Abstract
Autism spectrum disorder (ASD) is a group of highly genetic neurodevelopmental disorders characterized by language, social, cognitive, and behavioral abnormalities. ASD is a complex disorder with a heterogeneous etiology. The genetic architecture of autism is such that a variety of different rare mutations have been discovered, including rare monogenic conditions that involve autistic symptoms. Also, de novo copy number variants and single nucleotide variants contribute to disease susceptibility. Finally, autosomal recessive loci are contributing to our understanding of inherited factors. We will review the progress that the field has made in the discovery of these rare genetic variants in autism. We argue that mutation discovery of this sort offers an important opportunity to identify neurodevelopmental mechanisms in disease. The hope is that these mechanisms will show some degree of convergence that may be amenable to treatment intervention.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-015-0363-9) contains supplementary material, which is available to authorized users.
Keywords: Autism spectrum disorders, Intellectual disability, Autism genetics, Rare genetic variants, Neurodevelopment
Introduction
Autism spectrum disorder (ASD) is a group of neurodevelopmental disorders presenting genetic and phenotypic heterogeneity across the affected individuals. Within ASD, affected individuals exhibit a combination of abnormalities in adaptive and social functioning, language, and cognitive abilities [1].
Genetic studies indicate that numerous genes across the genome contribute to the risk of ASD. There has been substantial recent progress in the discovery of rare mutations. For example, many studies have identified a large number of copy number variants (CNVs) and single nucleotide variants (SNVs) to be associated with disease. De novo mutations associated with ASD were found to be rare and distributed over many genes across the genome [2–5]. Recessive loci in ASD have also been studied, reflecting the role of inherited variation [6–10].
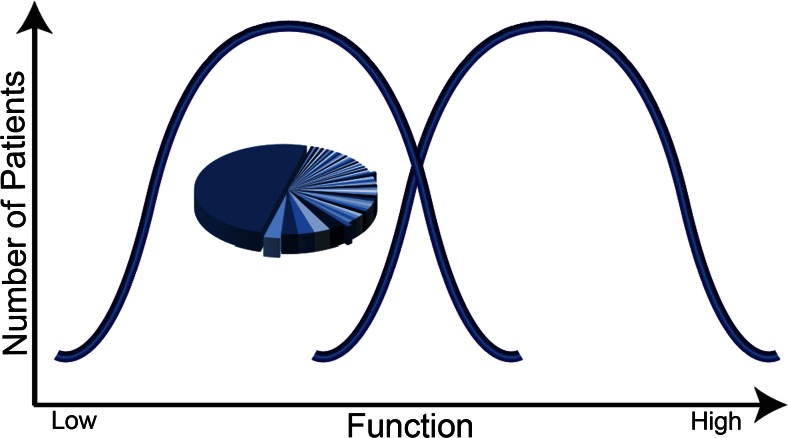
One hypothetical model with some support is that autism may reflect approximately 2 groups, the first involving lower-functioning patients and a second involving higher-functioning patients (Fig. 1). This model would further hypothesize that lower-functioning autism may be constituted by a genetic cause that reflects a smaller number of individually rare yet highly penetrant mutations [some even reflecting monogenic causes such as fragile X syndrome (FXS) and others], whereas higher-functioning autism might reflect a genetic architecture involving a large number of common susceptibility variants of minor effect. An example of support for this hypothesis is that some forms of rare variation appear to be enriched largely in lower-functioning patients [3, 9, 11–15]. Figure 1 represents a proposed model that may help guide approaches to dissecting the heterogeneity in autism, yet further work is necessary to provide more substantial proof for this model.
Fig. 1.
Model on genetic architecture of autism spectrum disorder (ASD; adapted from Morrow et al. [11]). The heterogeneity in autism may be dissected into 2 groups, approximately lower-functioning and higher-functioning groupings. One hypothesis with some support is that the genetic architecture of lower-functioning autism may involve a relatively smaller number per patient of rare and highly penetrant mutations. By contrast, the genetic architecture of higher-functioning autism may involve a large number of common variants with minor effect. The pie chart indicates the multitude of individually rare, highly penetrant genetic causes that may be identified in lower-functioning ASD. Of the cases of autism, at least 70 % have no known genetic mechanisms [16]. The data in the pie chart are hypothetical to reflect a large number of individually rare loci identified in cases; the largest piece of the pie represents a majority of cases in whom a genetic mutation is still not identified
In this review, we highlight the progress in discovery of rare ASD-associated mutations. There is also support for an important role for common variation in autism [17]. However, here we will highlight progress in discovery of rare mutations that can often pinpoint coding changes in specific genes. We argue that these discoveries provide critical traction for investigation of disease-relevant neurodevelopmental mechanisms. As convergent pathways emerge from the study of enough such rare variants, it is possible that some of these pathways may be targeted for new drug development.
Monogenic Disorders with Autistic Symptoms
There is a variety of monogenic disorders that include autistic symptoms. These disorders provide important models that may be informative about complex forms of autism and provide clues regarding the pathophysiology of autism symptoms. It is important to acknowledge, however, that these syndromes most often constitute a broader range of developmental anomalies than autism alone. Below we review a subset of these syndromes, with an emphasis on how they might inform the field to potential neurodevelopmental mechanisms. All monogenic disorders explained below are summarized in Table 1.
Table 1.
Monogenic disorders associated with autistic symptoms
| Monogenic disorder | Gene | Protein function | Role within brain development | References |
|---|---|---|---|---|
| Angelman syndrome (OMIM 105830) | UBE3A | Plays a role in the ubiquitination of proteins targeted for degradation | Affects circuit development and excitatory/inhibitory balance | [18–26] |
| Autism with macrocephaly (OMIM 605309) | PTEN | Inhibits PI3K/AKT signaling pathway | Affects development of neuronal and synaptic morphology | [27, 28, 30, 31, 34–37] |
| CHARGE syndrome (OMIM 214800) | CHD7 | Influences chromatin structure and gene expression to affect early embryonic development | Regulates genes involved in neural crest cell and axon guidance | [42–50] |
| Christianson syndrome (OMIM 300243) | SLC9A6 | Modulates intraendosomal pH through role as a sodium–proton exchanger | Affects neuronal arborization and synapse development through alterations in signaling and trafficking | [51–54] |
| Fragile X syndrome (OMIM 300624) | FMR1 | Affects protein translation by binding to RNA | Represses translation of specific RNAs at the postsynapse, affecting synaptic plasticity | [55–60] |
| Neurofibromatosis type 1 (OMIM 162200) | NF1 | Inhibits Ras/MAPK signaling pathway | Affects synaptic plasticity as a result of dendritic spine localization | [61–68] |
| Rett syndrome (OMIM 312750) | MECP2 | Binds to CpG sites and interacts with histone deacetylases to repress transcription | Affects circuit function and excitatory synaptic activity within cortex | [71–76] |
| Timothy syndrome (OMIM 601005) | CACNA1C | Mediates the flux of calcium ions through the Cav1.2 channel | Affects connectivity through influence on neuronal differentiation and synaptic plasticity | [78, 79] |
| Tuberous sclerosis (OMIM 191100) | TSC1/2 | Inhibit mTOR signaling when complexed together | Affects neuronal development, including cellular growth and size | [80–87] |
CHARGE = C, coloboma of the iris/retina; H, heart defects; A, atresia of the choanae; R, retardation of growth/development; G, genital abnormalities; and E, ear abnormalities; PI3K = phosphoinositide 3-kinase; AKT = protein kinase B; MAPK = mitogen-activated protein kinase; mTOR = mammalian target of rapamycin
Angelman Syndrome
First described in 1965, Angelman syndrome (AS) is a genetic form of intellectual disability (ID). Phenotypic presentation includes abnormal gait, frequent laughter, seizures, microcephaly, and craniofacial abnormalities. Despite the range of symptoms, individuals with AS are often recognized by a generally happy disposition [18, 19]. AS affects between 1 in 15,000 and 1 in 40,000 individuals [20, 21]. Some studies have provided support to indicate that approximately 60% of patients may also be diagnosed with autism [22].
AS is an example of a condition caused by genetic imprinting, as it results from loss-of-function mutations or deletions involving the maternal copy of UBE3A. UBE3A is located on human chromosome 15 and encodes ubiquitin-protein ligase E3A (UBE3A). UBE3A is involved in the ubiquitination of other proteins within the cell, thereby targeting them for degradation. Evidence suggests that UBE3A has 2 roles within the process of ubiquitination—the first in catalyzing complex formation and the second in recognizing substrate specificity [19, 23, 24].
Within the brain, UBE3A has been shown to affect circuit development and the regulation of excitatory/inhibitory balance. One UBE3A target, activity-regulated cytoskeleton-associated protein, mediates surface expression of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors (AMPARs) at excitatory synapses. In the absence of UBE3A expression, activity-regulated cytoskeleton-associated protein degradation is decreased, resulting in increased internalization of AMPARs and the development of impaired circuit function [25]. It has also been demonstrated that UBE3A plays a role in inhibitory synaptic function within the neocortex. Loss of maternally derived UBE3A leads to a vesicle cycling defect at inhibitory synapses causing a build-up of vesicles at the axon terminal and decreased synaptic transmission. As the inhibitory synaptic deficit is more severe than that of excitatory synapses, the ultimate consequence is an excitatory/inhibitory imbalance [26]. Interestingly, duplication of the same region of chromosome 15, involving UBE3A and additional genes, is also observed in another autism-like syndrome known as 15q duplication syndrome [29].
Autism with Macrocephaly
Autism with macrocephaly (also known as Cole–Hughes syndrome) is a monogenic form of autism with progressive postnatal macrocephaly [30–32]. Affected individuals can present with ID and language/social delays, as well as obesity, retarded bone growth, and facial anomalies [30, 32]. Of all individuals diagnosed with autism, it is estimated that 20% may have macrocephaly [33].
