Abstract
The heart is one of the most vital organs in the human body, which actively pumps the blood through the vascular network to supply nutrients to as well as to extract wastes from all other organs, maintaining the homeostasis of the biological system. Over the past few decades, tremendous efforts have been exerted in engineering functional cardiac tissues for heart regeneration via biomimetic approaches. More recently, progresses have been achieved towards the transformation of knowledge obtained from cardiac tissue engineering to building physiologically relevant microfluidic human heart models (i.e. heart-on-chips) for applications in drug discovery. The advancement in the stem cell technologies further provides the opportunity to create personalized in vitro models from cells derived from patients. Here starting from the heart biology, we review recent advances in engineering cardiac tissues and heart-on-a-chip platforms for their use in heart regeneration and cardiotoxic/cardiotherapeutic drug screening, and then briefly conclude with characterization techniques and personalization potential of the cardiac models.
1 Introduction
Being one of the most vital organs in the human body, the heart functions as a potent biological pump that actively delivers/recycles the blood towards/from all other organs through the vascular system. As a result, the capability to regenerate an injured or diseased heart has always been a focus and popular subject of research in tissue engineering and regenerative medicine. Comparing to other tissues, it is highly challenging to engineer functional cardiac substitutes due to the fact that mature cardiomyocytes exhibit limited proliferation potential, thus preventing spontaneous recovery of the damaged cardiac tissues [1]. With many years of endeavors, the field of cardiac tissue engineering has seen tremendous progress in fabricating functional cardiac tissues that largely recapitulate the biology of the heart [2–5], but challenges remain. For example, the alignment of the cardiomyocytes in native heart complicates the parameters required for engineering cardiac tissues, where factors that can induce and promote the alignment/bundling of the cardiomyocytes need to be incorporated into the design of the artificial substrates [6, 7]. Beating of the cardiomyocytes poses another obstacle. While cardiomyocytes beat synchronously in the heart, such capacity is easily lost during in vitro manipulation due to the relatively harsh environment and mismatching matrix properties that cardiomyocytes experience when they are isolated, processed, and combined with the matrices. Methods based on electric stimulation and inclusion of electroconductive materials that improve the spontaneous and synchronous beating of engineered heart tissues also require optimization [8, 9].
On the other hand, drug-induced cardiotoxicity has become a great concern in the drug development process and clinically [10, 11]. In the past four decades approximately 20% of drug recalls arose from cardiotoxicity, such as Fenphen, Micturin, and Seldane [12]. Recently, drug attrition rate in the drug development process has further increased with a substantial portion contributed by drug-induced cardiac toxicity. For instance, cardiac safety issues have accounted for half of the almost fifty drugs that have been retracted from the market since 1990s [13, 14]. This is mainly due to a lack of effective in vitro assays that accurately predict toxicity in early stages of drug development. Current paradigms for testing drug efficacy and toxicity are typically time-consuming, expensive, and yet often ineffective. Conventional two-dimensional (2D) static cultures of cardiomyocytes and animal models are the long-standing tools on which the field relies to study cardiotoxicity and cardiotherapeutic drug effects [15, 16]. While these over-simplified 2D models do not necessarily recapitulate their in vivo counterparts to provide accurate predictions, major disadvantages of animal models include their overall ineffectiveness to predict human response and associated ethical concerns arose in the past decade [17]. Fortunately, experiences accumulated from cardiac tissue engineering have already enabled researchers to build biologically relevant miniature human hearts in vitro [18, 19]. The combination with advanced microfluidic technologies has further expedited such a process by pushing forward the development of the heart-on-a-chip platforms, which mimic the biology and the physiology of their cardiac counterparts in vivo [20–22]. These realistic human heart models integrated with the microfluidic vasculature can be used to probe the systemic effects of drugs on the cardiac tissues and therefore better predict drug responses in the human body.
In this Review, we will start by describing human heart biology and physiology including the basic cellular/matrix components, structures, and architecture. Then we will discuss methods to engineer functional cardiac tissues via biomimicry, followed by recent advances in the construction of microfluidic human heart-on-a-chip systems and associated characterization techniques. Finally, we will briefly conclude with personalization approaches using the stem cells technologies.
2 Breaking down the heart
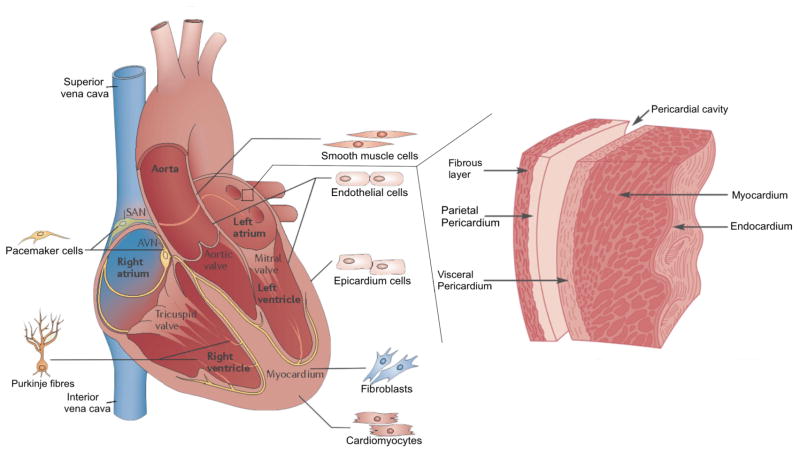
Human cardiogenesis occurs in the lateral mesodermal layer during the early stages of embryonic development [23]. Endodermic modulation of adjacent tissue, to the later mesoderm, guides proper cardiovascular development [24, 25]. During such development of the four main chambers of the heart (two atria and two ventricles) and their associated features (valves and heart wall), specific cell populations are organized in the different parts of the heart (Fig. 1). The myocardium, primarily composed of cardiomyocytes, works as the main motor of the entire cardiovascular engine [26]. These cells arrange themselves in a parallel manner bonded by gap junctions and Fascia adherens unions, forming the myocardial fibers, which are responsible for the contraction. Aggregated myocardial fibers share a distinguished direction and particular thickness, which significantly enhance the contractile force exerted to pump the blood through the vascular system [27].
Figure 1.
Schematics showing mature heart cell populations with geographical anatomy, and general histological representation of the heart wall. Adapted with permission from Ashley et al., 2004 and Xin et al., 2013 [29, 41].
During the development process, atrial and ventricular cardiomyocytes first form, followed by the cells of the mammalian cardiac conduction system [26], including pacemakers cells and Purkinje fibers, which have been phenotypically characterized as specialized cardiomyocytes [28]. These cells generate and conduct the electrical impulse from the left and right bundle branch to the myocardium of the ventricles. Notably, 50–60% of the entire cell population of the heart is comprised of cardiac fibroblasts that lay down the extracellular matrix (ECM) of the heart wall, with the rest of the population largely being cardiomyocytes that are responsible for the spontaneous contraction of the heart [26, 29].
While cardiomyocytes and cardiac fibroblasts occupy the majority of the cells in the heart, other types do exist and are located throughout the heart [30]. For example, the cell population on the endocardium as well as those lining the blood vessels and cardiac valves is mainly composed of endothelial cells. The pericardium parietal wall is rich in epicardium cells, which are also located in the coronary vasculature. Neural crest cells contribute to parts of the outflow tract and the septum, enhancing tissue oxygenation [26, 29]. Immune response in the heart is balanced by macrophages, although they also play a role in the control of cell population and maintenance of the ECM [31].
Although cell-cell interactions form the basis of the heart, autocrine and paracrine interactions have a major impact on the tissue responses through the secretion of a combination of different factors that modulate the molecular composition of the tissue microenvironment [32]. These factors include cytokines, growth factors, and ECM molecules that act either on the cells by which they are secreted or neighboring cells [32]. For example, bone morphogenetic protein (BMP), myocyte enhancer factor-2 (MEF2), and WNT-mediated signaling together control specialization of cardiomyocytes [26, 30, 33]. Such an organized orchestra of cells and tissue factors regulates, stabilizes, and reinforces the cardiac phenotype. It is the concerted interactions between cells, ECM, and signaling molecules all together that contribute to the development and maintenance of the functions of the heart.
3 Engineering myocardium via biomimetic approaches
Tissue engineering seeks to regenerate damaged or diseased tissues/organs via the development and integration of biological substitutes [42]. While majority of the approaches rely on the use of artificial matrices, tissues have also been successfully fabricated without involvement of scaffolds [43].
