Abstract
The mammalian neocortex has undergone repeated selection for increases and decreases in size and complexity, often over relatively short evolutionary time. But because probing developmental mechanisms across many species is experimentally unfeasible, it is unknown whether convergent morphologies in distantly related species are regulated by conserved developmental programs. In this work, we have taken advantage of the abundance of available mammalian genomes to find evidence of selection on genomic regions putatively regulating neurogenesis in large- versus small-brained species. Using published fetal human RNA-seq data, we show that the gene-neighborhood (i.e., microsynteny) of long intergenic non-coding RNAs (lincRNAs) implicated in cortical development is differentially conserved in large-brained species, lending support to the hypothesis that lincRNAs regulating neurogenesis are selectively lost in small-brained species. We provide evidence that this is not a phenomenon attributable to lincRNA expressed in all tissue types and is therefore likely to represent an adaptive function in the evolution of neurogenesis. A strong correlation between transcription factor-adjacency and lincRNA sequence conservation reinforces this conclusion.
Introduction
The mammalian neocortex is remarkably diverse. While it shows some general uniformity across species (e.g., a six-layered structure and division into functional areas), it is as varied as the adaptive behaviors it governs [1, 2]. Development of the neocortex, however, like most aspects of development [3], retains a much stricter pattern across species, involving a conserved arsenal of progenitor-types. Indeed, these major progenitor-types—apical radial glia (aRG) and basal radial glia (bRG), as well as apical and basal intermediate progenitors (IPs)—are putatively present in all mammals [4, 5, 6, 7]. But in those mammals with larger, convoluted neocortices (i.e., gyrencephalic species), a heterogeneity of bRGs is observed [8], and an increased proliferative potential in basally dividing progenitors is important for cortical size and folding [4, 6, 9, 10, 11, 12, 13]. In addition, recent work has shown that, both neuroanatomically and neurodevelopmentally, mammals may be segregated into two principal groups, delimited by a threshold gyrencephaly index (GI) value of 1.5 (corresponding to approximately one billion cortical neurons) [12]. Thus, we may define species as being high-GI or low-GI. But despite these categorical differences, species differences in cortical development at the genomic level have been given surprisingly little attention [14, 15, 16, 17, 18, 19] and virtually no attention across all mammalian orders [20, 21]. It is largely unknown, therefore, how neurogenesis has evolved in mammals to generate so many radical increases and decreases in neocortical size—or even whether any general principles of building bigger brains can be found across disparate clades.
Here, in order to assess the degree to which neocortical convergence, both in the generation of certain neural progenitor-types and the presentation of cortical growth and folding above a threshold value, is corroborated by convergence at the genomic level, we probed published RNA-seq data collected from human fetal neocortical germinal zones during neurogenesis [22]. Because lincRNAs show accelerated evolution in humans [23], have high levels of tissue- and age-specificity [24], and are potential developmental regulators [25, 26, 27], we limited our probes to lincRNAs. We did this, furthermore, because protein and transcript abundance are poorly correlated [28], at least among closely related species, thus making it difficult to interpret the significance of coding-gene expression for explaining interspecific phenotypic differences. We show that the ancestral gene-neighborhoods of lincRNAs implicated in cortical development (neurodevelopmental lincRNAs), in contrast to lincRNAs predominantly expressed in other tissues, are selectively lost in small-brained species. We argue that this supports not only a functional role for lincRNAs in mammalian neurogenesis [29], but an adaptive role for lincRNAs in neocortical evolution.