Autism with macrocephaly can be caused by dominant, partial loss-of-function mutations in PTEN, encoding the phosphatase and tensin homologue protein on chromosome 10 (PTEN) [31, 34, 35]. Although reports of the exact incidence vary, it is estimated that between 5% and 17% of individuals with autism and macrocephaly carry a PTEN mutation [31, 34]. PTEN is a tumor suppressor gene, and the PTEN protein functions as a dual-specificity phosphatase, capable of acting on both tyrosine and threonine/serine residues. Although it is active within several pathways involving cell growth, cell cycle arrest, and apoptosis, one key function is inhibition of phosphoinositide 3-kinase/protein kinase B signaling [31, 34, 36].
Within the brain, PTEN affects the development of neuronal and synaptic morphology. Analyses of PTEN-deficient neurons in mice revealed an increase in the size of neuronal cell bodies and arbors, as well as an increase in dendritic spine density. Synaptic structures throughout the cortex and cerebellum were enlarged, and axons showed evidence of severe myelin thickening. Loss of PTEN expression also resulted in the development of decreased synaptic functionality, or weakening of both synaptic transmission and synaptic plasticity within the hippocampus [37, 38].
PTEN mutations, often within the germline, have also been implicated in other disorders involving autistic symptoms, collectively known as PTEN hamartoma tumor syndromes. Notable examples of PTEN hamartoma tumor syndromes include Cowden syndrome and Bannayan–Riley–Ruvalcaba syndrome, and a defining feature of both disorders is the presence of benign tumors, known as hamartomas, throughout the body [39]. In Cowden syndrome, these hamartomas frequently become malignant, and affected individuals present with macrocephaly, ID, and autistic traits [40]. Characteristic features of Bannayan–Riley–Ruvalcaba syndrome also include macrocephaly, ID, and autistic traits, as well as thyroiditis, vascular malformations, and delayed motor development [31, 41].
CHARGE Syndrome
CHARGE, first described in 1979, is a syndrome encompassing a constellation of congenital anomalies. The term CHARGE is an acronym for the syndrome’s six core features: C, coloboma of the iris/retina; H, heart defects; A, atresia of the choanae; R, retardation of growth/development; G, genital abnormalities; and E, ear abnormalities. Occurring once in every 8500–10,000 live births [42, 43], CHARGE syndrome also involves a range of secondary features, including deafness, laryngomalacia, vestibulocochlear defects, facial nerve palsy, and oral clefts [44–47]. Many individuals with CHARGE syndrome are reported to exhibit autistic-like behaviors, with an estimated 27.5% meeting classification for autism [45].
Although an initial report implicated a mutation in SEMAPHORIN 3E [48], the majority of CHARGE syndrome cases have since been shown to result from dominant, loss-of-function mutations in CHD7, located on human chromosome 8 and coding for chromodomain helicase DNA-binding protein-7 (CHD7). Members of the CHD protein superfamily affect development of the early embryo through their influence on chromatin structure and gene expression. CHD7 is expressed ubiquitously throughout a range of tissue types, resulting in the variety of systems affected in CHARGE syndrome [44, 49].
Of particular relevance to the field of neurodevelopment is the role of CHD7 in the regulation of genes involved in the guidance of neural crest cells and axons. In the absence of CHD7 expression, several genes were shown to be misregulated, including members of the semaphorin and ephrin families [50].
Christianson Syndrome
Christianson syndrome (CS), first noted in 1999 in a 5-generation South African pedigree, is an X-linked recessive neurodevelopmental disorder found in males [51]. Although the exact prevalence is not known, CS is estimated to affect between 1 in 16,000 and 1 in 100,000 individuals [52]. A majority of CS individuals are nonverbal and present with epilepsy, ID, and ataxia. Additional core symptoms are postnatal microcephaly and hyperkinetic behavior. Additional symptoms may include eye movement dysfunction, gastroesophageal reflux disease, low muscle tone, cerebellar atrophy, and phenotypic regression [51–53]. In one study of phenotypic diversity within CS, 43% of affected individuals received a clinical diagnosis of autism, and a majority of patients present with autistic symptoms [52].
CS is the result of likely loss-of-function mutations in SLC9A6, encoding the Na+-H+ exchanger 6 (NHE6) protein. NHE6 functions as an endosomal sodium–proton exchanger, exchanging internal hydrogen ions for external sodium ions. NHE6 is localized to early, recycling, and late endosomes, and it has been shown to modulate intraendosomal pH [54].
Evidence reveals that NHE6 also plays a role in the regulation of neuronal circuit development. In the absence of NHE6 expression, brain-derived neurotrophic factor/tropomyosin receptor kinase B signaling is attenuated, which is associated with impoverished axonal and dendritic arborization. Additionally, loss of NHE6 causes reductions in both synapse number and strength, thereby diminishing functional connectivity [54].
FXS
FXS is an X-linked disorder characterized by cognitive, behavioral, and physical features. FXS occurs most often in males, with an approximate incidence of 1 in 4000 (1 in 8000 females) [55]. Cognitive symptoms include ID and language deficits, whereas behavioral symptoms include hyperactivity and social deficits. Prominent physical features are macro-orchidism, enlarged ears, and an elongated face [56]. According to results of a national parent survey, approximately 46% of males and 16% of females affected by FXS are also diagnosed with autism [57].
FXS is caused by mutation in FMR1, involving an expansion of CGG repeats and subsequent gene silencing by hypermethylation [58]. FMR1 is located at a fragile site on the X chromosome and codes for the fragile X mental retardation protein (FMRP) [58]. FMRP is an RNA-binding protein with robust expression in neurons; it is found at the postsynapse within dendrites, where it affects local protein translation [59]. Its loss has been shown to increase long-term depression in mouse hippocampus, suggesting that FMRP may affect protein synthesis by repressing translation of a specific group of mRNAs [60].
Neurofibromatosis, Type 1
Affecting as many as 1 in 2700 newborns [61], neurofibromatosis type 1 (NF1) is a monogenic disorder characterized by both physical and cognitive symptoms. Hallmarks of the syndrome include café-au-lait spots and benign nerve tumors known as neurofibromas. Additional physical symptoms include freckling of the groin and/or underarm, Lisch nodules of the iris, optic pathway gliomas, and bone lesions [62]. Approximately 80% of individuals with NF1 exhibit deficits in cognitive functioning [63]. Poor social skills, signs of attention deficit hyperactivity disorder (ADHD), and difficulties with executive function are also observed in affected individuals. In addition, it is estimated that at least 26% of individuals with NF1 meet the criteria for autism [64].
NF1 is the result of a dominant, loss-of-function mutation in NF1, located on human chromosome 17 and encoding the neurofibromin protein [65–67]. Neurofibromin negatively regulates the Ras–mitogen-activated protein kinase (MAPK) pathway, and mutations in NF1 lead to hyperactivation of the signaling cascade. As neurofibromin is located postsynaptically in dendritic spines, excessive activation of the pathway results in altered synaptic plasticity, thought to be the cause of the cognitive aspects of the NF1 phenotype [64, 68]. An additional form of neurofibromatosis is neurofibromatosis type 2, which is caused by mutations in NF2, encoding the merlin protein. Neurofibromatosis type 2 is generally characterized by tumors within the brain, cranial nerves, and spinal cord [69]. Although related, it is a disorder distinct from NF1, with few current publications addressing a potential association with autistic symptoms [70].
Rett Syndrome
Rett syndrome (RTT) is a progressive neurodevelopmental disorder affecting an estimated 1 in 10,000 individuals, almost all of whom are female [71, 72]. Individuals with RTT develop typically for the first 6–18 months of life, after which they experience a period of regression, losing speech and deliberate use of the hands. Phenotypic presentation of the syndrome also includes microcephaly, ataxia, epilepsy, breathing difficulties, and stereotyped hand movements [73], and up to 40% of individuals with RTT meet the criteria for autism [74].
In 1999, researchers identified dominant, loss-of-function mutations in the X-chromosome gene MECP2, encoding methyl-CpG-binding protein 2 (MeCP2), as the cause of RTT [71]. MECP2 undergoes X-inactivation, resulting in the range of phenotypic severity seen within the syndrome. MeCP2 consists of 2 functional domains—a methyl-CpG binding domain and a transcriptional repressor domain. When MeCP2 is bound to chromatin, the transcriptional repressor domain interacts with histone deacetylases, resulting in transcriptional repression of target genes containing CpG residues [71]. More recent evidence also supports the role of MeCP2 in gene activation [75].
Within the brain, MeCP2 is involved in circuit function and the balance between excitation and inhibition. Evidence demonstrates that in the absence of MeCP2 expression, excitatory synaptic activity within cortex is reduced. Cortical pyramidal neurons exhibit a decrease in both spontaneous firing rate and amplitude of miniature excitatory postsynaptic currents [76].
Just as loss of MeCP2 expression can lead to dysfunction, so too can MECP2 duplication. Increased MECP2 dosage results in a rare disorder distinct from RTT, known as MECP2 duplication syndrome. MECP2 duplication syndrome causes severe ID and progressive spasticity in all affected males. Additional clinical symptoms include impaired or absent speech, impaired or absent walking ability, seizures, and premature lethality [77].
Timothy Syndrome
Timothy syndrome (TS) is a monogenic disorder characterized primarily by cardiac arrhythmias and dysfunction of multiple organ systems. Symptoms include syndactyly, elongation of the cardiac QT interval, congenital heart defects, hypoglycemia, immune system deficiency, and developmental delays. TS and the associated heart defects are often lethal, with an average age of 2.5 years at death. In 1 study of surviving children, 60% met criteria for autism and 80% met criteria for ASD [78]. Owing to the syndrome’s rarity, the exact prevalence of TS is unknown.