With respect to cardiac tissue engineering, two main scaffold-free concepts have been adopted. The first concept focuses on the generation of sheets of confluent cells which may then be either directly used or further stacked together into multi-layer structures, the so called cell-sheet engineering [44, 45] (Fig. 2A). A cell sheet is obtained by first culturing cells on a substrate coated with temperature-sensitive polymers followed by the release of the confluent monolayer of cells at a lower temperature under which swollen molecules reduce the adhesion between the cells and the substrate [46]. Using such an approach, thick myocardium-like tissues have been assembled. Interestingly, vascularization through the myocardium layers was realized by sandwiching endothelial cell sheets in between two cardiomyocyte sheets [47]. The second concept of scaffold-free cardiac tissue engineering relies on the generation of multicellular aggregates termed spheroids. Fig. 2B illustrates how uniform embryoid bodies (EBs, i.e. spheroids composed of embryonic stem cells) have been obtained by seeding the cells onto substrates containing microwells made of cell-repellent materials such as poly(ethylene glycol) (PEG) and polydimethylsiloxane (PDMS) [48]. These EBs, whose sizes depend on both the seeding density and the dimension of the wells, are then able to differentiate into multiple cell types including cardiomyocytes, endothelial cells, osteoblasts, and neurons [49–51]. A following study further showed that cardiogenesis was promoted for EBs with a diameter of 450 μm compared with other sizes, as identified by the higher expression of sarcomeric α-actinin and cardiomyogenic genes [52] (Fig. 2B). The use of EBs is particularly important in mimicking cardiogenesis in vitro at early phases of embryonic development. Although the scaffold-free approaches have demonstrated their success to certain degrees, they barely provide directionality the native myocardium possesses, which is required for engineering cardiac tissue surrogates to recapitulate their in vivo functionality.
Figure 2.
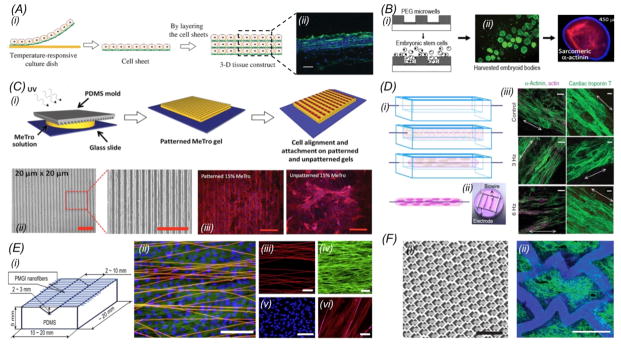
(A) (i) Fabrication of multi-layered cardiac cell sheets; (ii) Fluorescence image on the right shows Troponin T staining of the cardiac muscle in a construct containing 6 layers of cardiac cell sheets. Green: Troponin T; blue, nuclei. Scale bar: 20 μm. Adapted with permission from Masuda et al., 2008 and Sakaguchi et al., 2013 [44, 93]; (B) Fabrication of PEG microwells and formation of EBs of uniform sizes. Cardiogenesis was maximized in EBs of 450 μm in diameter. Adapted with permission from Karp et al., 2007 and Hwang et al., 2009 [48, 52]; (C) (i) Schematic diagram showing the fabrication of micropatterned hydrogels using a micromolding technique for cardiomyocytes alignment. (ii) Bright-field images of micropatterned MeTro with 20 × 20 μm (width × spacing) channels; (iii) Fluorescence images of aligned cardiomyocytes cultured on the micropatterned MeTro; scale bars: 200 μm. Adapted with permission from Annabi et al., 2013 [73]; (D) (i) The biowire approach where a surgical suture was used to induce compaction and alignment of cardiomyocytes in the surrounding hydrogel. (ii, iii) Cultured biowires under electrical stimulation improved the phenotype of cardiomyocytes. Adapted with permission from Nunes et al., 2013 [74]; (E) (i) Preparation of scaffolds with suspended electrospun nanofibers. (ii) A superimposed confocal image of (iii–v) showing cardiomycytes on aligned nanofibers where the nanofibers were stained red, f-actin green, and nuclei blue; (vi) image showing parallel alignment of sarcomeres, where nanofibers were stained bright pink and α-actin stained red. Scale bars: (ii) 100 μm, (iii) 50 μm, (iv) 50 μm, (v) 100 μm, and (vi) 10 μm. Adapted with permission from Orlova et al., 2011 [75]; (F) (i) Scaffolds with an accordion-like honeycomb structure resulted in anisotropic mechanical properties possessed by the native myocardium. (ii) Fluorescence image showing alignment of cardiomyocytes cultured on an accordion-like honeycomb scaffold. Green indicates F-actin. Scale bars: (i) 1 mm, and (ii) 200 μm. Adapted with permission from Engelmayr et al., 2008[76].
Scaffold-based approaches involve the creation of artificial matrices on which cardiomyocytes are grown. Polymers including those from both natural and synthetic sources are used to fabricate the scaffolds. Commonly used natural polymers include elastin, collagen, gelatin, fibrin, hyaluronic acid and alginate [53, 54]. One of the greatest advantages of these natural materials is that they typically possess specific ligands (e.g. RGD) that allow for and promote cell adhesion [55]. However, the poorly defined chemical composition and batch-to-batch variability have limited their applications in certain cases. In addition, their mechanical properties are not always sufficient to support many tissue types [56]. Some attempts have been made in order to overcome such mechanical deficiency. For example, static compression of collagen gels has been adopted to increase the biomaterial densification and improve the mechanical properties [57–60]. Acellular patches developed from compressed collagen have been grafted on infarcted myocardium, demonstrating to significantly improve the cardiac function [60]. Another approach to enhance the mechanical properties of natural polymers is to generate hybrid systems. For this purpose, Brigham et al. have fabricated semi-interpenetrating networks of photocrosslinkable hyaluronic acid (MeHA) and collagen to achieve mechanical properties that far surpass those obtainable with collagen or MeHA alone [61]. Chemical modifications can also be considered in order to tune the mechanical strength of natural polymers. As an example, the methacrylamide modification degree of gelatin methacrylate (GelMA) can be modified to obtain scaffolds with desired mechanical properties that are suitable for various tissue engineering applications [62].
As an alternative to natural polymers, many synthetic materials have been designed and developed, such as the biodegradable polycaprolactone (PCL), poly(lactic acid) (PLA), poly(glycolic acid) (PGA), and PLGA, the co-polymer of PLA and PGA [63–66]. Unlike natural polymers, the properties of these synthetic materials can be easily tuned for different applications. For example, the degradation rate of PLGA, as determined by the ratio of the lactic acid to glycolic acid segments, the molecular weight and crystallinity, is anywhere between 1 month to years [67–70]. This tunable degradability allows the use of the scaffolds to initially populate the cells, but gradually degrade as the tissues develop, eventually leading to the generation of fully integrated functional tissues [71, 72].
With these natural and synthetic materials, researchers have devised a number of methods to induce and enhance the alignment of the engineered myocardium. Weiss et al. combined methacrylate-modified tropoelastin (MeTro) hydrogels with microfabrication techniques to produce substrates with aligned ridges/grooves via photopatterning [73]. After seeding, neonatal rat cardiomyocytes attached and aligned along the long axis of the micropatterns, evoking a cellular organization similar to the native myocardium (Fig. 2C). Alternatively, Radisic and co-workers recently proposed an innovative biowire concept, where a surgical suture functioning as the directional guidance was surrounded by a PDMS channel filled with collagen type I encapsulating cardiomyocytes derived from human induced pluripotent stem cells (iPSCs). The biowire structure was able to enhance cell alignment along the direction of the suture, whereas electrical stimulation further induced the maturation of the myocardium (Fig. 2D) [74]. Similarly, the Orlova Group fabricated electrospun nanofiber scaffolds with an aligned topography for culture of cardiomyocytes. An enhancement in the alignment of the cardiomyocytes was observed when the distance between adjacent nanofibers was reduced (i.e. increased fiber density), showing nearly parallel distribution of α-actin filaments in the cells and a faster propagation of the electrical stimulation wave along the direction of the fibers compared to the perpendicular direction (Fig. 2E) [75]. Moreover, the architecture of a porous scaffold can guide volumetric cell distribution in three dimensions (3D) into parallel-aligned blocks in order to mimic the bundled structure of cardiac tissues. Freed et al. used poly(glycerol sebacate) (PGS) scaffolds assuming an accordion-like honeycomb microstructure for engineering cardiac tissues based on the observation that the myocardium is characterized by preferentially oriented muscle fibers surrounded by a honeycomb-like network of collagen (Fig. 2F). The alignment of cardiomyocytes in the honeycomb scaffolds was significantly promoted as indicated by the orientation index approximately 37% higher than that of the control scaffolds with an isotropic structure. The engineered myocardium also exhibited mechanical properties comparable to the ones of adult rat right ventricular myocardium [76].