Materials and Methods
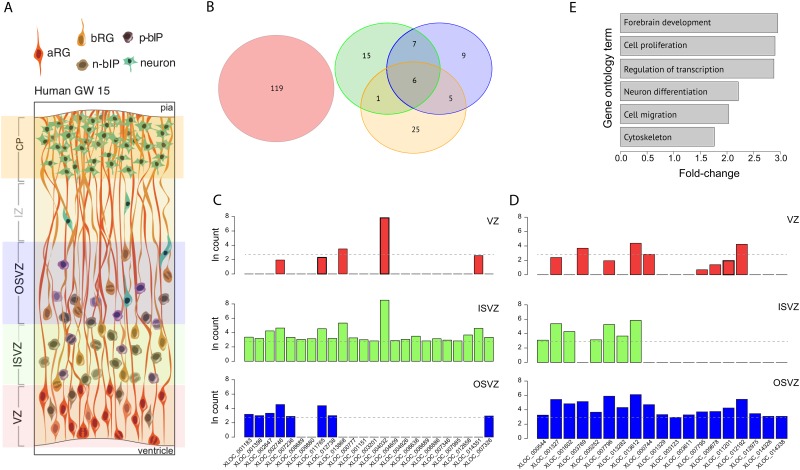
We used previously published RNA-seq data (Series GSE38805) collected from the ventricular zone (VZ), inner subventricular zone (ISVZ), outer subventricular zone (OSVZ), and cortical plate (CP) of human fetal neocortex at gestation week (GW)13–16 [22] and employed the lincRNA discovery pipeline outlined by [30] to identify 187 lincRNA differentially expressed during human neocortical neurogenesis (Fig 1A–1D, S1 Table). We aligned those lincRNAs found in our dataset with those deposited at the Human Body Map lincRNAs, so that all XLOC IDs are preserved from [30]. Of these, 161 were differentially upregulated in a germinal zone, including 43 overexpressed in the ISVZ and/or OSVZ compared to the VZ (Fig 1B–1D). Fifty-seven of the 161 lincRNAs are also relatively highly expressed in the adult brain [30]; and, by comparing names and/or start-sites for our lincRNAs with those identified in recent work on lincRNA expression in tetrapods, we found 52 to be putatively conserved across primates or eutheria according to sequence similarity [25]. All differential expression analyses were run with the R package DESeq [31].
Fig 1. Germinal zone-specific transcript levels of lincRNAs in GW13–16 human neocortex as determined by RNA-seq.
(A) Schematic of human germinal zones dissected for RNA-seq [22], depicting aRG, bRG, neurogenic basal intermediate progenitors (n-bIPs), proliferative basal intermediate progenitors (p-bIPs), and neurons. Adapted from [9]. (B) Number of differentially expressed lincRNAs in each germinal zone (VZ, red; ISVZ, green; OSVZ, blue; CP, orange). Analyses were run for each layer against the VZ. (C,D) LincRNAs differentially overexpressed in the (C) ISVZ and (D) OSVZ. The dashed grey line shows the mean transcript level for lincRNAs overexpressed in the VZ. (E) LincRNAs expressed during human neurogenesis tend to have gene-adjacent neighbors involved in neocortical development. Shown are fold-enrichments of Gene Ontology (GO) terms for adjacent protein-coding gene neighbors of the 142 lincRNAs expressed during human neurogenesis (see S1 Table). GO terms are listed if they are over-represented in the protein-coding gene set (FDR < 0.05). Fold differences for enriched GO terms were analyzed using DAVID (http://david.abcc.ncifcrf.gov/summary.jsp) with the entire set of genes expressed in the fetal brain [22] as a base.
Previous work has shown that sequence conservation is a poor predictor of functional conservation in non-coding RNA [32] and that long non-coding RNAs are often functionally—or at least transcriptionally—linked to adjacently located protein-coding genes [33, 34]. Therefore, we analyzed lincRNA conservation as a function of gene-neighborhood (i.e., microsynteny). For each lincRNA, we defined its gene-neighborhood as the immediately flanking protein-coding genes and discarded any lincRNAs which did not have at least one flanking gene expressed during neurogenesis. Orthologous flanking genes were identified using Orthomam v8.0 [35]; if they could not be found there, then 1-to-1 orthologs were identified in Ensembl.
The greatest distance between a lincRNA and its nearest neighbor was 22.3 Mb (XLOC_000380), although the median distance was 46 kb. The final list included 142 lincRNAs, whose gene-neighborhoods were collectively enriched for Gene Ontology terms related to forebrain development and cell proliferation (S1 Table; Fig 1E).