Timothy syndrome is caused by gain-of-function dominant, frequently de novo mutations in CACNA1C, located on human chromosome 12 and encoding the α-1 subunit of the Cav1.2 L-type calcium channel. The Cav1.2 channel is voltage-dependent and is expressed in the heart and throughout the brain. When CACNA1C is mutated, prolonged abnormal calcium current occurs as the result of a failure in channel inactivation [78].
CACNA1C mutations have multiple functional consequences within the central nervous system. Loss of Cav1.2 mRNA expression in the forebrain leads to a loss of late long-term potentiation, as well as decreased activation of signaling pathways such as MAPK/extracellular regulated kinase. Additionally, the CACNA1C mutation seen in TS causes defects in differentiation of projection neurons, resulting in the development of impaired connectivity [79].
Tuberous Sclerosis Complex
Tuberous sclerosis complex (TSC) is an autosomal dominant disorder marked by benign tumors throughout multiple organ systems. These tumors, known as hamartomas, have an unpredictable distribution, but are often found within the brain, the skin, the heart, the kidneys, and the lungs [80, 81]. TSC is associated with a variety of cognitive and developmental deficits, including ID, ADHD, and epilepsy. TSC is estimated to affect as many as 1 in 5800 newborns [82], approximately 40% of whom are diagnosed with ASD [83, 84].
TSC is the result of a loss-of-function mutation in 1 of 2 TSC genes—TSC1, located on human chromosome 9, and TSC2, located on human chromosome 16. TSC1 encodes the hamartin protein, whereas TSC2 encodes a protein known as tuberin, both of which are suggested to function as tumor suppressors [80, 81]. The TSC phenotype in individuals with germline TSC mutations may be explained by the “second hit theory”. Sporadic somatic mutation of the remaining copy of TSC leads to a loss of heterozygosity in a given cell, resulting in tumor formation [85].
As the phenotypic presentation of TSC is similar regardless of which gene is mutated, hamartin and tuberin were presumed to affect the same cellular pathway [81]. The 2 proteins have since been shown to form a complex responsible for inhibition of the mammalian target of rapamycin signaling pathway [86]. In the absence of hamartin/tuberin, there is overactivation of the mammalian target of rapamycin pathway, resulting in abnormal development of neurons, including cellular enlargement [87].
With regard to all of the above monogenic syndromes, one concern about the association of these disorders with autism is that it can be challenging to diagnose autistic symptoms in the setting of severe ID. However, in addition to the presentation of autistic symptoms in the context of these monogenic disorders, there is other evidence that supports a link between these monogenic disorders and idiopathic autism likely due to complex inheritance. For one example, FMRP binds to the mRNA of genes in which mutations have been found in idiopathic autism [88]. Additionally, mutations in CHD7 and its binding partner CHD8, for example, have also been found in idiopathic autism [3]. Rare variants in CHD7 are thought to confer increased autism risk, even in the absence of a CHARGE syndrome diagnosis [89]. Further still, mutations in genes highly related to MECP2, such as MBD5, have been found in idiopathic autism [90].
CNVs in ASD
CNVs have been essential to our understanding of autism genetics. Advances in genomic microarray technology coupled with decreasing cost have allowed for greater testing of individuals and identification of many CNVs in autism [91–93]. CNVs refer to deletions or duplications of genomic segments. They are either inherited from a parent or occur de novo owing to germline alterations. Although CNVs are a source of normal human genetic variation [94, 95], a seminal study in 2007 demonstrated autism cases have an increased burden of de novo CNVs [96]. The authors found simplex families (i.e., only 1 affected child) had a significantly greater rate of de novo CNVs (10%) compared with multiplex families (i.e., multiple affected children) and control families (3% and 1%, respectively). Recent studies of simplex families using microarrays with greater resolution found a burden rate of de novo CNVs from 5.8–7.9% in probands to 1.7–2.0% in unaffected siblings [97, 98]. Furthermore, an increased CNV burden was identified in the same loci encompassing balanced chromosomal abnormalities, such as translocations and inversions, in an autism/neurodevelopmental disorder cohort [99].
Large case–control studies have been employed to elucidate specific loci associated with autism susceptibility. This research strategy compares the frequency of specific CNVs across the genome in probands with autism to unaffected controls. Notable recurrent CNVs include 1q21.1 [98, 100, 101], 7q11.23 [98], 15q13.3 [98], 16p11.2 [98, 102–104], 17q12 [101, 105], and 22q11.2 [98, 106]. Recurrent CNVs may arise as a result of nonallelic homologous recombination whereby regions flanked by segmental duplications may be prone to deletions and duplications during meiosis [107, 108]. There is considerable overlap among recurrent CNVs in autism and other cognitive disorders such as ID, schizophrenia, and epilepsy [109]. This suggests potential shared pathologic mechanisms underlying various neurodevelopmental disorders.
It is important to consider gene expression changes that may result from CNVs. In fact, CNVs usually represent polygenic loci whereby a series of contiguous genes are likely altered by gene dosage, that is, there is a reduction or increase in transcript and protein levels. Therefore, the regulation of the precise level of a series of genes is altered. In this way, while a single gene within a CNV may be informative with regard to mechanisms, it is most likely that these genes implicated by multigenic CNVs act interactively. Recently, there has also been increased focus on the possibility that microRNAs found within CNVs (which also regulate a gene expression program in trans) may be another important part of the polygenic mechanisms resulting from changes in gene copy number [110]. Another interesting feature of CNV disorders is that in some cases, deletions and duplications at the same locus can cause overlapping, reciprocal or quite distinct phenotypes [102, 104, 111–112]. Finally, some recurrent ASD CNVs exhibit incomplete penetrance whereby they are observed in unaffected siblings and control populations [113].
Recurrent CNV Disorders in Autism
1q21.1
CNVs in the 1q21.1 locus are enriched in individuals with autism [98, 100, 101, 114]. Deletions and duplications have been reported in approximately 0.2% (25 in 5218 and 27 in 16,557 subjects) and 0.1% (9 in 5218 and 17 in 16,557 subjects), respectively, of patients from large cohorts with ID, ASD, and congenital anomalies [100, 114]. The 1q21 locus is roughly 1.35 Mb, consisting of at least 7 genes [100]. Probands with deletions are characterized with mild-to-moderate developmental delay, microcephaly, joint hypermobility, congenital heart abnormalities, hypotonia, seizures, cataracts, and dysmorphic features (e.g., frontal bossing and bulbous nose) [100, 114]. Probands with the reciprocal duplication are characterized with mild-to-moderate developmental delay, autistic behaviors, relative macrocephaly and dysmorphic features (e.g., frontal bossing and hypertelorism). The 1q21.1 CNV is incompletely penetrant with variable expressivity as parents and siblings may also have the same CNV yet display none of the associated symptoms [100, 114].
Dosage of the HYDIN paralog located in this region may be important for influencing head size and growth [114]. Additionally, increased dosage of the CON1 subtype of the DUFF1220 protein domain is correlated with greater autism severity [115].
7q11.23
Duplications in 7q11.23 are significantly associated with autism susceptibility in a simplex cohort with a frequency of 0.09% in 3816 ASD probands [98]. These de novo duplications spanned 1.37 Mb, encompassing 22 genes. Individuals with 7q11.23 duplications exhibit a highly heterogeneous phenotype. Common features include ASD, cognitive deficits, severe expressive language delay, facial dysmorphisms (e.g., short philtrum and thin lips), anxiety, behavioral problems, and hyperactivity [98, 116, 117].
This CNV locus is especially interesting because the reciprocal deletion causes Williams–Beuren syndrome (WBS), a genomic syndrome notable for excessively social behavior. WBS is characterized by a hypersocial personality, cognitive deficits (particularly in visuospatial skills), anxiety, facial dysmorphisms (e.g., flat nasal bridge, upturned nose, and delicate chin), cardiovascular disease, growth retardation, and connective tissue abnormalities [118]. The deleted region is referred to as the WBS chromosome region 1.5 Mb (95%) or 1.8 Mb (5%) [119].
Using induced pluripotent stem cell (iPSC) methods, Adamo et al. [120] found that GTF2I mediates the dose-dependent transcription of 7q11.23. These researchers also identified other gene candidates, such as PDLIM1, which is associated with neurite outgrowth; MYH14, associated with hearing impairment; and BEND4, a transcription factor containing a BEN domain. Transcriptional analysis using lymphocytes from the Simons Simplex Collection (SSC) identified misregulated genes, including BCL7B, EIF4H, and LAT2 [121]. Pathway analysis of misregulated genes was notable for forebrain development, determination of bilateral symmetry, and hippocampal development.
15q11-q13
15q11-13 duplications constitute one of the most common genetic findings in autism [98, 104, 106, 122]. The 15q11-q13 locus contains 5 recurrent breakpoints (BP1–BP5), with BP2–BP3 being particularly associated with autism [98, 122]. This region is also notable for imprinting. Deletions in BP2–BP3 are associated with either Prader–Willi syndrome (PWS; paternal) or AS (maternal), depending on parental origin of the affected chromosome [123]. The phenotype is heterogeneous and may include ASD, hypotonia, speech and motor delays, anxiety, emotional lability, and hyperactivity [124]. As described above, one of the strongest gene candidates for autism in this locus is UBE3A. Some studies have reported that PWS caused by maternal uniparental disomy was more likely to be associated with autism symptomatology than PWS caused by paternal deletions [125, 126]. This suggests increased dosage of genes in the maternal locus, such as UBE3A, may increase the risk of autism. However truncating mutations of a gene in the PWS paternal-only expressed region, MAGEL2, was recently found in 4 patients diagnosed with ASD [127].