Additionally, surface coatings have been employed in engineering cardiac tissues by providing a more in vivo like microenvironment. For instance, PGA scaffolds coated with laminin were found to increase cell size and induce a change in the electrical properties of a membrane populated with cardiomyocytes [77]. Other studies demonstrated that laminin improves the adhesion and alignment of cardiomyocytes [78], whereas Grosberg and colleagues developed brick wall pattern of fibronectin to promote the generation of anisotropically oriented cardiac muscle stripes [20].
Although promising results have been obtained by combining cardiomyocytes with polymeric scaffolds, current strategies relying on engineered scaffolds may still not be able to fully recapitulate the physiological cell-matrix interactions [79]. Accordingly, decellularized xenogenic or allogenic ECM-based scaffolds have been devised to better mimic the natural microenvironment and provide accurate biophysical stimuli and signaling molecules [80]. The process is based on the removal of cellular content from the tissue or organ through extensive treatment with a surfactant or enzyme such as sodium dodecyl sulfate (SDS), Triton X-100, or trypsin [81–83], followed by repopulation with autogenic cells. The resulting construct is potentially non-immunogenic, yet the chemical and enzymatic degradation process during decellularization must ensure completely elimination of the cellular material [84], without disintegrating the ECM composition and integrity [83]. Considering the complexity of the cardiac tissue, a decellularized heart would be beneficial in preserving the integrity of its 3D anatomical structure and architecture [82]. Ott et al. reseeded decellularized rat hearts with rat cardiomyocytes and endothelial cells, enabling the recovery of the cardiac function and pumping capacity [81]. More recently, decellularized mouse hearts have been repopulated with human iPSCs-derived multipotential cardiovascular progenitor cells, which exhibited spontaneous migration, proliferation, and differentiation into cardiomyocytes, smooth muscle cells, and endothelial cells [85]. Successful as these examples were, controlling precise cell positioning during the repopulation process that reproduces the heterogeneity of the native heart tissues and the attainment of an adequate mechanical force to pump blood are still challenges that remain to be addressed [79, 85, 86].
Interestingly, recent technological advancement on 3D bioprinting has opened up an entirely new avenue to construct cardiac tissues due to the need to mimic their physiological counterparts with specific patterns in cell alignment, ECM distribution, and vasculature. This technique is based on a combination of rapid prototyping and the use of bioinks to directly print cardiac tissues with well-defined architectures, cell types, biomolecules, and growth factors [87]. Although hydrogels are the most widely used materials for bioprinting [88, 89], some researchers have recently exploited the possibility of using decellularized ECM as an ideal bioink to reproduce the natural microenvironment that cells experience in their native tissue [86, 90]. For example, the Cho Lab developed a decellularized ECM bioink that is liquid at temperatures below 15 °C to enable easy printing while gelation occurs at above 37 °C. Structures fabricated using the decellularized ECM bioink induced better differentiation of the embedded stem cells towards cardiac lineages, when compared with pure collagen and alginate bioinks [90].
In our opinion, bioprinting possesses a number of advantages for the fabrication of functional cardiac tissues including precise control over the geometry of the constructs and spatial distribution of multiple types of cells, high reproducibility, as well as the possibility to generate customized geometries. Despite convenience, challenges still persist for this versatile technique, including resolution and deposition speed [91]. High precision can be potentially achieved by reducing the diameter of the nozzles, but it is necessary to prevent cell death due to the high shear stress that cells might undergo during the printing process [89]. Proper storage of the cells inside the bioink over the lengthy printing process poses another issue. The printing speed needs to be increased, likely by employing multiple nozzles for simultaneously printing of complex biological structures [92].
4 Simulating electrophysiology of the heart
During the development, maintenance, and regeneration of tissues, intercellular electrical mechanisms play a key role. This is particularly true for certain types of tissues including myocardium, skeletal muscles, and neurons that express strong electrophysiological behaviors in the body. Therefore, through the application of proper external electrical stimulation that simulates the in vivo signals, researchers have demonstrated better control over cellular adhesion, growth, maturation, and orientation, significantly enhancing the quality of engineered cardiac tissues (Fig. 3) [74, 94–96]. Electrical stimuli can be either monophasic or biphasic, in the form of sinusoidal or square waves, and delivered in pulses or continuously [97]. The most commonly used approaches for applying the electrical stimulation are based on the use of a pair of carbon or platinum electrodes that provides a uniform electric field in between. Although efficient, microscale precision with local stimulation has cannot be achieved with these simplified electrode pairs. A possible solution to overcome such limitation is given by the use of multi-electrode arrays (MEAs). MEAs are fabricated by lithographic techniques, while circuitry for stimulation, recording and processing of the signals obtained can be integrated directly into their designs [98, 99]. MEAs permit single-cell resolution stimulation, that is, MEAs can generate electric fields over only two or few microelectrodes in the array instead of applying a field across the large area of a cardiac tissue. Recently, techniques for metal electrode deposition on glass substrates have been developed to further advance the fabrication of microelectrodes analogous to MEAs [100]. Nevertheless, these 2D electrodes are rather limited to their use in stimulating planar tissues.
Figure 3.
Schematics illustrating methods of conducting electrical stimulation and enhancing the electrical properties of the matrices for cardiomyocytes.
In order to apply electrical stimulation to thick tissue constructs, several types of 3D electrodes have been microfabricated. For example, an interdigitated (IDE) array of platinum electrodes was recently developed to induce cell alignment that was shifted from the direction of the underlying GelMA micropatterns [101]. Besides the external stimulation, the electrical properties of the scaffolds themselves are equally important. Comparing to the native myocardium, the polymeric scaffolds possess very limited electrical conductivity as insulating materials. Lately, a few studies have sought to address these drawbacks by the integration of nanomaterials. For example, Khademhosseini et al. demonstrated that the electrical activity of cardiomyocytes cultured on hybrid carbon nanotubes (CNTs)-GelMA hydrogels was significantly enhanced compared with those residing in pure GelMA hydrogels [102]. In a sense, the CNTs added into the hydrogels function as a conductive bridge among adjacent cardiomyocytes for relaying electrical signals. In addition, Kharaziha et al. developed hybrid scaffolds composed of PGS:gelatin (PG) electrospun nanofibers and CNTs. The composite nanofibers scaffolds resulted in greater alignment as well as enhanced better synchronous synchronized beating compared to those cultured on PG scaffold without CNTs [103]. Similarly, gold nanostructures have been incorporated into 3D scaffolds to improve the electrical conductivity of the cellular network and to increase the electrical communication between neighboring cardiomyocytes. Dvir et al. demonstrated that cardiac cells co-embedded with gold nanowires within alginate scaffolds could better align and contract when compared with those grown in pure alginate. Moreover, the same group showed that electrical stimulation increased the levels of proteins involved in muscle contraction [104]. In a recent work, coiled electrospun nanofibers integrating gold nanoparticles were fabricated. Cardiomyocytes cultured on these conductive nanofibrous matrices assumed organization into aligned and elongated tissues generating low excitation threshold and high contraction rate [105].
5 Cardiac bioreactors and heart-on-a-chip
The recently developed organs-on-a-chip technology has integrated tissue models within a microfluidic platform simulating the circulation system that mimic both the biology and physiology of the human (multi)-organ systems [115–131]. Using these organs-on-a-chip platforms, multiple organoids can be maintained under a well-defined microenvironment containing tightly controlled biochemical and biophysical cues to study inter-organoid interactions, perform drug screening, analyze developing, healthy, and diseased tissues, as well as to optimize treatments for personalized medicine [132, 133]. The construction of physiologically relevant organ models is the central theme in the organs-on-a-chip research. Technologies in tissue engineering have been adapted that allow for precise control of cell distribution, adhesion, morphology, and behavior [134, 135] in the microfluidic devices through micropatterning [136], surface chemistry [137], and 3D scaffolds with defined composition, structures, and geometries [138]. Cyclic mechanical strain [139], shear stresses [140], and electrical stimulation [20, 141] may also be applied inside the microfluidic chambers to expose cells to cues mimicking those they experience under physiological conditions to promote in vitro tissue maturation. Moreover, hypoxia/normoxia conditions can be set up to mimic normal/pathological conditions [142] and study cell behaviors [142, 143].