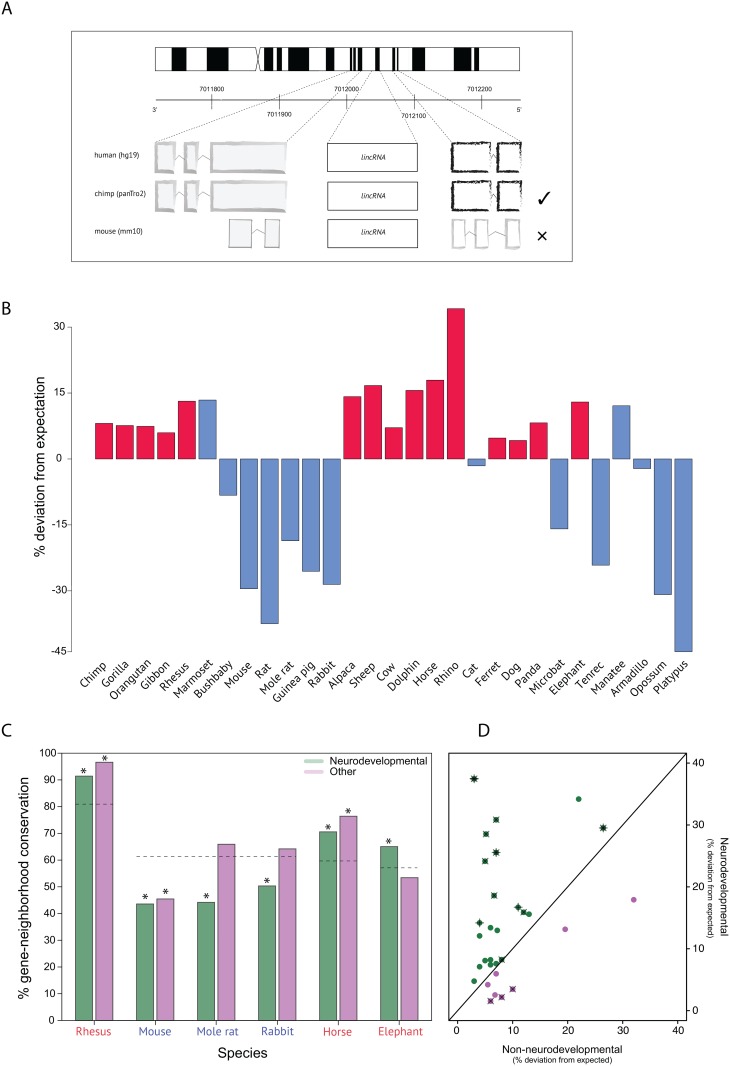
We assessed lincRNA gene-neighborhood conservation for the 142 lincRNAs in humans and 30 other species (see S2 Table) by BLASTing the lincRNA sequence retrieved from the human RNA-seq data and visually inspecting (using the UCSC Genome Browser) the gene-neighborhood for each species (Fig 2A). Regions with the maximum BLASTn alignment score, E-value < 1 × 10−4, and query cover > 20% were selected. When BLASTn was unsuccessful in returning any sequences matching these criteria, discontiguous megablast was used. To increase the likelihood of finding an orthologous region in non-human species, we used both the entire lincRNA sequence, as well as, when available, only the region of the lincRNA showing signs of transcriptional activity in human as evidenced by ENCODE data on chromatin-level information [36]. Both sequences consistently identified the same region in non-human species. Due to considerable scaffolding at the time of the analysis, we were only able to assess a portion of the lincRNAs (66) for the dolphin.
Fig 2. LincRNA gene-neighborhood conservation in neurodevelopmental and non-neurodevelopmental tissue.
(A) Schematic of the protocol used for determining lincRNA gene-neighborhood conservation across species. In the example given, only chimp is scored as conserved. Chromosomal synteny is not a condition for gene-neighborhood conservation. (B) Gene-neighborhood conservation of 142 lincRNAs expressed during human neurogenesis across 29 mammalian species (S2 Table). Gene-neighborhood conservation is shown to be above null phylogenetic expectations in high-GI species (red) and below expectations for low-GI species (blue). The two exceptions are the marmoset, a low-GI primate, and the manatee, a large-brained lissencephalic Afrotherian; both of these show lincRNA gene-neighborhood conservation considerably above null expectations The chicken is not shown. (C) Gene-neighborhood conservation for neurodevelopmental lincRNAs (green) and lincRNAs showing maximum levels of expression in non-brain human tissue (lilac) for three high-GI (red) and three low-GI (blue) species. Conservation in the naked mole rat, rabbit, and elephant are significantly different for neurodevelopmental compared to non-brain (Other) lincRNAs, while similar levels of conservation are observed for both classes of lincRNAs in macaque, mouse, and horse. Predicted conservation values for each species (dashed lines) were calculated from null evolutionary models based on divergence times with human. Asterisks denote significantly different values from predicted (P < 0.05). (D) Plot of neurodevelopmental versus non-neurodevelopmental (absolute) conservation deviation scores based on a phylogenetic expectation model (see Materials and Methods) for 29 mammalian species. From the regression slope delineating no deviation from expected scores, it is clear that neurodevelopmental lincRNAs deviate more frequently and more sizeably from conservation expectations than non-neurodevelopmental lincRNAs. Crosses and Xs indicate negative deviations for neurodevelopmental and non-neurodevelopmental lincRNAs, respectively. (B) is adapted from [20].