16p11.2
Deletions and duplications of 16p11.2 have been associated with autism and related neurodevelopmental disorders [98, 102, 103], yet are incompletely penetrant [102, 128]. This 593-kb segment encompasses 25 genes and accounts for approximately 1% of cases of ASD [98, 102]. Both deletions and duplications are associated with language delays, ASD, behavioral problems, congenital anomalies, and seizures [129–131], although the deletion may appear more commonly in autism [102, 132], and the duplication appears to be more common in schizophrenia [112]. Contrasting phenotypes are observed: deletions are associated with increased brain volume/macrocephaly and increased body mass index/obesity, while duplications are associated with decreased brain volume/microcephaly and decreased body mass index [129, 133, 134]. Structural brain alterations were also found in a dosage-dependent manner based on whether the 16p11.2 locus was deleted or duplicated [135]. Brain regions involved in reward circuitry such as the striatum, mediodorsal thalamus, orbitofrontal cortex, and insula had significantly greater gray matter volume in individuals with deletions compared with duplications. However, individuals with duplications had significantly greater gray matter volume in language regions such as the middle, superior temporal gyrus, and caudate.
Overexpresssing the human transcript of a gene located in 16p11.2, KCTD13, in zebrafish resulted in a microcephaly phenotype, whereas knocking down its expression resulted in a macrocephaly phenotype [136]. Genome-wide transcriptome analysis revealed a significant positive correlation between the 16p11.2 locus and expression of MAPK3, YPEL3, CORO1A, and KCTD13 [121]. Head circumference was also highly correlated with expression of TAOK2, CORO1A, KCTD13, and QPRT. Another transcriptome analysis of 16p11.2 deletions and duplications in 1) cortical mouse tissue and 2) human lymphoblasts from multiplex autism families found dysregulated gene expression in synaptic functioning (e.g., NRXN1, NRXN3), chromatin modification (e.g., CHD8, EHMT1, MECP2), transcriptional regulation (e.g., TCF4, SATB2), and ID (e.g., FMR1) [137].
22q11.2
The 22q11 locus is one of the most robust CNV findings in neurodevelopmental genetics. Deletions and duplications of 22q11.2 are strongly associated with autism across multiple large case–control studies [98, 104, 106]. However, CNVs in this region are associated with a range of phenotypes and are incompletely penetrant [113]. 22q11 deletion syndrome (22q11DS), also referred to as Velocardiofacial or DiGeorge syndrome, represents one of the strongest genetic risk factors for neuropsychiatric disorders. Historically, 22q11DS constitutes a major risk factor for a schizophrenia-like condition [138]. Approximately 85% of individuals with 22q11DS have the recurrent 3-Mb deletion, encompassing roughly 60 genes [139]. In the largest study of individuals with 22q11DS, Schneider et al. [140] examined the psychiatric profile of 1402 participants (aged 6–68 years) throughout their lifespan. The prevalence of schizophrenia spectrum disorders greatly increased from early adulthood (23.5%) to adulthood (41.7%). ASD prevalence was highest in adolescents (26.5%). A subset of this cohort available for cognitive testing showed a mean full-scale IQ of 71.25, with almost half with a full-scale IQ > 70. Deficits in executive functioning have also been identified, specifically related to working memory and attention [141].
There are a number of genes in this locus potentially contributing to autism and other phenotypes. COMT may influence cognitive functioning and psychosis [142–144]. Epistasis may also occur between COMT and PRODH, and is likely to affect behavior [139]. Heterozygous Tbx1 mice displayed impairments in social interactions, ultrasonic vocalizations, and working memory [145]. Haploinsufficiency of Tbx1 and Gnb1l in a 22q11DS mouse model appears to be responsible for decreased prepulse inhibition, a sensorimotor gating measure associated with psychiatric disorders [146]. GNB1L is also associated with ASD [147]. Ranbp1 is associated with microcephaly and important for cortical progenitor proliferations in layer 2/3 [148]. Cxcr4 potentially regulates cortical interneuron migration and placement [149]. There are also a number of microRNA-related loci in this region, including miR-185 and DiGeorge syndrome chromosomal (or critical) region 8, a component of the microprocessor complex that is involved in the initial step of miRNA biogenesis [110].
22q13
22q13 deletion syndrome, also known as Phelan–McDermid syndrome, is characterized by global developmental delay, absent or severely delayed speech, ASD, neonatal hypotonia, normal or accelerated growth, and dysmorphic features [150, 151]. Dysmorphic facial features include dolicocephaly, flat midface, wide nasal bridge, and bulbous nose. Cerebellar vermis hypoplasia, enlarged posterior fossa and ventricles, and thin corpus collosum and white matter are common neuroimaging findings [152].
Haploinsufficiency of SHANK3 is thought to be an important locus in Phelan–McDermid syndrome [153, 154]. Furthermore, SHANK3 is also considered a major effect locus in autism [153, 154], with deletions found in at least 0.5% of patients with autism [155]. SHANK3 encodes a master scaffolding protein localized to the postsynaptic density in excitatory synapses [156, 157]. Shank3-null mice exhibit deficits in synaptic functioning such as impaired AMPAR trafficking, glutamatergic transmission, and long-term potentiation [158, 159]. These mice recapitulate aberrant symptoms in social behavior, communication, repetitive behaviors, motor functioning, and learning/memory [158–160]. However, SHANK3 is not the only gene responsible for this 22q13 deletion syndrome [161]. Mice lacking IB2, also known as MAPK8IP2, have abnormal Purkinje cell morphology, and decreased AMPA and increased N-methyl-D-aspartate cerebellar glutamatergic transmission [162]. These mice also exhibit deficits in social, motor, and cognitive functioning. PLXNB2 may also mediate pathological cerebellar symptoms [152].
Other Recurrent CNVs in ASD and CNVs that Identify Individual Genes
A number of other recurrent CNVs are enriched in autism cohorts; also, some recurrent CNVs have identified individual genes. 15q13.3 deletions have been reported in autism [98, 104, 163, 164], with FAN1 being implicated as a putative gene candidate [165]. 17q12 deletions have been identified in individuals with autism [101, 105]. ACACA may be responsible for the ASD phenotype in this CNV [101]. Some CNVs identified in large case–control studies consist of only 1 or a few genes. These include deletions in 1p33 (AGBL4) [104], 2p16.3 (NRXN1) [106], 3p26.2 (SUMF1) [106], 10q23.2 (GRID1) [106], 19q13.33 (CLEC11A, SHANK1, SYT3) [166], and Xp22.1 (DDX53, PTCHD1) [104], as well as duplications in 3p26.3 (CNTN4) [106] and 3q26.31 (NLGN1) [106]. Among the most highly recurrent single-gene CNVs are NRXN1 mutations. Interestingly, both NRXN1 and the postsynaptic binding partners NLGN1-4 have been strongly implicated in autism and reviewed elsewhere [106, 167–170]. MBD5, a methyl-CpG-binding domain gene, has been identified as the causal gene in the 2q23.1 locus, which is characterized by ASD and ID [171, 172]. Deletions encompassing the C-terminus of AUTS2 are associated with autistic features and developmental delay [173]. Finally, deletions involving ASTN2 and TRIM32 at the 9q33.1 locus are enriched in male patients exhibiting a range of neurodevelopmental phenotypes such as ASD, ADHD, obsessive–compulsive disorder, and speech/language delay [174]. Recurrent CNVs are summarized in Table 2.
Table 2.
Recurrent copy number variants in autism
| Locus | Deletion/duplication | Position (hg18) | Number of genes | Reference |
|---|---|---|---|---|
| 1p33 | Deletion | chr 1: 49,685,647–49,770,826 | 1 (AGBL4) | [104] |
| 1q21.1 | Deletion | chr 1: 145,013,719–146,293,282 | 14 | [98*, 104, 175] |
| Duplication | chr 1: 145,013,719–146,293,282 | 14 | [98*, 101, 104, 175] | |
| 2p16.3 | Deletion | chr 2: 51,002,576–51,157,742 | 1 (NRXN1) | [98*, 101, 106] |
| 3p26.2 | Deletion | chr 3: 4,199,731–4,236,304 | 1 (SUMF1) | [106] |
| 3p26.3 | Duplication | chr 3: 2,548,148–2,548,531 | 1 (CNTN4) | [106] |
| 3q26.31 | Duplication | chr 3: 174,754,378–174,771,975 | 1 (NLGN1) | [106] |
| 7q11.23 | Duplication | chr 7: 72,411,506–73,782,113 | 22 | [98*, 104] |
| 8p23.3 | Duplication | chr 8: 710,491–1,501,580 | 1 (DLGAP2) | [98*, 103, 104] |
| 10q23.2 | Deletion | chr 10: 87,941,666–87,949,029 | 1 (GRID1) | [106] |
| 11q13.3 | Deletion | chr 11: 70,154,458–70,187,872 | 1 (SHANK2) | [98*, 104] |
| 15q11-13 | Duplication | chr 15: 21,240,037–26,095,621 | 12 | [98*, 104, 106] |
| 15q13.2 -15q13.3 | Deletion | chr 15: 28,723,577–30,231,488 | 6 | [98*, 104] |
| 16p11.2 | Deletion | chr 16: 29,578,715–30,001,681 | 26 | [98, 104, 106] |
| Duplication | chr 16: 29,578,715–30,001,681 | 26 | [98*, 104, 106] | |
| 17q12 | Deletion | chr 17: 31,893,783–33,277,865 | 15 | [98*, 104, 105] |
| 19q13.33 | Deletion | chr 19: 55,808,307–55,935,995 | 3 (CLEC11A, SHANK1, SYT3) | [166] |
| 22q11.2 | Deletion | chr 22: 17,265,500–19,786,200 | 56 | [98*, 104, 175] |
| Duplication | chr 22: 17,265,500–19,786,200 | 56 | [98*, 104, 106] | |
| 22q13 | Deletion | chr 22: 49,243,247–49,465,88 | 16 | [98] |
| Xp22.1 | Deletion | chr X: 22,829,183–23,214,712;chr X: 23,116,188–23,280,628 | 2 (DDX53, PTCHD1) | [104] |
Chr = chromosome
*Denotes study reporting the copy number variant position and number of genes. For copy number variants with both deletions and duplications, the “combined” coordinates from Sanders et al. [98] are reported
De Novo SNVs in ASD
Advances in sequencing technologies have been providing substantial information on de novo SNVs in ASD. Recent studies have focused on whole-exome sequencing (WES) of simplex pedigrees to identify de novo SNVs. Those studies have reported hundreds of candidate genes with coding variants; however, for the vast majority of these candidates only a single mutation was identified in most genes (the “n = 1 problem”). Such single-hit genes provide important data, but it is difficult to interpret whether they are truly associated with ASD without recurrent hits [3–5, 176]. Sanders et al. [4] developed a model suggesting that the likelihood of observing 2 nonsense/splice site de novo mutations in the same gene in probands is not by chance. By using the same permutation test, it was indicated that a candidate gene with ≥ 2 disruptive (nonsense, frameshift, or splice site) mutations has a higher probability of being associated with ASD than a gene with 1 disruptive de novo mutation. Based on this approach, a growing number of genes have been implicated to date through the identification of recurrent, deleterious de novo SNVs in pedigree-based WES. For example, sequencing was performed on 2446 families with autistic patients from SSC to identify 44 candidate genes. In 6 genes, including CHD8, DYRK1A, GRIN2B, TBR1, PTEN, and TBL1XR1, recurrent disruptive mutations were detected (Table 3) [177]. Following this study, almost 3500 probands and their unaffected siblings were resequenced for 64 candidate genes in a case–control design. CHD2, ADNP, SYNGAP1, TRIP12, and PAX5 were found to have recurrent mutations in multiple probands [15]. Recurrent de novo mutations (i.e., ≥ 2 in the same gene), which include protein truncating and/or deleterious missense, to date found to be associated with ASD are summarized in Table 3.