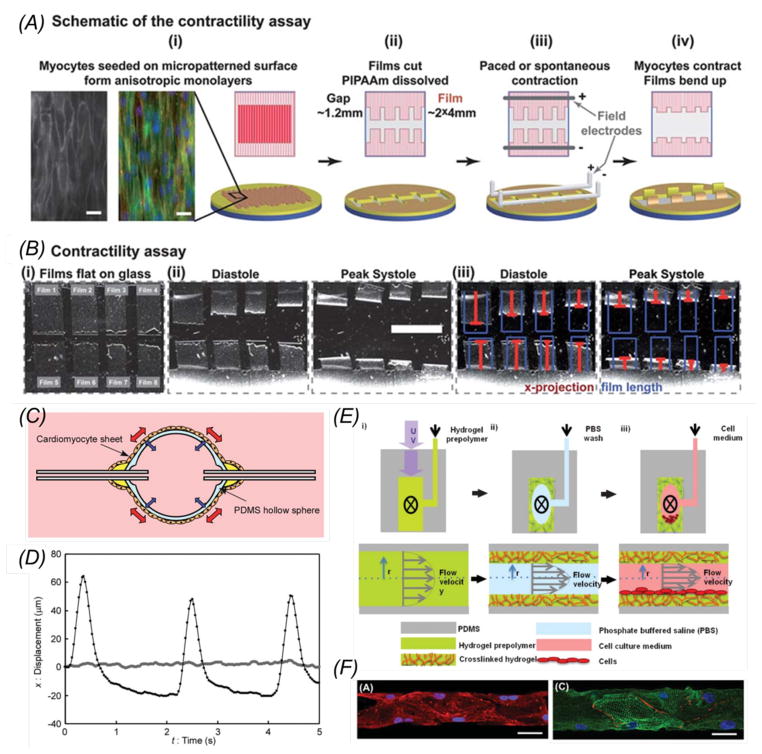
Recently, significant advancement has been made in building heart-on-a-chip platforms with the development of microscale tissue constructs that reproduce the structure and function of the myocardium. The Parker group has designed muscular thin films (MTFs) and subsequently employed them for pharmacological and electrophysiological studies [144]. As examples, biohybrid tissue constructs made of anisotropically organized cardiomyocytes and elastomeric thin films were microfabricated (Fig. 4A) to measure contractility (Fig. 4B), cytoskeletal organization, and action potential propagation, demonstrating that the sarcomere organization and cellular alignment influence the diastolic and systolic MTF stress peak [20]. The first version of such heart-on-a-chip platform was then modified to introduce controls over injection of drugs, embedded electrodes, a metallic base to maintain physiological temperature, and a transparent cover to visualize MTF deformations [22]. The same system was successfully applied to investigate genetic, structural, and functional aspects of the failing myocardium [38]. Radisic and co-workers developed a platform to perform pharmacological tests on cardiac bundles. They embedded a poly(tetrafluoroethylene) (PTFE) tubing within a bioreactor to provide contact guidance for cardiomyocytes to adhere and elongate. The platform was further integrated with electrodes for parallel or perpendicular stimulation of the cardiac bundles, inducing a mature phenotype of cultured cardiomyocytes. The system was then tested with nitric oxide showing a decrease in the beating rate, demonstrating its capability to be employed for modeling in vitro cardiac tissue development and disease [145].
Figure 4.
Heart-on-a-chip models. (A) Schematics illustrating the fabrication process of contracting heart stripes; (B) Time-lapse bright-field images showing contracting myocardium stripes. Scale bar: 5 mm. Adapted with permimssion from Grosberg et al., 2011 [20]; (C) Schematic cross-sectional view of a heart-on-a-chip pump made of a PDMS elastomer hollow sphere covered with a sheet of pulsating cardiomyocytes. The contraction of the cell monolayer squeezes the sphere and pumps the fluid through the connected microchannel. (D) Displacement over time of a particle caused by the pulsation. The light grey and black plots indicate the movement of the particle before and after transplantation of the cardiomyocytes monolayer, respectively. Adapted with permission from Tanaka et al., 2007 [147]; (E) Schematic showing the heart-in-a-channel model, where the inner surface of a microfluidic channel was coated with MeTro and seeded with a monolayer of beating cardiomyocytes; (F) Confocal images showing cardiomyocytes stained with troponin I (red) and nuclei (blue) in the left panel, and α-actinin (green), connexin-43 (red), and nuclei (blue) in the right panel. Scale bar: 50 μm. Adapted with permission from Annabi et al., 2013 [150].
An intriguing series of studies was carried out by the Kitamori group that developed biomicroactuators embedding beating cardiomyocytes to bend PDMS micropillars [146]. Later on, the same group designed a heart-on-a-chip pump, harnessing the cooperative contractile forces produced by synchronous pulsatile cardiomyocytes organized in a cell sheet to pump fluids through a microfluidic channel [147]. Rather than using a push-bar structure and a diaphragm to transmit the pulsatile contraction to the fluid, the same team developed a microspherical heart-like pump powered by a contracting cardiomyocytes sheet wrapped around the external surface (Fig. 4C and D). Significantly, despite simpler design compared to the previous pump, this system was able to move fluids within connected microcapillaries for up to 5 days. Moreover, the heart-like pump does not require an electrical interface and can be potentially embedded within medical implant devices requiring an internal actuator [147]. The Parker group designed another cardiac bioactuator that combined PDMS elastomer and cardiomyocytes to develop a ’medusoid’ construct mimicking the shape, kinematics, and interaction with fluids of a jellyfish. Noteworthy, computational modeling was successfully coupled with experimental tests to dissect a complex system into basic components, which were subsequently optimized and connected with each other to mimic the natural behavior of a living organism based on simple materials and basic cell-material interactions [148].
Besides microfluidic heart-on-a-chip systems for pharmacological tests, other studies attempted to develop more physiologically relevant cardiac tissue constructs that recapitulate the cell-cell or cell/matrix interactions in vivo. Particularly, the Simmons group developed a bi-layered microfluidic device embedded with valvular interstitial and endothelial cells [149]. Interstitial cells were cultured in a 3D GelMA hydrogel while endothelial cells were exposed to physiological shear stress. Although simplified, this model proved to be effective in demonstrating that endothelial cells either alone or exposed to shear-reduced interstitial cell de-differentiation towards myofibroblasts. On the other hand, the Khademhosseini group focused on cell-matrix interactions to develop heart-in-a-channel platform (Fig. 4, E and F). Particularly, they found that MeTro coating of microfluidic channels significantly improved adhesion and spontaneous beating of cardiomyocytes compared to surface treatment by gelatin-based materials [150].
Meanwhile, it is of paramount importance to assess the functionality of the engineered cardiac constructs and heart-on-a-chip systems, including maturation, viability, and beating. Cardiomyocytes express an array of contractile proteins such as sarcomeric α-actinin, myosin, troponin I, troponin T, and tropomyosin, as well as disease markers such as creatine kinase when the cardiomyocytes undergo damages [112, 113]. Most of these biomarkers can be easily detected through conventional approaches based on molecular biology, immunostaining, and enzyme-linked immunosorbant assay (ELISA) by sampling the medium or the tissues. Nevertheless, these methods are typically invasive, meaning that only end-point assays can be conducted for individual samples, greatly limiting the use of the heart models. Recent development in microfluidic technologies now allows for direct integration of biosensors for analyzing secreted biomarkers in situ or on-chip in a non-invasive and continuous manner [107–110]. For example, Wikswo and colleagues integrated automated pumping and fluid handling systems into their microphysiometry platform to conduct kinetic analysis of metabolites (e.g. lactate and glucose) secreted by the organoids as well as to monitor the physical environment (e.g. pH, oxygen) of the platform [111].