Conservation scores for each lincRNA of a given species were tallied as either conserved (1; at least one conserved neighbor) or not conserved (0) (S2 Table). Expectation scores were then calculated under an Ornstein-Uhlenbeck model with a single optimum based on phylogenetic generaliized least squares [37] using three 102-species pruned mammalian supertrees [12, 38] and R package geiger [39]. The percentage deviations of actual from expected scores for each species are presented in Fig 2B. GI values were collected from [12].
178 lincRNAs showing their highest levels of expression in non-brain human tissue were collected from [30] and analyzed as above for 30 species (Fig 2C and 2D). Rates of molecular evolution for different species were collected from [40] and used to assess the degree to which different molecular rates across species might confound adaptive explanations for lincRNA gene-neighborhood evolution (S1 Fig).
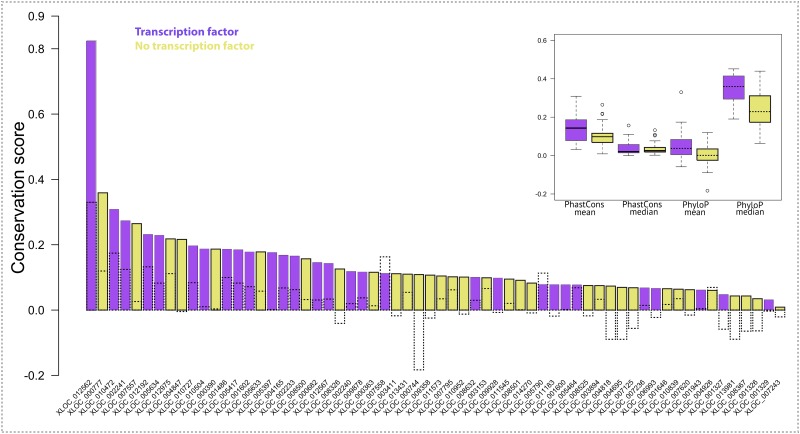
Because sequence conservation was likely to be more constrained in closely related species, we computed PhyloP and PhastCons sequence conservation scores for primates for lincRNAs showing gene-neighborhood conservation in primates but not in mouse (S3 Table) [41]. These were used to assess the sequence-level conservation in primates for lincRNAs with and without transcription factor-adjacency (Fig 3).
Fig 3. Sequence conservation among primates for lincRNAs expressed during human neurogenesis tends to be higher for lincRNAs flanked by at least one transcription factor.
PhastCons scores are shown for 62 lincRNAs, whose gene-neighborhoods are not conserved in mouse (S2 Table). PhyloP scores for the same lincRNAs are shown as dotted bars. (Inset) Boxplot of the mean and median PhyloP and PhastCons scores for primates for the 62 lincRNAs. Mean conservation scores between lincRNAs with (purple) and without (brown) at least one adjacent transcription factor were significantly different for both PhastCons (T = -2.371, P = 0.023) and PhyloP (T = -3.513, P = 0.001), but median scores were significantly different only for PhyloP (T = -5.211, P < 0.001).
Results
We identified 187 lincRNAs differentially expressed in a germinal zone or the cortical plate (CP) of the embryonic human neocortex (Fig 1A–1D, S1 Table). Of these, we shortlisted 142, which had at least one adjacent protein-coding gene also expressed during human neurogenesis (Fig 1E). We then determined whether the genes immediately flanking the lincRNA in the human genome (defined as the lincRNA gene-neighborhood; Fig 2A) were similarly flanking the orthologous genomic sequence in 30 other species (29 mammals plus chicken). We found, firstly, that lincRNA gene-neighborhood conservation could not be explained by phylogenetic relatedness (Fig 2A–2C). By calculating the number of lincRNAs expected to be conserved in each species based on phylogenetic relatedness to human, we could determine which species fell below and which above null expectations. We found that all low-GI species fell below and all high-GI species above phylogenetic expectations (Fig 2B), with two exceptions: the marmoset, a near-lissencephalic primate that is hypothesized to have recently evolved from a gyrencephalic ancestor and therefore may still be in the process of purging unneeded neurodevelopmental lincRNAs [42]; and the manatee, a large-brained (382g), albeit lissencephalic species [43]. Importantly, we found no similar pattern of conservation with non-neurodevelopmental lincRNAs [30] (Fig 2C and 2D); no significant correlation between rate of molecular evolution and neurodevelopmental lincRNA gene-neighborhood conservation (P > 0.1); and GI to be a stronger predictor (R 2 = 0.68, P < 0.001) than body weight (Z = 1.967, P = 0.025, one-tailed) or longevity (Z = 2.105, P = 0.018, one-tailed) of gene-neighborhood conservation. We therefore provide evidence for a possible genomic correlate of the GI threshold [12] in the disproportionate conservation of neurodevelopmental lincRNAs in high- versus low-GI species.