Table 3.
De novo recurrent mutations (≥2) in genes associated with autism spectrum disorder. These mutations included protein truncating and/or deleterious missense
| Gene | Protein | Protein function* | OMIM phenotype | References |
|---|---|---|---|---|
| ADNP | Activity-dependent neuroprotector homeobox protein | Contributes to cell survival during modulation of p53 activity, may function as a transcription factor | Mental retardation, autosomal dominant 28, OMIM 615873 | [15, 88, 178] |
| ANK2 | Ankyrin 2 | Connects integral membrane proteins to the underlying spectrin–actin cytoskeleton | Cardiac arrhythmia, ankyrin-b-related, OMIM 600919 | [5, 88, 178, 193] |
| ANKRD11 | Ankyrin repeat domain containing protein | Acts as an inhibitor for ligand-dependent activation of transcription | KBG syndrome, OMIM 148050 | [13, 88] |
| APH1A | Gamma-secretase subunit APH-1A | Cleaves integral membrane proteins as part of the gamma-secretase complex | NA | [176, 178] |
| ARID1B | AT-rich interaction domain containing protein 1B | Has roles in transcriptional activation and repression of certain genes by chromatin remodeling | Mental retardation, autosomal dominant 12, MRD12 OMIM 614562 |
[88, 178] |
| ASH1L | Histone-lysine N-methyltransferase ASH1L | Methylates histone H3 | NA | [178, 193] |
| ASXL3 | Putative polycomb group protein ASXL3 | Acts as part of multiprotein complexes to regulate transcriptionally repressive state of homeotic genes | Bainbridge–Ropers syndrome, OMIM 615485 | [178] |
| BCL11A | B-cell CLL lymphoma/leukemia 11A | Acts as a myeloid and B-cell proto-oncogene | Leukemia, chronic lymphocytic susceptibility, OMIM 109543 | [5, 178] |
| BRCA2 | Breast cancer type 2 susceptibility protein | Has roles in DNA repair and homologous recombination | Glioblastoma 3, OMIM 613029 | [176] |
| CACNA2D3 | Voltage-dependent calcium channel subunit alpha-2/delta-3 | Mediates the influx of calcium ions | NA | [5, 178] |
| CDC42BPB | Serine/threonine-protein kinase MRCK beta | Has roles in the regulation of cytoskeleton reorganization and cell migration | NA | [3, 178] |
| CHD2 | Chromodomain helicase DNA binding protein 2 | Functions as a chromatin remodeling factor | Epileptic encephalopathy, Childhood onset, OMIM 615369 | [15, 88] |
| CHD8 | Chromodomain-helicase-DNA-binding protein 8 | Represses transcription by remodeling chromatin structure | Autism, susceptibility to 18, OMIM 615032 | [3, 15, 88, 177, 178] |
| CTTNBP2 | Cortactin binding protein 2 | Involved in dendritic spinogenesis and spine maintenance through cortactin regulation | NA | [5, 178] |
| CUL3 | Cullin-3 | Has roles in ubiquitination and degradation of certain proteins | Pseudohypoaldosteronism, type IIE OMIM 614496 |
[3, 178, 179] |
| DIP2A | DIP2 disco interacting protein homolog A | Functions in axon patterning in the central nervous system | NA | [5, 88] |
| DSCAM | Down syndrome cell adhesion protein | Has roles in neuronal self-avoidance and axon guidance during development | NA | [88] |
| DYRK1A | Dual specificity tyrosine-phosphorylation-regulated kinase 1A | Influences regulation of nuclear functions of cell proliferation | Down syndrome, OMIM 190685 | [88, 177, 178] |
| ETFB | Electron transfer flavoprotein, beta polypeptide | Has roles in mitochondrial fatty acid and amino acid catabolism as part of the electron transfer flavoprotein | Glutaric acidemia IIB, OMIM 231680 | [178] |
| FAT1 | Protocadherin Fat 1 | Is involved in cellular polarization, cell migration and cell–cell contact | NA | [176] |
| FOXP1 | Forkhead box P1 | Acts as a transcriptional repressor | Mental retardation with language impairment with or without autistic features, OMIM 613670 | [88] |
| GABRB3 | Gamma-aminobutyric acid (GABA) receptor subunit beta-3 | Has roles in GABA-mediated neuronal inhibition as subunit of the GABA-A receptor | Epilepsy, childhood absence susceptibility to; 5, OMIM 612269 | [178] |
| GIGYF1 | GRB10-interacting GYF protein 1 | Has roles in regulation of tyrosine kinase receptor signaling | NA | [88] |
| GRIN2B | Glutamate receptor ionotropic, NMDA 2B | Is involved in excitatory synaptic transmission and plasticity as a subunit of the NMDA receptor | Epileptic encephalopathy, early infantile 27, OMIM 616139 | [15, 88, 177, 178] |
| KATNAL2 | Katanin p60 ATPase-containing subunit A-like 2 | Has roles in reorganization of cellular microtubule arrays | Autism, OMIM 209850 | [3, 4, 88, 178] |
| KCNMA1 | Calcium-activated potassium channel subunit alpha-1 | Has roles in regulation of smooth muscle tone and neuronal excitability as subunit of potassium channels | Generalized epilepsy and paroxysmal dyskinesia, OMIM 609446 | [176] |
| KDM5B | Lysine (K)-specific demethylase 5B | Demethylates H3K4me2 and H3K4me3 | NA | [88] |
| KDM6B | Lysine-specific demethylase 6B | Demethylates histone H3 | NA | [5, 88] |
| KMT2C (MLL3) | Histone-lysine N-methyltransferase MLL3 | Has roles in transcriptional coactivation | Kleefstra syndrome, OMIM 610253 | [3, 178] |
| KMT2E (MLL5) | Histone-lysine N-methyltransferase 2E | Mono- and dimethylates histone H3 | NA | [13, 88] |
| MBD5 | Methyl-CpG binding domain protein 5 | Likely involved in formation or function of heterochromatin | Mental retardation, autosomal dominant 1, OMIM 156200 | [3] |
| MED13L | Mediator of RNA polymerase II transcription subunit 13-like | Functions as part of the mediator complex | Transposition of the great arteries, dextro-looped 1, OMIM 608808 | [5, 88] |
| MIB1 | Mindbomb E3 ubiquitin protein ligase 1 | Functions as an E3 ubiquitin ligase | Left ventricular noncompaction-7, OMIM 615092 | [178] |
| MYO9B | Unconventional myosin-IXb | Has roles in intracellular movements | Celiac disease, susceptibility to, 4, OMIM 609753 | [178] |
| MYT1L | Myelin transcription factor 1-like protein | Functions as a transcription factor associated with neuronal differentiation | NA | [178] |
| NAA15 | N-alpha-acetyltransferase 15, NatA auxiliary subunit | Has roles in N-terminal acetyltransferase activity | NA | [178] |
| NCKAP1 | NCK-associated protein | Functions as part of the WAVE complex | NA | [5, 88] |
| NR3C2 | Mineralocorticoid receptor | Functions as a receptor for mineralocorticoids and glucocorticoids | Pseudohypoaldosteronism, type 1, autosomal dominant, OMIM 177735 | [178] |
| NTNG1 | Netrin G1 | Has roles in axon guidance | Autism, OMIM 209850 | [3] |
| PAX5 | Paired box protein 5 | Has roles in B-cell differentiation, neural development, and spermatogenesis | Bipolar disorder, OMIM 190685 | [3, 15] |
| PHF2 | PHD finger protein 2 | Functions as a lysine demethylase | NA | [88] |
| PHF3 | PHD finger protein 3 | Functions as a transcription factor | NA | [4, 13] |
| POGZ | Pogo transposable element with ZNF domain | Plays a role in mitotic cell cycle progression | NA | [5, 88, 176, 178] |
| PTEN | Phosphatase and tensin homolog | Inhibits PI3K/AKT signaling pathway | Macrocephaly/autism syndrome, OMIM 605309 | [15, 177, 178] |
| RELN | Reelin | Controls cell–cell interactions | Lissencephaly 2, OMIM 257320 | [176, 178] |
| RIMS1 | Regulating synaptic membrane exocytosis protein 1 | Involved in exocytosis | Cone-rod dystrophy 7, OMIM 603649 | [5, 13, 88] |
| SCN2A | Sodium channel protein type 2 subunit α isoform 1 | Mediates the voltage-dependent sodium ion permeability of excitable membranes | Epileptic encephalopathy, early infantile 11, OMIM 613721 | [4, 88, 178, 180] |
| SETD5 | SET domain-containing protein 5 | Likely has roles in encoding a methyltransferase | Mental retardation, autosomal dominant 23, OMIM 615761 | [5, 176, 178, 180] |
| STXBP1 | Syntaxin-binding protein 1 | Has roles in release of neurotransmitters | Epileptic encephalopathy, OMIM 612164 | [180] |
| SUV420H1 | Suppressor of variegation 4-20, homolog 1 | Trimethylates histone H4 | NA | [4, 5, 178] |
| SYNGAP1 | Ras/Rap GTPase-activating protein | Major component of the postsynaptic density | Mental retardation, autosomal dominant 5, OMIM 612621 | [13, 15, 178, 180] |
| TBL1XR1 | F-box-like/WD repeat-containing protein | Functions as a transcription activator | Autism, OMIM 209850 | [177] |
| TBR1 | T-box brain protein 1 | Functions as a transcription factor involved in the regulation of developmental processes | Medulloblastoma, OMIM 155255 | [15, 88, 177, 178] |
| TCF7L2 | Transcription factor 7-like 2 | Functions as a transcription factor in the Wnt signaling pathway | Colorectal cancer, OMIM 114500 | [88] |
| TNRC6B | Trinucleotide repeat containing 6B protein | Plays a role in RNA-mediated gene silencing | NA | [13, 88] |
| TRIO | Trio rho guanine nucleotide exchange factor | Promotes the exchange of GDP by GTP | Schizophrenia, OMIM 181500 | [3, 4, 178] |