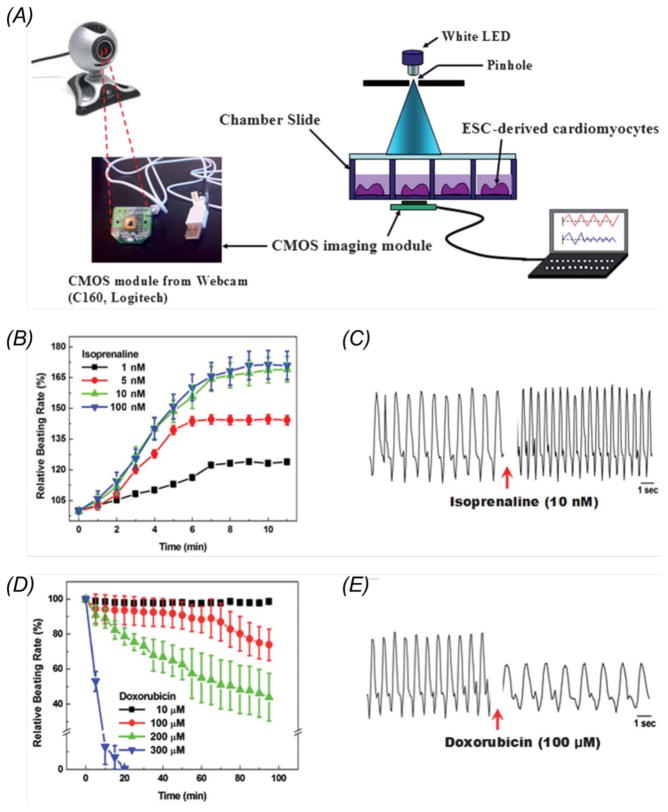
In order to specifically match the needs in monitoring the heart-on-chips, the Khademhosseini Lab has developed a cost-effective, lensless miniature imager based on webcams that can be conveniently mounted at the bottom of the microfluidic chambers to follow the real-time beating behaviors of the cardiomyocytes (Fig. 5A). The beating rate was successfully analyzed using custom-written programs, and it was shown that the rate of dropped when the drug isoproterenol was introduced into the system (Fig. 5, B and C) whereas it increased upon treatment with doxorubicin (Fig. 5, D and E), both in a dose-dependent manner. Higher sensitivity of the mini-microscope can be achieved by utilizing webcams with a more advance frame detection system and image processing [106]. Overall such a device has been demostrated to possess much more flexibility compared with the conventional bench-top microscope. Multiple units can be easily integrated with an array of heart-on-a-chip platforms to achieve high-throughput drug screening.
Figure 5.
(A) Experimental setup showing on-chip monitoring of cardiomyocytes beating using a webcam-based lens-less microscope; (B) Relative beating rate of cardiomyocytes treated with doxorubicin (1, 5, 10, 100 nM); (C) Representative pattern of the beating signal recorded from cardiomyocytes showing a clear increase in the beating frequency following isoprenaline (10 nM) treatment; (D) Relative beating rate of cardiomyocytes treated with doxorubicin (10, 100, 200, and 300 μM); (E) Representative pattern of the beating signal recorded from cardiomyocytes showing a clear decrease in the beating frequency following doxorubicin (100 μM) treatment. Adapted with permision from Kim et al., 2011 [106].
6 Conclusions and perspective
Overall, significant advances have been made during the past years in engineering functional cardiac tissues for heart regeneration. Several factors have been proven to play a key role in adhesion, growth, and alignment of cardiomyocytes, as well as in their phenotypical maturation and organization in complex cardiac tissues. Among them, structural properties of the scaffold, surface treatment, mechanical properties, and electrical stimulation represent critical elements. Moreover, the integration of these parameters with the co-culture of multiple cell types comprising of the heart appears fundamental to achieve functional cardiac tissues. Based on the knowledge obtained from cardiac tissue engineering together with advancements in microfluidic technologies, more physiologically relevant cardiac tissue models termed the heart-on-a-chip platforms have been further developed recently. These heart-on-a-chip platforms are integrated with perfusable microfluidic networks simulating the vasculature, which not only provide nutrients but also actively contribute to the functional maturation of the cardiac tissues through the delivery of soluble biomolecules as well as the interactions among other organs.
While most earlier studies have focused on animal-derived cardiomyocytes due to their easy accessibility, the paradigm is now starting to shift towards the use of cardiomyocytes of human origin and particularly those derived from iPSCs [38]. The iPSCs hold strong potential in engineering heart-on-a-chip platforms as they are widely available and possess pluripotent capacity to differentiate into most cell lineages in the body (e.g. cardiomyocytes, hepatocytes, and endothelial cells) [39]. Clinically, individual variability among patients has greatly hindered the drug discovery process and reduced treatment efficacy. The fact that human iPSCs are derived in a patient-matched manner has made them a superb source to construct human heart models that can be used for personalized drug screening as well as for understanding patient-specific fundamentals of diseases [40]. As we thrive to achieve personalized medicine it should be acknowledged that not every heart disease is the same, and a categorization of the diseases exists. For example, the number one heart disease in the United States is coronary heart disease [114]. In such a case the construction of a healthy heart model coupled with a defined diseased vascular microenvironment seems more important over disease heart models. The congenital heart diseases such as ventricular-septal defects and atrial-septal defects, are an opposite example where disease heart models are required to develop better treatments. The healthy and diseased heart models derived from human iPSCs, when combined with advanced microfluidic technologies, would become highly valuable human heart-on-a-chip platforms for personalized medicine and therapeutics.
Acknowledgments
The authors gratefully acknowledge funding by the Defense Threat Reduction Agency (DTRA) under Space and Naval Warfare Systems Center Pacific (SSC PACIFIC) Contract No. N66001-13-C-2027. The authors also acknowledge funding from the Office of Naval Research Young National Investigator Award, the National Institutes of Health (EB012597, AR057837, DE021468, HL099073, R56AI105024), and the Presidential Early Career Award for Scientists and Engineers (PECASE). The publication of this material does not constitute approval by the government of the findings or conclusions herein.
References
- 1.van Berlo JH, Molkentin JD. Nature medicine. 2014;20:1386–1393. doi: 10.1038/nm.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiu LL, Iyer RK, Reis LA, Nunes SS, Radisic M. Frontiers in bioscience (Landmark edition) 2011;17:1533–1550. doi: 10.2741/4002. [DOI] [PubMed] [Google Scholar]
- 3.Radisic M, Christman KL. Mayo Clinic Proceedings. Vol. 88. Elsevier; 2013. Materials science and tissue engineering: repairing the heart; pp. 884–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiu L, Radisic M. Current Opinion in Chemical Engineering. 2013;2:41–52. [Google Scholar]
- 5.Zhang B, Xiao Y, Hsieh A, Thavandiran N, Radisic M. Nanotechnology. 2011;22:494003. doi: 10.1088/0957-4484/22/49/494003. [DOI] [PubMed] [Google Scholar]
- 6.Camci-Unal G, Annabi N, Dokmeci MR, Liao R, Khademhosseini A. NPG Asia Materials. 2014;6:e99. [Google Scholar]
- 7.Ahmed M, Yildirimer L, Khademhosseini A, Seifalian AM. Journal of nanoscience and nanotechnology. 2012;12:4775–4785. doi: 10.1166/jnn.2012.4884. [DOI] [PubMed] [Google Scholar]
- 8.Tandon N, Cannizzaro C, Chao PG, Maidhof R, Marsano A, Au HTH, Radisic M, Vunjak-Novakovic G. Nature protocols. 2009;4:155–173. doi: 10.1038/nprot.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu LL, Iyer RK, King J, Radisic M. Tissue Engineering Part A. 2011;17:1465–1477. doi: 10.1089/ten.tea.2007.0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menna P, Salvatorelli E, Minotti G. Chemical research in toxicology. 2008;21:978–989. doi: 10.1021/tx800002r. [DOI] [PubMed] [Google Scholar]
- 11.Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, Schacht AL. Nature reviews Drug discovery. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 12.Piccini JP, Whellan DJ, Berridge BR, Finkle JK, Pettit SD, Stockbridge N, Valentin J, Vargas HM, Krucoff MW. American heart journal. 2009;158:317–326. doi: 10.1016/j.ahj.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 13.Shah RR. 2006 [Google Scholar]
- 14.Schiller LR, Johnson DA. The American journal of gastroenterology. 2008;103:815–819. doi: 10.1111/j.1572-0241.2008.01818.x. [DOI] [PubMed] [Google Scholar]