Secondly, we found that, in primates, sequence conservation was highest for gene-neighborhoods containing at least one transcription factor (Fig 3). Mean conservation scores between lincRNAs with and without at least one adjacent transcription factor were significantly different for both PhastCons and PhyloP measures of sequence conservation. These results are in line with increasing evidence for the role of lincRNAs in neurodevelopment as important transcriptional regulators (e.g., [44]). Among large-brained primates, interspecific discrepancies in the timing of neurogenesis are largely a matter of scale, rather than a rearrangement of transcriptional events [45]. It is possible that, at this close phylogenetic range, there has been a strong selection pressure to conserve lincRNAs, even at the primary sequence level, which act as regulatory elements of transcription factors during neurogenesis.
Discussion
The adaptation of proliferative basal progenitors may be tantamount to a relaxation of constraints along lineages leading to larger-brained species [46]. However, in light of the evidence presented here, we think that convergent gain-of-function along lineages leading to large-brained species is unrealistic. Rather, our analysis of lincRNA gene-neighborhood conservation suggests that the selective loss of genomic elements regulating neurogenesis may be responsible for the evolution of smaller brains in mammals. This means that the genomic developmental toolbox necessary for adapting proliferative basal progenitors, leading to increases in neocortical size and folding, is ancestral to eutherian mammals. Our definition of conservation in terms of microsyteny, rather than primary sequence similarity, allows for such conservation of lincRNAs over extended evolutionary time periods, despite the well-known phenomenon of rapid sequence changes in lincRNAs [47, 48]. Of course, it could be argued that the loss of lincRNAs in low-GI species, which are typically small-bodied, may simply be caused by a higher rate of meiotic recombination in low-GI species, resulting in more frequent meiotic errors and thereupon loss of lincRNAs. However, several lines of evidence presented here make this unlikely to be the case: GI is a better predictor (R2 = 0.68, P < 0.001) than lifespan (R 2 = 0.44, P < 0.001) or body weight (R 2 = 0.39, P < 0.001) of lincRNA gene-neighborhood conservation [20]; in non-neurodevelopmental lincRNAs, gene-neighborhood conservation can generally be explained by phylogenetic relatedness (Fig 2C and 2D); and rates of molecular evolution are not typically faster in low-GI species compared to high-GI species (S1 Fig). Nonetheless, we cannot entirely rule out faster microsynteny evolution in smaller-brained species as a contributing factor—and, indeed, we would expect it to make some contribution—to neurodevelopmental lincRNA conservation in large-brained species. It is also worth noting that a considerable fraction of lincRNAs overlap enhancer regions [30], which allows for the possibility that enhancer-associated RNAs, rather than lincRNAs, are driving the microsynteny conservation we observe. However, because enhancer-associated RNAs are not enriched (Z = 0.213, P = 0.584) in the neurodevelopmental over the non-neurodevelopmental set of lincRNAs (S2 Table), even though both sets are more enriched than average (Z > 2, P < 0.05), we are confident that the observed effect is driven primarily by selection on lincRNA microsyteny. Finally, while we analysed all mammalian species for which genomic data were available, our sample constitutes a minor fraction (< 1%) of all mammalian species, and so we cannot conclusively say that the confidence intervals for GI, lifespan, and body weight as predictors of lincRNA conservation would hold if the other 99% of mammals were analysed.
We think that, because broadly non-functional heritable sequence mutations are more frequent in lincRNAs compared to protein-coding genes, sequence similarity may not be a reliable measure for functional conservation in lincRNAs between distantly related species. Rather, the microsynteny of a lincRNA, particularly if it is cis-acting, may be a better indicator across species of its functional conservation [32].
We therefore hypothesize, given the results presented here, that the selective loss or retention of neurodevelopmental lincRNAs is relevant for neocortical development and evolution. We think this is a first step in uncovering how convergent evolution of a complex structure may have been achieved at the genomic level. Evidence for lincRNA transcriptional activity in other mammalian species will be crucial for taking this hypothesis forward. Ultimately, how such genomic elements function towards neocortical growth and folding will require investigations into the molecular mechanisms of the differentially conserved lincRNAs identified here.