| TRIP12 | E3 ubiquitin-protein ligase TRIP12 | Has roles in ubiquitin fusion degradation pathway and DNA repair | NA | [15] |
| VIL1 | Villin | Functions as a structural component of the brush border cytoskeleton | NA | [178] |
| WAC | WW domain containing adaptor with coiled-coil | Links gene transcription to histone 2B monoubiquitination | Leukemia, acute myeloid, OMIM 601626 | [88] |
| WDFY3 | WD repeat and FYVE domain-containing protein 3 | Functions as a conductor for aggregate clearance by autophagy | NA | [5, 88, 178] |
NMDA = N-methyl-D-aspartate; PI3K = phosphoinositide 3-kinase; AKT = protein kinase B; GDP = guanosine diphosphate; GTP = guanosine triphosphate; NA = not available
*Protein function information is based on Gene Cards (www.genecards.org)
Further, the contribution of disruptive SNVs to the ASD risk was shown by burden analysis on SSC pedigrees such as excess number of frameshift indels and loss-of-function mutations in cases compared with controls [3–5, 13]. In a recent study on WES data of 3871 cases of ASD and 9937 ancestry-matched or parental controls, more loss-of-function mutations were observed in cases than in controls. Using the transmission and de novo association statistical model, 7 novel genes, including ASH1L, MLL3, ETFB, NAA15, MYO9B, MIB1, and VIL1, were identified [178].
Understanding how SNVs in implicated genes affect the phenotypic characteristics of ASD is crucial as it is a complex disorder with a variety of presentations across probands. In a recent study on SSC pedigrees, the average number of truncating SNVs was found to be higher in probands with IQs < 100 than in those with IQs ≥ 100. This observation suggests that truncating SNVs likely contribute to lower-functioning ASD [12]. Similarly, a significant decrease in nonverbal IQ was identified in probands with an increased number of truncating mutations [3], and the presence of de novo frameshift indels was observed to be associated with a lower IQ in SSC pedigrees [13]. Interestingly, these studies of de novo SNV burden have shed light on possible epidemiologic factors that may contribute to autism risk. Specifically, multiple studies demonstrated a correlation between increased paternal age and the number of de novo SNVs [3, 13, 179].
Recessive Loci in ASD
There is substantial progress demonstrating the role of de novo CNVs and point mutations in ASD, as explained above. However, the contribution of inherited loci in autism remains to be clarified. The Homozygosity Mapping Collaborative for Autism embarked on studies of recessive loci in pedigrees with recent shared ancestry. This project identified an enrichment of implicated loci with genes that were regulated by neuronal activity [6]. These studies also identified a number of mutations in these pedigrees, for example the endosomal Na+/H+ exchanger 9 (NHE9), glycine metabolic genes, and SYNE1 [6, 181]. Some of the genes implicated showed comparable mutations in autism pedigrees without recent shared ancestry. In addition, some pathways showed evidence supportive of transheterozygous mutations, that is, heterozygous mutations in 2 genes in a common pathway [181]. This genetic mechanism is commonly encountered in model organisms [182]; however, this mechanism of inheritance has not been exhaustively studied.
Homozygosity mapping was also used to identify novel rare, recessive loci in syndromes including autism symptoms. These include a protein truncating mutation in CC2D1A in 9 consanguineous families with severe autosomal recessive nonsyndromic mental retardation [183–185]. CC2D1A is a putative signal transducer involved in positive regulation of the I-κB kinase/nuclear factor-κB cascade. Wild-type protein in lymphoblastoid cells was shown to be absent in the affected subjects [183]. Another example includes homozygous mutations truncating BCKDK, identified in autism with epilepsy and ID in pedigrees with recent shared ancestry. Branched-chain ketoacid dehydrogenase kinase has an important role of phosphorylating and inactivating the E1α subunit of branched-chain ketoacid dehydrogenase complex. Mutations in the subunits of this complex cause accumulation of branched-chain amino acids, leading to maple syrup urine disease [186].
As an extension of traditional homozygosity mapping, another approach is homozygous haplotype mapping, which is used to identify haplotypes within shared runs of homozygosity (ROH) regions [7]. ROHs are regions of the genome with consecutive homozygous single nucleotide polymorphisms. Therefore, ROH reflects coinheritance of a segment of the genome derived from an ancestor shared by the parents and, in this way, they may be a marker of recessive loci. The length of the segment reflects the number of generations back wherein the ancestor was in common such that longer ROH blocks reflect a more recently shared ancestor. Increased ROHs were found to be associated with ID in autism in a population of 2108 families with autistic children from SSC [9]. ROH blocks were identified widely distributed across the genome. An increased burden of ROH was observed in probands with IQs ≤ 70 compared with their unaffected matched siblings, but not in probands with IQs > 70. The authors also identified an association of increased ROH burden for female sex and lower IQ across both sexes in autism [9]. Similarly, in another study on 612 affected children with ID, larger homozygous regions were found in the severe ID case group compared with the nonsevere ID case group. In addition, larger ROH blocks accounted for 20% of homozygosity in individuals within the severe ID case group and only for 6% in individuals within the nonsevere case group [187]. In yet another study in a Taiwanese Han population, ROH was found to be associated with speech delay [188].
In some studies the combination of analysis of ROHs and WES has led to the identification of new candidate gene mutations. For example, in the study by Chahrour et al. [8] ROH analysis and WES on the patients from the selected families showed 4 candidate autism-related genes (in 4 of the 16 probands): UBE3B, CLTCL1, NCKAP5L, and ZNF18. Similar to Morrow et al. [6], the 4 candidate genes were found to be neuronal depolarization dependent, supporting the role of disrupted synaptic transmission in ASD [8]. Lim et al. [14] also showed increased rare complete knockouts, including compound heterozygous and homozygous variants, in cases compared with controls by using whole-exome analysis, further suggesting a significant contribution of recessive loci to ASD risk [14]. In conclusion, studies to date on recessive loci of ASD have indicated that multiple genetic variants distributed across the genome and inherited in a recessive fashion appear to contribute to the risk of ASD. This may be an approach that can be capitalized on to a greater extent in future larger studies.
Autism Genetic Studies May Point to Convergent Pathways: The Road from Genetics to Therapeutics
As described above, studies have identified many autism risk candidate genes. Additionally, transcriptome studies in postmortem autism brain have identified gene networks disrupted in autism. Despite the vast genetic heterogeneity, accumulating evidence suggests that there are common pathways disrupted in the autistic brain. Discovery of these convergent pathways may be critical to identifying potential therapeutic targets.
Large-scale WES efforts in autism have pointed to several convergent pathways. Functional clustering of de novo SNVs in SSC identified enrichment of FMRP target genes, chromatin modifiers, and genes involved in embryonic development [88]. In another study, analysis of rare coding variants in almost 4000 autism cases identified a set of high-confidence autism risk genes. These genes were enriched for synaptic and postsynaptic genes, targets of the RNA binding proteins FMRP and RBFOX, histone modifiers, and chromatin remodeling genes [178]. Investigations of CNVs in autism have pointed to some similar pathways as were detected in exome sequencing studies. Large-scale studies of rare CNVs found that genes involved in ubiquitination and neuronal cell adhesion were enriched in autism associated CNVs [106]. In addition, deletions were enriched for genes involved in neuronal function and guanosine triphosphatase/Ras signaling [104]. Further studies found an enrichment of genes involved in MAPK signaling and neuronal development in autism-associated deletions [189]. Also, analysis of CNVs identified from WES data found an enrichment of cytoskeletal and autophagy genes in small CNVs in autism [190].
