherited variant on chromosome 11q23 increases susceptibility to IDH-mutated but not IDH-normal gliomas regardless of grade or histology. Neuro Oncol. 2013;15:535–41. doi: 10.1093/neuonc/nos324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tseng YY, Moriarity BS, Gong W, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82–6. doi: 10.1038/nature13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim T, Cui R, Jeon YJ, et al. Long-range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARLo-5. Proc Natl Acad Sci U S A. 2014;111:4173–8. doi: 10.1073/pnas.1400350111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schrijver I. 2013 Surveys and anatomic pathology education programs. Northfield, IL: American College of Pathologists; 2013. Genetics and molecular pathology; pp. 203–9. [Google Scholar]
- 31.Louis DN, Perry A, Burger P, et al. International Society Of Neuropathology — Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol. 2014;24:429–35. doi: 10.1111/bpa.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.