Supporting Information
lincRNA conservation scores plotted against paired differences between species-level and mammalian-average molecular rates, as reported by [40]. Separate regression analyses for high-GI species (R2 = -0.06, P = 0.52) and low-GI species (R2 = 0.03, P = 0.30) are also not significant.
(EPS)
(XLSX)
(XLSX)
(XLS)
Acknowledgments
We would like to thank Holger Brandl for help implementing the lincRNA pipeline, and Farhath Badsha for helpful discussion. E.L. would like to thank Evan Charles for helpful discussion.
Data Availability
All data contained in the paper are available as supporting information with the paper or their sources are detailed in the paper.
Funding Statement
W.B.H. was supported by grants from the DFG (SFB 655, A2) and the ERC (250197), by the DFG-funded Center for Regenerative Therapies Dresden, and by the Fonds der Chemischen Industrie. EL was supported by the Max Planck Gesellschaft. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Krubitzer L, Kaas J. The evolution of the neocortex in mammals: how is phenotypic diversity generated? Current Opinion in Neurobiology. 2005. August;15(4):444–453. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16026978. 10.1016/j.conb.2005.07.003 [DOI] [PubMed] [Google Scholar]
- 2. Molnár Z. Evolution of cerebral cortical development. Brain, Behavior and Evolution. 2011;78(1):94–107. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21691047. 10.1159/000327325 [DOI] [PubMed] [Google Scholar]
- 3. Kalinka AT, Tomancak P. The evolution of early animal embryos: conservation or divergence? Trends in Ecology & Evolution. 2012. July;27(7):385–393. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22520868. 10.1016/j.tree.2012.03.007 [DOI] [PubMed] [Google Scholar]
- 4. Borrell V, Calegari F. Mechanisms of brain evolution: regulation of neural progenitor cell diversity and cell cycle length. Neuroscience Research. 2014. September;86:14–24. 10.1016/j.neures.2014.04.004 [DOI] [PubMed] [Google Scholar]
- 5. Molnár Z, Clowry G. Cerebral cortical development in rodents and primates. Progress in brain research. 2012;195:45–70. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22230622. [DOI] [PubMed] [Google Scholar]
- 6. Florio M, Huttner WB. Neural progenitors, neurogenesis and the evolution of the neocortex. Development (Cambridge, England). 2014. June;141(11):2182–2194. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24866113. 10.1242/dev.090571 [DOI] [PubMed] [Google Scholar]
- 7. Taverna E, Götz M, Huttner WB. The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annual review of cell and developmental biology. 2014;30:465–502. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25000993. 10.1146/annurev-cellbio-101011-155801 [DOI] [PubMed] [Google Scholar]
- 8. Betizeau M, Cortay V, Patti D, Pfister S, Gautier E, Bellemin-Ménard A, et al. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron. 2013. October;80(2):442–457. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24139044. 10.1016/j.neuron.2013.09.032 [DOI] [PubMed] [Google Scholar]
- 9. Lewitus E, Kelava I, Huttner WB. Conical expansion of the outer subventricular zone and the role of neocortical folding in evolution and development. Frontiers in human neuroscience. 2013;7:424 Available from: http://www.ncbi.nlm.nih.gov/pubmed/23914167. 10.3389/fnhum.2013.00424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun T, Hevner RF. Growth and folding of the mammalian cerebral cortex: from molecules to malformations. Nature reviews Neuroscience. 2014. March;15(4):217–232. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24646670. 10.1038/nrn3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stahl R, Walcher T, De Juan Romero C, Pilz GA, Cappello S, Irmler M, et al. Trnp1 regulates expansion and folding of the mammalian cerebral cortex by control of radial glial fate. Cell. 2013. April;153(3):535–549. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23622239. 10.1016/j.cell.2013.03.027 [DOI] [PubMed] [Google Scholar]
- 12. Lewitus E, Kelava I, Kalinka AT, Tomancak P, Huttner WB. An adaptive threshold in mammalian neocortical evolution. PLoS biology. 2014. November;12(11):e1002000 Available from: http://www.ncbi.nlm.nih.gov/pubmed/25405475. 10.1371/journal.pbio.1002000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011. July;146(1):18–36. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21729779. 10.1016/j.cell.2011.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Enard W, Przeworski M, Fisher SE, Lai CSL, Wiebe V, Kitano T, et al. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002. August;418(6900):869–872. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12192408. 10.1038/nature01025 [DOI] [PubMed] [Google Scholar]