- 15.Bhadriraju K, Chen CS. Drug discovery today. 2002;7:612–620. doi: 10.1016/s1359-6446(02)02273-0. [DOI] [PubMed] [Google Scholar]
- 16.Liang P, Lan F, Lee AS, Gong T, Sanchez-Freire V, Wang Y, Diecke S, Sallam K, Knowles JW, Nguyen PK. Circulation. 2013 doi: 10.1161/CIRCULATIONAHA.113.001883. CIRCULATIONAHA–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hackam DG, Redelmeier DA. Jama. 2006;296:1727–1732. doi: 10.1001/jama.296.14.1731. [DOI] [PubMed] [Google Scholar]
- 18.Novakovic GV, Eschenhagen T, Mummery C. Cold Spring Harbor perspectives in medicine. 2014;4:a014076. doi: 10.1101/cshperspect.a014076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cimetta E, Godier-Furnémont A, Vunjak-Novakovic G. Current opinion in biotechnology. 2013;24:926–932. doi: 10.1016/j.copbio.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grosberg A, Alford PW, McCain ML, Parker KK. Lab on a chip. 2011;11:4165–4173. doi: 10.1039/c1lc20557a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, Yuan H, Jiang D, Zhang D, Zangi L. Nature medicine. 2014;20:616–623. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agarwal A, Goss JA, Cho A, McCain ML, Parker KK. Lab on a chip. 2013;13:3599–3608. doi: 10.1039/c3lc50350j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buckingham M, Meilhac S, Zaffran S. Nature Reviews Genetics. 2005;6:826–837. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 24.Cripps RM, Olson EN. Developmental biology. 1998;203:106–115. doi: 10.1006/dbio.1998.9040. [DOI] [PubMed] [Google Scholar]
- 25.Ocorr K, Perrin L, Lim H, Qian L, Wu X, Bodmer R. Trends in cardiovascular medicine. 2007;17:177–182. doi: 10.1016/j.tcm.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olson EN. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandez-Teran M, Hurle J. The Anatomical Record. 1982;204:137–147. doi: 10.1002/ar.1092040207. [DOI] [PubMed] [Google Scholar]
- 28.Srivastava D. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Xin M, Olson EN, Bassel-Duby R. Nature reviews Molecular cell biology. 2013;14:529–541. doi: 10.1038/nrm3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mercola M, Ruiz-Lozano P, Schneider MD. Genes & development. 2011;25:299–309. doi: 10.1101/gad.2018411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Epelman S, Lavine KJ, Randolph GJ. Immunity. 2014;41:21–35. doi: 10.1016/j.immuni.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baraniak PR, McDevitt TC. Regenerative medicine. 2010;5:121–143. doi: 10.2217/rme.09.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP. Proceedings of the National Academy of Sciences. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J. Nature genetics. 1997;15:30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- 35.Merscher S, Funke B, Epstein JA, Heyer J, Puech A, Lu MM, Xavier RJ, Demay MB, Russell RG, Factor S. Cell. 2001;104:619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 36.Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K. Nature. 2003;424:443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- 37.Olson EN, Schneider MD. Genes & development. 2003;17:1937–1956. doi: 10.1101/gad.1110103. [DOI] [PubMed] [Google Scholar]
- 38.McCain ML, Sheehy SP, Grosberg A, Goss JA, Parker KK. Proceedings of the National Academy of Sciences. 2013;110:9770–9775. doi: 10.1073/pnas.1304913110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masumoto H, Ikuno T, Takeda M, Fukushima H, Marui A, Katayama S, Shimizu T, Ikeda T, Okano T, Sakata R. Scientific reports. 2014;4 doi: 10.1038/srep06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamura K, Hirano K, Wu SM. Journal of cardiovascular translational research. 2013;6:46–53. doi: 10.1007/s12265-012-9413-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.staff B. Wikiversity Journal of Medicine 2014 [Google Scholar]
- 42.Langer R, Vacanti JP. Science. 1993;260 doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 43.Stevens KR, Pabon L, Muskheli V, Murry CE. Tissue Engineering Part A. 2008;15:1211–1222. doi: 10.1089/ten.tea.2008.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakaguchi K, Shimizu T, Horaguchi S, Sekine H, Yamato M, Umezu M, Okano T. Sci Rep. 2013;3:1316. doi: 10.1038/srep01316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haraguchi Y, Shimizu T, Matsuura K, Sekine H, Tanaka N, Tadakuma K, Yamato M, Kaneko M, Okano T. Methods Mol Biol. 2014;1181:139–55. doi: 10.1007/978-1-4939-1047-2_13. [DOI] [PubMed] [Google Scholar]
- 46.Okano T, Yamada N, Sakai H, Sakurai Y. Journal of biomedical materials research. 1993;27:1243–1251. doi: 10.1002/jbm.820271005. [DOI] [PubMed] [Google Scholar]
- 47.Sekine H, Shimizu T, Sakaguchi K, Dobashi I, Wada M, Yamato M, Kobayashi E, Umezu M, Okano T. Nature communications. 2013;4:1399. doi: 10.1038/ncomms2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karp JM, Yeh J, Eng G, Fukuda J, Blumling J, Suh KY, Cheng J, Mahdavi A, Borenstein J, Langer R, Khademhosseini A. Lab Chip. 2007;7:786–94. doi: 10.1039/b705085m. [DOI] [PubMed] [Google Scholar]
- 49.Mummery C, Ward-van Oostwaard D, Doevendans P, Spijker R, van den Brink S, Hassink R, van der Heyden M, Opthof T, Pera M, de la Riviere AB, Passier R, Tertoolen L. Circulation. 2003;107:2733–40. doi: 10.1161/01.CIR.0000068356.38592.68. [DOI] [PubMed] [Google Scholar]
- 50.Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M, Gepstein A, Livne E, Binah O, Itskovitz-Eldor J, Gepstein L. J Clin Invest. 2001;108:407–14. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kehat I. Circulation Research. 2002;91:659–661. doi: 10.1161/01.res.0000039084.30342.9b. [DOI] [PubMed] [Google Scholar]
- 52.Hwang YS, Chung BG, Ortmann D, Hattori N, Moeller HC, Khademhosseini A. Proc Natl Acad Sci US A. 2009;106:16978–83. doi: 10.1073/pnas.0905550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Christman KL, Lee RJ. J Am Coll Cardiol. 2006;48:907–13. doi: 10.1016/j.jacc.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 54.Zimmermann W, Melnychenko I, Eschenhagen T. Biomaterials. 2004;25:1639–1647. doi: 10.1016/s0142-9612(03)00521-0. [DOI] [PubMed] [Google Scholar]
- 55.Shachar M, Cohen S. Heart failure reviews. 2003;8:271–276. doi: 10.1023/a:1024729919743. [DOI] [PubMed] [Google Scholar]
- 56.Ben-Yishay A, Grande DA, Schwartz RE, Menche D, Pitman MD. Tissue engineering. 1995;1:119–133. doi: 10.1089/ten.1995.1.119. [DOI] [PubMed] [Google Scholar]
- 57.Neel EAA, Cheema U, Knowles JC, Brown RA, Nazhat SN. Soft Matter. 2006;2:986–992. doi: 10.1039/b609784g. [DOI] [PubMed] [Google Scholar]
- 58.Serpooshan V, Quinn TM, Muja N, Nazhat SN. Soft Matter. 2011;7:2918–2926. [Google Scholar]
- 59.Serpooshan V, Quinn TM, Muja N, Nazhat SN. Acta biomaterialia. 2013;9:4673–4680. doi: 10.1016/j.actbio.2012.08.031. [DOI] [PubMed] [Google Scholar]
- 60.Serpooshan V, Zhao M, Metzler SA, Wei K, Shah PB, Wang A, Mahmoudi M, Malkovskiy AV, Rajadas J, Butte MJ. Biomaterials. 2013;34:9048–9055. doi: 10.1016/j.biomaterials.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brigham MD, Bick A, Lo E, Bendali A, Burdick JA, Khademhosseini A. Tissue Engineering Part A. 2008;15:1645–1653. doi: 10.1089/ten.tea.2008.0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A. Biomaterials. 2010;31:5536–5544. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bouten CV, Dankers PY, Driessen-Mol A, Pedron S, Brizard AM, Baaijens FP. Adv Drug Deliv Rev. 2011;63:221–41. doi: 10.1016/j.addr.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 64.Chen Q, Harding SE, Ali NN, Lyon AR, Boccaccini AR. Materials Science and Engineering: R: Reports. 2008;59:1–37. [Google Scholar]