Analyses of co-expression networks in autism have also discovered convergent pathways in autism. Differential gene expression analysis of autism cortex transcriptome data revealed decreased expression of genes functioning at the synapse and increased expression of immune genes [191]. An extended transcriptome study of autism cortex also found upregulation of immune genes in autism specifically identifying increased expression of activated microglia genes. This study also identified misexpression of synaptic activity genes in autism [192]. To narrow down autism heterogeneity to the pathology of a specific development time point, cell type, and brain region, Willsey et al. [193] analyzed expression data from several brain regions at many stages of development. Co-expression networks seeded with genes recurrently mutated in autism were found to converge with networks in deep layer projection neurons of the prefrontal and primary motor–somatosensory cortex during midfetal development [193]. Parikshak et al. [194] mapped autism risk genes to transcriptional networks throughout development and identified glutamatergic neurons in upper cortical layers as a critical cell type and brain region in autism. Undoubtedly, pathway and related network analyses have identified some degree of convergence with regard to potential mechanisms in which genes may participate, but, at present, these mechanisms remain fairly broad. One limitation of this approach is that gene pathway methods are clearly limited by a very incomplete understanding of the function of specific molecules in circuit development and function. One hope is that investigation of specific molecules with disease-associated mutations may be a path forward to a more in-depth understanding of cellular and neurocircuitry abnormalities.
iPSC-derived neurons may provide a new, valuable preclinical model for understanding autism-related cellular dysfunction and for predicting treatment outcomes. For example, iPSC-derived neurons were generated from patients diagnosed with the autism-associated disorder TS, which results from mutations in the L-type calcium channel Cav1.2 and leads to decreased channel inactivation. TS neurons show dysregulation of genes involved in catecholamine synthesis, which can be reversed by treatment with an L-type-channel blocker, roscovitine [195]. In another study, disruption of calcium signaling, changes in gene expression, and decreased neurite growth were observed in iPSC-derived neurons from a nonsyndromic patient with autism with a translocation disrupting the cation channel gene TRPC6. Some of the defective phenotypes could be reversed by treatment with insulin-like growth factor-1 or a TRPC6 agonist, hyperforin [196]. The use of iPSC-derived neurons to connect gene variants to autism-related cellular phenotypes provides a new path to identifying and screening potential therapeutics for autism. Future studies to expand on our understanding of the convergent pathways identified by these genetic studies may lead to mechanism-based treatments for autism; however, this approach will also rely on the identification of biomarkers that indicate an autism subtype defined by the disruption of a specific pathway. For example, some forms of idiopathic autism with macrocephaly may reflect pathobiology related to overactive signaling, even though the patient may not have an apparent PTEN mutation. In this case, the combinations of neuroimaging and perhaps measures of peripheral blood signaling could be tested to predict response to treatment for this specific autism subtype.
Conclusion
The last decade of research has established an unprecedented number of rare mutations in autism and autism-related neurodevelopmental conditions. Extension of these gene discovery approaches will be important to discover a great number of genes and alleles, and thereby convergent mechanisms with a higher resolution of mechanistic understanding. However, now is also an important time to expand our efforts in mechanistic studies. This endeavor may be helped by the recent emergence of iPSC methods; however, in vivo studies will remain important. Such in vivo studies will rely on animal models, and new animal models and/or approaches to study circuitry are warranted. In addition, the field may also benefit from larger neuroimaging studies of genetically characterized patients. In conclusion, discovery of rare genetic mutations has provided an unprecedented opportunity for an important transition to studies that may now elucidate disease-relevant mechanisms in autism.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 1224 kb)
Acknowledgments
This work was supported by the National Institutes of Health grants 2T32 MH019927-2 (EDG), T32NS0624443 (MFP), P20 GM103645-01 (EMM), R01 MH105442-01 (EMM), and R01 MH102418. EMM is also funded by Burroughs Wellcome Fund (1006815.01, Career Award for Medical Scientists). This work was supported by a grant from the Simons Foundation (286756, EMM). LNS is funded by an Autism Speaks Dennis Weatherstone Predoctoral Fellowship (#9169).
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Footnotes
E.D. Gamsiz and L.N. Sciarra contributed equally to this manuscript and are listed alphabetically.
References
- 1.Geschwind DH. Genetics of autism spectrum disorders. Trends Cogn Sci. 2011;15:409–416. doi: 10.1016/j.tics.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levy D, Ronennus M, Yamrom B, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 3.O'Roak BJ, Vives L, Girirajan S, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanders SJ, Murtha MT, Gupta AR, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iossifov I, Ronemus M, Levy D, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrow EM, Yoo SY, Flavell SW, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casey JP, Magalhaes T, Conroy JM, et al. A novel approach of homozygous haplotype sharing identifies candidate genes in autism spectrum disorder. Hum Genet. 2012;131:565–579. doi: 10.1007/s00439-011-1094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chahrour MH, Yu TW, Lim ET, et al. Whole-exome sequencing and homozygosity analysis implicate depolarization-regulated neuronal genes in autism. PLoS Genet. 2012;8:e1002635. doi: 10.1371/journal.pgen.1002635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gamsiz ED, Viscidi EW, Frederick AM, et al. Intellectual disability is associated with increased runs of homozygosity in simplex autism. Am J Hum Genet. 2013;93:103–109. doi: 10.1016/j.ajhg.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim ET, Raychaudhuri S, Sanders SJ, et al. Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron. 2013;77:235–242. doi: 10.1016/j.neuron.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrow EM, Walsh CA. Isolate populations and rare variation in autism spectrum disorders. Autism spectrum disorders. New York: Oxford University Press; 2011. [Google Scholar]
- 12.Chang J, Gilman SR, Chiang AH, Sanders SJ, Vitkup D. Genotype to phenotype relationships in autism spectrum disorders. Nat Neurosci. 2015;18:191–198. doi: 10.1038/nn.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong S, Walker MF, Carriero NJ, et al. De novo insertions and deletions of predominantly paternal origin are associated with autism spectrum disorder. Cell Rep. 2014;9:16–23. doi: 10.1016/j.celrep.2014.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim ET, Raychaudhuri S, Sanders SJ, et al. Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron. 2013;77:235–242. doi: 10.1016/j.neuron.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Roak BJ, Stessman HA, Boyle EA, et al. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat Commun. 2014;5:5595. doi: 10.1038/ncomms6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaaf CP, Zoghbi HY. Solving the autism puzzle a few pieces at a time. Neuron. 2011;70:806–808. doi: 10.1016/j.neuron.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 17.Gaugler T, Klei L, Sanders SJ, et al. Most genetic risk for autism resides with common variation. Nat Genet. 2014;46:881–885. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams CA, Frias JL. The Angelman ("happy puppet") syndrome. Am J Med Genet. 1982;11:453–460. doi: 10.1002/ajmg.1320110411. [DOI] [PubMed] [Google Scholar]
- 19.Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 20.Stromme P. Aetiology in severe and mild mental retardation: a population-based study of Norwegian children. Develop Med Child Neurol. 2000;42:76–86. doi: 10.1017/s0012162200000165. [DOI] [PubMed] [Google Scholar]
- 21.Thomson AK, Glasson EJ, Bittles AH. A long-term population-based clinical and morbidity profile of Angelman syndrome in Western Australia: 1953–2003. Disabil Rehabil. 2006;28:299–305. doi: 10.1080/09638280500190631. [DOI] [PubMed] [Google Scholar]
- 22.Bonati MT, Russo S, Finelli P, et al. Evaluation of autism traits in Angelman syndrome: a resource to unfold autism genes. Neurogenetics. 2007;8:169–178. doi: 10.1007/s10048-007-0086-0. [DOI] [PubMed] [Google Scholar]
- 23.Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 24.Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci U S A. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greer PL, Hanayama R, Bloodgood BL, et al. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140:704–716. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wallace ML, Burette AC, Weinberg RJ, Philpot BD. Maternal loss of Ube3a produces an excitatory/inhibitory imbalance through neuron type-specific synaptic defects. Neuron. 2012;74:793–800. doi: 10.1016/j.neuron.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cole TR, Hughes HE. Autosomal dominant macrocephaly: benign familial macrocephaly or a new syndrome? Am J Med Genet. 1991;41:115–124. doi: 10.1002/ajmg.1320410128. [DOI] [PubMed] [Google Scholar]