- 15. Ali F, Meier R. Positive selection in ASPM is correlated with cerebral cortex evolution across primates but not with whole-brain size. Molecular biology and evolution. 2008. November;25(11):2247–2250. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18718919. 10.1093/molbev/msn184 [DOI] [PubMed] [Google Scholar]
- 16. Fedrigo O, Pfefferle AD, Babbitt CC, Haygood R, Wall CE, Wray GA. A potential role for glucose transporters in the evolution of human brain size. Brain, behavior and evolution. 2011;78(4):315–326. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21986508. 10.1159/000329852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Charrier C, Joshi K, Coutinho-Budd J, Kim J, Lambert N, de Marchena J, et al. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell. 2012. May;149(4):923–935. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22559944. 10.1016/j.cell.2012.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Montgomery SH, Mundy NI. Evolution of ASPM is associated with both increases and decreases in brain size in primates. Evolution; international journal of organic evolution. 2012. March;66(3):927–932. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22380452. 10.1111/j.1558-5646.2011.01487.x [DOI] [PubMed] [Google Scholar]
- 19. Xu S, Chen Y, Cheng Y, Yang D, Zhou X, Xu J, et al. Positive selection at the ASPM gene coincides with brain size enlargements in cetaceans. Proceedings Biological sciences / The Royal Society. 2012. November;279(1746):4433–4440. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22977148. 10.1098/rspb.2012.1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lewitus E, Kalinka AT. Neocortical development as an evolutionary platform for intragenomic conflict. Frontiers in Neuroanatomy. 2013;7(2). Available from: http://www.ncbi.nlm.nih.gov/pubmed/23576960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Enard W. Comparative genomics of brain size evolution. Frontiers in Human Neuroscience. 2014;8(345). Available from: http://www.frontiersin.org/human_neuroscience/10.3389/fnhum.2014.00345/abstract. 10.3389/fnhum.2014.00345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fietz SA, Lachmann R, Brandl H, Kircher M, Samusik N, Schröder R, et al. Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proceedings of the National Academy of Sciences of the United States of America. 2012. July;109(29):11836–11841. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22753484. 10.1073/pnas.1209647109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hubisz MJ, Pollard KS. Exploring the genesis and functions of Human Accelerated Regions sheds light on their role in human evolution. Genetics of human evolution. 2014. December;29(0):15–21. Available from: http://www.sciencedirect.com/science/article/pii/S0959437X14000781. [DOI] [PubMed] [Google Scholar]
- 24. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012. September;22(9):1775–1789. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22955988. 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014. January;505(7485):635–640. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24463510. 10.1038/nature12943 [DOI] [PubMed] [Google Scholar]
- 26. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013. July;154(1):26–46. 10.1016/j.cell.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Washietl S, Kellis M, Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome research. 2014. April;24(4):616–628. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24429298. 10.1101/gr.165035.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Khan Z, Ford MJ, Cusanovich DA, Mitrano A, Pritchard JK, Gilad Y. Primate transcript and protein expression levels evolve under compensatory selection pressures. Science (New York, NY). 2013. November;342(6162):1100–1104. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24136357. 10.1126/science.1242379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ng S, Lin L, Soh BS, Stanton LW. Long noncoding RNAs in development and disease of the central nervous system. Trends in genetics: TIG. 2013. August;29(8):461–468. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23562612. 10.1016/j.tig.2013.03.002 [DOI] [PubMed] [Google Scholar]
- 30. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011. September;25(18):1915–1927. 10.1101/gad.17446611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11(10):R106 Available from: http://genomebiology.com/2010/11/10/R106. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chodroff RA, Goodstadt L, Sirey TM, Oliver PL, Davies KE, Green ED, et al. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol. 2010;11(7):R72 10.1186/gb-2010-11-7-r72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009. August;5(8):e1000617 10.1371/journal.pgen.1000617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Basu S, Müller F, Sanges R. Examples of sequence conservation analyses capture a subset of mouse long non-coding RNAs sharing homology with fish conserved genomic elements. BMC bioinformatics. 2013;14 Suppl 7:S14 Available from: http://www.ncbi.nlm.nih.gov/pubmed/23815359. 10.1186/1471-2105-14-S7-S14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Douzery EJP, Scornavacca C, Romiguier J, Belkhir K, Galtier N, Delsuc F, et al. OrthoMaM v8: A Database of Orthologous Exons and Coding Sequences for Comparative Genomics in Mammals. Molecular Biology and Evolution. 2014. July;31(7):1923–1928. Available from: http://mbe.oxfordjournals.org/content/31/7/1923.abstract. 10.1093/molbev/msu132 [DOI] [PubMed] [Google Scholar]