- 65.Shin H, Jo S, Mikos AG. Biomaterials. 2003;24:4353–4364. doi: 10.1016/s0142-9612(03)00339-9. [DOI] [PubMed] [Google Scholar]
- 66.Prabhakaran MP, Venugopal J, Kai D, Ramakrishna S. Materials Science and Engineering: C. 2011;31:503–513. [Google Scholar]
- 67.Kulkarni RK, Moore EG, Hegyeli AF, Leonard F. Journal of biomedical materials research. 1971;5:169–181. doi: 10.1002/jbm.820050305. [DOI] [PubMed] [Google Scholar]
- 68.Park TG. Journal of Controlled Release. 1994;30:161–173. [Google Scholar]
- 69.Park TG. Biomaterials. 1995;16:1123–1130. doi: 10.1016/0142-9612(95)93575-x. [DOI] [PubMed] [Google Scholar]
- 70.Zolnik BS, Burgess DJ. Journal of Controlled Release. 2007;122:338–344. doi: 10.1016/j.jconrel.2007.05.034. [DOI] [PubMed] [Google Scholar]
- 71.Prabhakaran MP, Kai D, Ghasemi-Mobarakeh L, Ramakrishna S. Biomedical Materials. 2011;6:055001. doi: 10.1088/1748-6041/6/5/055001. [DOI] [PubMed] [Google Scholar]
- 72.Ji W, Yang F, Seyednejad H, Chen Z, Hennink WE, Anderson JM, van den Beucken J, Jansen JA. Biomaterials. 2012;33:6604–6614. doi: 10.1016/j.biomaterials.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 73.Annabi N, Tsang K, Mithieux SM, Nikkhah M, Ameri A, Khademhosseini A, Weiss AS. Adv Funct Mater. 2013;23 doi: 10.1002/adfm.201300570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nunes SS, Miklas JW, Liu J, Aschar-Sobbi R, Xiao Y, Zhang B, Jiang J, Masse S, Gagliardi M, Hsieh A, Thavandiran N, Laflamme MA, Nanthakumar K, Gross GJ, Backx PH, Keller G, Radisic M. Nat Methods. 2013;10:781–7. doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Orlova Y, Magome N, Liu L, Chen Y, Agladze K. Biomaterials. 2011;32:5615–24. doi: 10.1016/j.biomaterials.2011.04.042. [DOI] [PubMed] [Google Scholar]
- 76.Engelmayr GCJ, Cheng M, Bettinger CJ, Borenstein JT, Langer R, Freed LE. Nat Mater. 2008;7:1003–10. doi: 10.1038/nmat2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ozawa T, Mickle DA, Weisel RD, Koyama N, Wong H, Ozawa S, Li R. The Journal of thoracic and cardiovascular surgery. 2002;124:1157–1164. doi: 10.1067/mtc.2002.127449. [DOI] [PubMed] [Google Scholar]
- 78.McDevitt TC, Angello JC, Whitney ML, Reinecke H, Hauschka SD, Murry CE, Stayton PS. Journal of biomedical materials research. 2002;60:472–479. doi: 10.1002/jbm.1292. [DOI] [PubMed] [Google Scholar]
- 79.Marga F, Jakab K, Khatiwala C, Shephard B, Dorfman S, Forgacs G. 5th European Conference of the International Federation for Medical and Biological Engineering; Springer; 2012. pp. 27–30. [Google Scholar]
- 80.Moroni F, Mirabella T. American journal of stem cells. 2014;3:1. [PMC free article] [PubMed] [Google Scholar]
- 81.Ott HC, Matthiesen TS, Goh S, Black LD, Kren SM, Netoff TI, Taylor DA. Nature medicine. 2008;14:213–221. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 82.Song JJ, Ott HC. Trends in molecular medicine. 2011;17:424–432. doi: 10.1016/j.molmed.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 83.Oberwallner B, Brodarac A, Choi Y, Saric T, Anić P, Morawietz L, Stamm C. Journal of Biomedical Materials Research Part A. 2014;102:3263–3272. doi: 10.1002/jbma.35000. [DOI] [PubMed] [Google Scholar]
- 84.Choi YC, Choi JS, Kim BS, Kim JD, Yoon HI, Cho YW. Tissue Engineering Part C: Methods. 2012;18:866–876. doi: 10.1089/ten.tec.2012.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lu T, Lin B, Kim J, Sullivan M, Tobita K, Salama G, Yang L. Nature communications. 2013;4:2307. doi: 10.1038/ncomms3307. [DOI] [PubMed] [Google Scholar]
- 86.Paulsen SJ, Miller JS. Developmental Dynamics. 2015 doi: 10.1002/dvdy.24254. [DOI] [PubMed] [Google Scholar]
- 87.Murphy SV, Atala A. Nature Biotechnology. 2014;32:773–785. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- 88.Fedorovich NE, Alblas J, de Wijn JR, Hennink WE, Verbout AJ, Dhert WJ. Tissue Eng. 2007;13:1905–25. doi: 10.1089/ten.2006.0175. [DOI] [PubMed] [Google Scholar]
- 89.Malda J, Visser J, Melchels FP, Jüngst T, Hennink WE, Dhert WJ, Groll J, Hutmacher DW. Advanced Materials. 2013;25:5011–5028. doi: 10.1002/adma.201302042. [DOI] [PubMed] [Google Scholar]
- 90.Pati F, Jang J, Ha D, Kim SW, Rhie J, Shim J, Kim D, Cho D. Nature communications. 2014;5:3935. doi: 10.1038/ncomms4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dababneh AB, Ozbolat IT. Journal of Manufacturing Science and Engineering. 2014;136:061016. [Google Scholar]
- 92.Song S, Choi J, Park Y, Lee J, Hong S, Sun K. Artificial organs. 2010;34:1044–1048. doi: 10.1111/j.1525-1594.2010.01143.x. [DOI] [PubMed] [Google Scholar]
- 93.Masuda S, Shimizu T, Yamato M, Okano T. Advanced Drug Delivery Reviews. 2008;60:277–85. doi: 10.1016/j.addr.2007.08.031. [DOI] [PubMed] [Google Scholar]
- 94.Robinson KR. The Journal of cell biology. 1985;101:2023–2027. doi: 10.1083/jcb.101.6.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Radisic M, Park H, Shing H, Consi T, Schoen FJ, Langer R, Freed LE, Vunjak-Novakovic G. Proceedings of the National Academy of Sciences. 2004;101:18129–18134. doi: 10.1073/pnas.0407817101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Markx GH. Organogenesis. 2008;4:11–17. doi: 10.4161/org.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Balint R, Cassidy NJ, Cartmell SH. Tissue Engineering Part B: Reviews. 2012;19:48–57. doi: 10.1089/ten.TEB.2012.0183. [DOI] [PubMed] [Google Scholar]
- 98.Kapoor N, Liang W, Marbán E, Cho HC. Nature biotechnology. 2013;31:54–62. doi: 10.1038/nbt.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kujala K, Ahola A, Pekkanen-Mattila M, Ikonen L, Kerkelä E, Hyttinen J, Aalto-Setälä K. International journal of biomedical science: IJBS. 2012;8:109. [PMC free article] [PubMed] [Google Scholar]
- 100.Tandon N, Marsano A, Maidhof R, Numata K, Montouri-Sorrentino C, Cannizzaro C, Voldman J, Vunjak-Novakovic G. Lab on a Chip. 2010;10:692–700. doi: 10.1039/b917743d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ahadian S, Ramón-Azcón J, Ostrovidov S, Camci-Unal G, Hosseini V, Kaji H, Ino K, Shiku H, Khademhosseini A, Matsue T. Lab on a Chip. 2012;12:3491–3503. doi: 10.1039/c2lc40479f. [DOI] [PubMed] [Google Scholar]
- 102.Shin S, Jung SM, Zalabany M, Kim K, Zorlutuna P, Kim S, Nikkhah M, Khabiry M, Azize M, Kong J. ACS nano. 2013;7:2369–2380. doi: 10.1021/nn305559j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kharaziha M, Shin S, Nikkhah M, Topkaya SN, Masoumi N, Annabi N, Dokmeci MR, Khademhosseini A. Biomaterials. 2014;35:7346–7354. doi: 10.1016/j.biomaterials.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dvir T, Timko BP, Brigham MD, Naik SR, Karajanagi SS, Levy O, Jin H, Parker KK, Langer R, Kohane DS. Nature nanotechnology. 2011;6:720–725. doi: 10.1038/nnano.2011.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fleischer S, Shevach M, Feiner R, Dvir T. Nanoscale. 2014;6:9410–9414. doi: 10.1039/c4nr00300d. [DOI] [PubMed] [Google Scholar]
- 106.Kim SB, Bae H, Cha JM, Moon SJ, Dokmeci MR, Cropekd DM, Khademhosseini A. Lab on a Chip. 2011;11:1801–1807. doi: 10.1039/c1lc20098d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Eklund SE, Snider RM, Wikswo J, Baudenbacher F, Prokop A, Cliffel DE. Journal of Electroanalytical Chemistry. 2006;587:333–339. [Google Scholar]
- 108.Qureshi A, Gurbuz Y, Niazi JH. Sensors and Actuators B: Chemical. 2012;171:62–76. [Google Scholar]
- 109.Weltin A, Slotwinski K, Kieninger J, Moser I, Jobst G, Wego M, Ehret R, Urban GA. Lab on a Chip. 2014;14:138–146. doi: 10.1039/c3lc50759a. [DOI] [PubMed] [Google Scholar]
- 110.Zhang J, Kruss S, Hilmer AJ, Shimizu S, Schmois Z, De La Cruz F, Barone PW, Reuel NF, Heller DA, Strano MS. Advanced healthcare materials. 2014;3:412–423. doi: 10.1002/adhm.201300033. [DOI] [PubMed] [Google Scholar]
- 111.Wikswo J, Prokop A, Baudenbacher F, Cliffel D, Csukas B, Velkovsky M. Engineering challenges of bionems: the integration of microfluidics, micro-and nanodevices, models and external control for systems biology. Iee Proceedings-Nanobiotechnology. 2006;153:81–101. doi: 10.1049/ip-nbt:20050045. (IET) [DOI] [PubMed] [Google Scholar]
- 112.Hazeltine LB, Simmons CS, Salick MR, Lian X, Badur MG, Han W, Delgado SM, Wakatsuki T, Crone WC, Pruitt BL. International journal of cell biology. 2012;2012 doi: 10.1155/2012/508294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Russo V, Young S, Hamilton A, Amsden BG, Flynn LE. Biomaterials. 2014;35:3956–3974. doi: 10.1016/j.biomaterials.2014.01.075. [DOI] [PubMed] [Google Scholar]
- 114.Murphy SL, Xu J, Kochanek KD. National vital statistics reports: from the Centers for Disease Control and Prevention, National Center for Health Statistics, National Vital Statistics System. 2013;61:1–117. [PubMed] [Google Scholar]
- 115.Griffith LG. Annals of the New York Academy of Sciences. 2002;961:83–95. doi: 10.1111/j.1749-6632.2002.tb03056.x. [DOI] [PubMed] [Google Scholar]
- 116.Borenstein JT, Weinberg EJ, Orrick BK, Sundback C, Kaazempur-Mofrad MR, Vacanti JP. Tissue engineering. 2007;13:1837–1844. doi: 10.1089/ten.2006.0156. [DOI] [PubMed] [Google Scholar]
- 117.Esch MB, King TL, Shuler ML. Annual review of biomedical engineering. 2011;13:55–72. doi: 10.1146/annurev-bioeng-071910-124629. [DOI] [PubMed] [Google Scholar]
- 118.Huh D, Hamilton GA, Ingber DE. Trends in cell biology. 2011;21:745–754. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Moraes C, Mehta G, Lesher-Perez SC, Takayama S. Annals of biomedical engineering. 2012;40:1211–1227. doi: 10.1007/s10439-011-0455-6. [DOI] [PubMed] [Google Scholar]
- 120.Ghaemmaghami AM, Hancock MJ, Harrington H, Kaji H, Khademhosseini A. Drug Discovery Today. 2012;17:173–181. doi: 10.1016/j.drudis.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lesher-Perez SC, Frampton JP, Takayama S. Biotechnology journal. 2013;8:180–191. doi: 10.1002/biot.201200206. [DOI] [PubMed] [Google Scholar]
- 122.Selimović Š, Dokmeci MR, Khademhosseini A. Current opinion in pharmacology. 2013;13:829–833. doi: 10.1016/j.coph.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 123.Wikswo JP, Block FE, Cliffel DE, Goodwin CR, Marasco CC, Markov DA, McLean DL, McLean JA, McKenzie JR, Reiserer RS. IEEE Trans Biomed EngCurrent opinion in chemical biology. 2013;60:682–690. doi: 10.1109/TBME.2013.2244891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bhatia SN, Ingber DE. Nature biotechnology. 2014 doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 125.Bhise NS, Ribas J, Manoharan V, Zhang YS, Polini A, Massa S, Dokmeci MR, Khademhosseini A. Journal of Controlled Release. 2014;190:82–93. doi: 10.1016/j.jconrel.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang YS, Khademhosseini A. Nanomedicine 2015 [Google Scholar]
- 127.Ebrahimkhani MR, Young CL, Lauffenburger DA, Griffith LG, Borenstein JT. Drug discovery today. 2014;19:754–762. doi: 10.1016/j.drudis.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Moya ML, George SC. Current opinion in chemical engineering. 2014;3:102–111. doi: 10.1016/j.coche.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Polini A, Prodanov L, Bhise NS, Manoharan V, Dokmeci MR, Khademhosseini A. Expert opinion on drug discovery. 2014;9:335–352. doi: 10.1517/17460441.2014.886562. [DOI] [PubMed] [Google Scholar]
- 130.Sung JH, Srinivasan B, Esch MB, McLamb WT, Bernabini C, Shuler ML, Hickman JJ. Experimental Biology and Medicine. 2014 doi: 10.1177/1535370214529397. 1535370214529397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wikswo JP. Experimental Biology and Medicine. 2014;239:1061–1072. doi: 10.1177/1535370214542068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bhatia SN, Ingber DE. Nat Biotechnol. 2014;32:760–72. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 133.Bersini S, Jeon JS, Moretti M, Kamm RD. Drug Discov Today. 2014;19:735–42. doi: 10.1016/j.drudis.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shin Y, Han S, Jeon JS, Yamamoto K, Zervantonakis IK, Sudo R, Kamm RD, Chung S. Nat Protoc. 2012;7:1247–59. doi: 10.1038/nprot.2012.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bersini S, Jeon JS, Dubini G, Arrigoni C, Chung S, Charest JL, Moretti M, Kamm RD. Biomaterials. 2014;35:2454–61. doi: 10.1016/j.biomaterials.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Amadio S, De Ninno A, Montilli C, Businaro L, Gerardino A, Volonte C. BMC Neurosci. 2013;14:121. doi: 10.1186/1471-2202-14-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Schutte J, Freudigmann C, Benz K, Bottger J, Gebhardt R, Stelzle M. Lab Chip. 2010;10:2551–8. doi: 10.1039/c005307d. [DOI] [PubMed] [Google Scholar]
- 138.Ye X, Lu L, Kolewe ME, Park H, Larson BL, Kim ES, Freed LE. Biomaterials. 2013;34:10007–15. doi: 10.1016/j.biomaterials.2013.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kim HJ, Huh D, Hamilton G, Ingber DE. Lab Chip. 2012;12:2165–74. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 140.Jang KJ, Mehr AP, Hamilton GA, McPartlin LA, Chung S, Suh KY, Ingber DE. Integr Biol (Camb) 2013;5:1119–29. doi: 10.1039/c3ib40049b. [DOI] [PubMed] [Google Scholar]
- 141.Scott A, Weir K, Easton C, Huynh W, Moody WJ, Folch A. Lab Chip. 2013;13:527–35. doi: 10.1039/c2lc40826k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Parvin A, Pranap R, Shalini U, Devendran A, Baker JE, Dhanasekaran A. PLoS One. 2014;9:e107453. doi: 10.1371/journal.pone.0107453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Grayson WL, Zhao F, Izadpanah R, Bunnell B, Ma T. J Cell Physiol. 2006;207:331–9. doi: 10.1002/jcp.20571. [DOI] [PubMed] [Google Scholar]
- 144.Feinberg AW, Feigel A, Shevkoplyas SS, Sheehy S, Whitesides GM, Parker KK. Science. 2007;317:1366–70. doi: 10.1126/science.1146885. [DOI] [PubMed] [Google Scholar]
- 145.Xiao Y, Zhang B, Liu H, Miklas JW, Gagliardi M, Pahnke A, Thavandiran N, Sun Y, Simmons C, Keller G, Radisic M. Lab Chip. 2014;14:869–82. doi: 10.1039/c3lc51123e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tanaka Y, Morishima K, Shimizu T, Kikuchi A, Yamato M, Okano T, Kitamori T. Lab Chip. 2006;6:230–5. doi: 10.1039/b512099c. [DOI] [PubMed] [Google Scholar]
- 147.Tanaka Y, Sato K, Shimizu T, Yamato M, Okano T, Kitamori T. Lab Chip. 2007;7:207–12. doi: 10.1039/b612082b. [DOI] [PubMed] [Google Scholar]
- 148.Nawroth JC, Lee H, Feinberg AW, Ripplinger CM, McCain ML, Grosberg A, Dabiri JO, Parker KK. Nat Biotechnol. 2012;30:792–7. doi: 10.1038/nbt.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chen MB, Srigunapalan S, Wheeler AR, Simmons CA. Lab Chip. 2013;13:2591–8. doi: 10.1039/c3lc00051f. [DOI] [PubMed] [Google Scholar]
- 150.Annabi N, Selimovic S, Acevedo Cox JP, Ribas J, Afshar Bakooshli M, Heintze D, Weiss AS, Cropek D, Khademhosseini A. Lab Chip. 2013;13:3569–77. doi: 10.1039/c3lc50252j. [DOI] [PMC free article] [PubMed] [Google Scholar]