- 36. Raney BJ, Cline MS, Rosenbloom KR, Dreszer TR, Learned K, Barber GP, et al. ENCODE whole-genome data in the UCSC genome browser (2011 update). Nucleic Acids Res. 2011. January;39(Database issue):D871–875. 10.1093/nar/gkq1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grafen A. The Phylogenetic Regression. Philosophical Transactions of the Royal Society of London B, Biological Sciences. 1989. December;326(1233):119–157. Available from: http://rstb.royalsocietypublishing.org/content/326/1233/119.short. 10.1098/rstb.1989.0106 [DOI] [PubMed] [Google Scholar]
- 38. Bininda-Emonds ORP, Cardillo M, Jones KE, MacPhee RDE, Beck RMD, Grenyer R, et al. The delayed rise of present-day mammals. Nature. 2007. March;446(7135):507–512. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17392779. 10.1038/nature05634 [DOI] [PubMed] [Google Scholar]
- 39. Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W. GEIGER: investigating evolutionary radiations. Bioinformatics. 2008. January;24(1):129–131. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18006550. 10.1093/bioinformatics/btm538 [DOI] [PubMed] [Google Scholar]
- 40. Bininda-Emonds ORP. Fast genes and slow clades: comparative rates of molecular evolution in mammals. Evol Bioinform Online. 2007;3:59–85. [PMC free article] [PubMed] [Google Scholar]
- 41. Hubisz MJ, Pollard KS, Siepel A. PHAST and RPHAST: phylogenetic analysis with space/time models. Briefings in bioinformatics. 2011. January;12(1):41–51. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21278375. 10.1093/bib/bbq072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kelava I, Reillo I, Murayama AY, Kalinka AT, Stenzel D, Tomancak P, et al. Abundant occurrence of basal radial glia in the subventricular zone of embryonic neocortex of a lissencephalic primate, the common marmoset Callithrix jacchus. Cerebral cortex (New York, NY: 1991). 2012. February;22(2):469–481. PMID: 22114084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kelava I, Lewitus E, Huttner WB. The secondary loss of gyrencephaly as an example of evolutionary phenotypical reversal. Frontiers in neuroanatomy. 2013;7:16 Available from: http://www.ncbi.nlm.nih.gov/pubmed/23805079. 10.3389/fnana.2013.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kamm GB, Pisciottano F, Kliger R, Franchini LF. The developmental brain gene NPAS3 contains the largest number of accelerated regulatory sequences in the human genome. Molecular biology and evolution. 2013. May;30(5):1088–1102. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23408798. 10.1093/molbev/mst023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Somel M, Rohlfs R, Liu X. Transcriptomic insights into human brain evolution: acceleration, neutrality, heterochrony. Current opinion in genetics & development. 2014. September;29C:110–119. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25233113. 10.1016/j.gde.2014.09.001 [DOI] [PubMed] [Google Scholar]
- 46. Boddy AM, McGowen MR, Sherwood CC, Grossman LI, Goodman M, Wildman DE. Comparative analysis of encephalization in mammals reveals relaxed constraints on anthropoid primate and cetacean brain scaling. Journal of Evolutionary Biology. 2012;25(5):981–994. Available from: 10.1111/j.1420-9101.2012.02491.x. 10.1111/j.1420-9101.2012.02491.x [DOI] [PubMed] [Google Scholar]
- 47. Kapusta A, Feschotte C. Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends in genetics: TIG. 2014. October;30(10):439–452. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25218058. 10.1016/j.tig.2014.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kostka D, Hahn MW, Pollard KS. Noncoding sequences near duplicated genes evolve rapidly. Genome biology and evolution. 2010;2:518–533. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20660939. 10.1093/gbe/evq037 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lincRNA conservation scores plotted against paired differences between species-level and mammalian-average molecular rates, as reported by [40]. Separate regression analyses for high-GI species (R2 = -0.06, P = 0.52) and low-GI species (R2 = 0.03, P = 0.30) are also not significant.
(EPS)
(XLSX)
(XLSX)
(XLS)
Data Availability Statement
All data contained in the paper are available as supporting information with the paper or their sources are detailed in the paper.