- 28.Kotulska K, Jozwiak S. Autism in monogenic disorders. Eur J Paediatr Neurol. 2011;15:177–80. doi: 10.1016/j.ejpn.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Chamberlain SJ, Lalande M. Neurodevelopmental disorders involving genomic imprinting at human chromosome 15q11-q13. Neurobiol Dis. 2010;39:13–20. doi: 10.1016/j.nbd.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 30.Naqvi S, Cole T, Graham JM., Jr Cole-Hughes macrocephaly syndrome and associated autistic manifestations. Am J Med Genet. 2000;94:149–152. doi: 10.1002/1096-8628(20000911)94:2<149::aid-ajmg7>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 31.Butler MG, Dasouki MJ, Zhou XP, et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cole TRP, Hughes HE. Autosomal dominant macrocephaly—benign familial macrocephaly or a new syndrome. Am J Med Genet. 1991;41:115–124. doi: 10.1002/ajmg.1320410128. [DOI] [PubMed] [Google Scholar]
- 33.Fombonne E, Roge B, Claverie J, Courty S, Fremolle J. Microcephaly and macrocephaly in autism. J Autism Develop Disord. 1999;29:113–119. doi: 10.1023/a:1023036509476. [DOI] [PubMed] [Google Scholar]
- 34.McBride KL, Varga EA, Pastore MT, et al. Confirmation study of PTEN mutations among individuals with autism or developmental delays/mental retardation and macrocephaly. Autism Res. 2010;3:137–141. doi: 10.1002/aur.132. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez-Escudero I, Oliver MD, Andres-Pons A, et al. A comprehensive functional analysis of PTEN mutations: implications in tumor- and autism-related syndromes. Hum Mol Genet. 2011;20:4132–4142. doi: 10.1093/hmg/ddr337. [DOI] [PubMed] [Google Scholar]
- 36.Maehama T, Dixon JE. PTEN: a tumour suppressor that functions as a phospholipid phosphatase. Trends Cell Biol. 1999;9:125–128. doi: 10.1016/s0962-8924(99)01519-6. [DOI] [PubMed] [Google Scholar]
- 37.Fraser MM, Bayazitov IT, Zakharenko SS, Baker SJ. Phosphatase and tensin homolog, deleted on chromosome 10 deficiency in brain causes defects in synaptic structure, transmission and plasticity, and myelination abnormalities. Neuroscience. 2008;151:476–488. doi: 10.1016/j.neuroscience.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwon CH, Luikart BW, Powell CM, et al. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eng C. PTEN: one gene, many syndromes. Hum Mutat. 2003;22:183–198. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- 40.Goffin A, Hoefsloot LH, Bosgoed E, Swillen A, Fryns JP. PTEN mutation in a family with Cowden syndrome and autism. Am J Med Genet. 2001;105:521–524. doi: 10.1002/ajmg.1477. [DOI] [PubMed] [Google Scholar]
- 41.Lynch NE, Lynch SA, McMenamin J, Webb D. Bannayan–Riley–Ruvalcaba syndrome: a cause of extreme macrocephaly and neurodevelopmental delay. Arch Dis Child. 2009;94:553–554. doi: 10.1136/adc.2008.155663. [DOI] [PubMed] [Google Scholar]
- 42.Issekutz KA, Graham JM, Jr, Prasad C, Smith IM, Blake KD. An epidemiological analysis of CHARGE syndrome: preliminary results from a Canadian study. Am J Med Genet A. 2005;133a:309–317. doi: 10.1002/ajmg.a.30560. [DOI] [PubMed] [Google Scholar]
- 43.Blake KD, Davenport SL, Hall BD, et al. CHARGE association: an update and review for the primary pediatrician. Clin Pediatr. 1998;37:159–173. doi: 10.1177/000992289803700302. [DOI] [PubMed] [Google Scholar]
- 44.Vissers LE, van Ravenswaaij CM, Admiraal R, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 45.Hartshorne TS, Grialou TL, Parker KR. Autistic-like behavior in CHARGE syndrome. Am J Med Genet A. 2005;133a:257–261. doi: 10.1002/ajmg.a.30545. [DOI] [PubMed] [Google Scholar]
- 46.Aramaki M, Udaka T, Kosaki R, et al. Phenotypic spectrum of CHARGE syndrome with CHD7 mutations. J Pediatr. 2006;148:410–414. doi: 10.1016/j.jpeds.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 47.Jongmans MC, Admiraal RJ, van der Donk KP, et al. CHARGE syndrome: the phenotypic spectrum of mutations in the CHD7 gene. J Med Genet. 2006;43:306–314. doi: 10.1136/jmg.2005.036061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lalani SR, Safiullah AM, Molinari LM, et al. SEMA3E mutation in a patient with CHARGE syndrome. J Med Genet. 2004;41:e94. doi: 10.1136/jmg.2003.017640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Janssen N, Bergman JE, Swertz MA, et al. Mutation update on the CHD7 gene involved in CHARGE syndrome. Hum Mutat. 2012;33:1149–1160. doi: 10.1002/humu.22086. [DOI] [PubMed] [Google Scholar]
- 50.Schulz Y, Wehner P, Opitz L, et al. CHD7, the gene mutated in CHARGE syndrome, regulates genes involved in neural crest cell guidance. Hum Genet. 2014;133:997–1009. doi: 10.1007/s00439-014-1444-2. [DOI] [PubMed] [Google Scholar]
- 51.Christianson AL, Stevenson RE, van der Meyden CH, et al. X linked severe mental retardation, craniofacial dysmorphology, epilepsy, ophthalmoplegia, and cerebellar atrophy in a large South African kindred is localised to Xq24-q27. J Med Genet. 1999;36:759–766. doi: 10.1136/jmg.36.10.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pescosolido MF, Stein DM, Schmidt M, et al. Genetic and phenotypic diversity of NHE6 mutations in Christianson syndrome. Ann Neurol. 2014;76:581–593. doi: 10.1002/ana.24225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gilfillan GD, Selmer KK, Roxrud I, et al. SLC9A6 mutations cause X-linked mental retardation, microcephaly, epilepsy, and ataxia, a phenotype mimicking Angelman syndrome. Am J Hum Genet. 2008;82:1003–1010. doi: 10.1016/j.ajhg.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ouyang Q, Lizarraga SB, Schmidt M, et al. Christianson syndrome protein NHE6 modulates TrkB endosomal signaling required for neuronal circuit development. Neuron. 2013;80:97–112. doi: 10.1016/j.neuron.2013.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song FJ, Barton P, Sleightholme V, Yao GL, Fry-Smith A. Screening for fragile X syndrome: a literature review and modelling study. Health Technol Assess. 2003;7:1–106. doi: 10.3310/hta7160. [DOI] [PubMed] [Google Scholar]
- 56.Hagerman RJ, McBogg P, Hagerman PJ. The fragile X syndrome: history, diagnosis, and treatment. J Dev Behav Pediatr. 1983;4:122–130. doi: 10.1097/00004703-198306000-00009. [DOI] [PubMed] [Google Scholar]
- 57.Bailey DB, Jr, Raspa M, Olmsted M, Holiday DB. Co-occurring conditions associated with FMR1 gene variations: findings from a national parent survey. Am J Med Genet A. 2008;146a:2060–2069. doi: 10.1002/ajmg.a.32439. [DOI] [PubMed] [Google Scholar]
- 58.Verkerk AJ, Pieretti M, Sutcliffe JS, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 59.Darnell JC, Jensen KB, Jin P, et al. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 60.Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 61.Evans DG, Howard E, Giblin C, et al. Birth incidence and prevalence of tumor-prone syndromes: estimates from a UK family genetic register service. Am J Med Genet A. 2010;152a:327–332. doi: 10.1002/ajmg.a.33139. [DOI] [PubMed] [Google Scholar]
- 62.Williams VC, Lucas J, Babcock MA, et al. Neurofibromatosis type 1 revisited. Pediatrics. 2009;123:124–133. doi: 10.1542/peds.2007-3204. [DOI] [PubMed] [Google Scholar]
- 63.Hyman SL, Shores A, North KN. The nature and frequency of cognitive deficits in children with neurofibromatosis type 1. Neurology. 2005;65:1037–1044. doi: 10.1212/01.wnl.0000179303.72345.ce. [DOI] [PubMed] [Google Scholar]
- 64.Plasschaert E, Descheemaeker MJ, Van Eylen L, et al. Prevalence of autism spectrum disorder symptoms in children with neurofibromatosis type 1. Am J Med Genet B. 2015;168:72–80. doi: 10.1002/ajmg.b.32280. [DOI] [PubMed] [Google Scholar]
- 65.Xu GF, O'Connell P, Viskochil D, et al. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990;62:599–608. doi: 10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]
- 66.Sabbagh A, Pasmant E, Imbard A, et al. NF1 molecular characterization and neurofibromatosis type I genotype-phenotype correlation: the French experience. Hum Mutat. 2013;34:1510–1518. doi: 10.1002/humu.22392. [DOI] [PubMed] [Google Scholar]
- 67.Wallace MR, Marchuk DA, Andersen LB, et al. Type 1 neurofibromatosis gene: identification of a large transcript disrupted in three NF1 patients. Science. 1990;249:181–186. doi: 10.1126/science.2134734. [DOI] [PubMed] [Google Scholar]
- 68.Oliveira AF, Yasuda R. Neurofibromin is the major ras inactivator in dendritic spines. J Neurosci. 2014;34:776–783. doi: 10.1523/JNEUROSCI.3096-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Asthagiri AR, Parry DM, Butman JA, et al. Neurofibromatosis type 2. Lancet. 2009;373:1974–1986. doi: 10.1016/S0140-6736(09)60259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Denayer E, Brems H, de Cock P, et al. Pathogenesis of vestibular schwannoma in ring chromosome 22. BMC Med Genet. 2009;10:97. doi: 10.1186/1471-2350-10-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Amir RE, Van den Veyver IB, Wan M, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 72.Laurvick CL, de Klerk N, Bower C, et al. Rett syndrome in Australia: a review of the epidemiology. J Pediatr. 2006;148:347–352. doi: 10.1016/j.jpeds.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 73.Hagberg B, Aicardi J, Dias K, Ramos O. A progressive syndrome of autism, dementia, ataxia, and loss of purposeful hand use in girls: Rett's syndrome: report of 35 cases. Ann Neurol. 1983;14:471–479. doi: 10.1002/ana.410140412. [DOI] [PubMed] [Google Scholar]
- 74.Moss J, Howlin P. Autism spectrum disorders in genetic syndromes: implications for diagnosis, intervention and understanding the wider autism spectrum disorder population. J Intellect Disabil Res. 2009;53:852–873. doi: 10.1111/j.1365-2788.2009.01197.x. [DOI] [PubMed] [Google Scholar]
- 75.Chahrour M, Jung SY, Shaw C, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dani VS, Chang Q, Maffei A, et al. Reduced cortical activity due to a shift in the balance between excitation and inhibition in a mouse model of Rett syndrome. Proc Natl Acad Sci U S A. 2005;102:12560–12565. doi: 10.1073/pnas.0506071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Van Esch H, Bauters M, Ignatius J, et al. Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. Am J Hum Genet. 2005;77:442–453. doi: 10.1086/444549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Splawski I, Timothy KW, Sharpe LM, et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 79.Berger SM, Bartsch D. The role of L-type voltage-gated calcium channels Cav1.2 and Cav1.3 in normal and pathological brain function. Cell Tissue Res. 2014;357:463–476. doi: 10.1007/s00441-014-1936-3. [DOI] [PubMed] [Google Scholar]
- 80.European Chromosome 16 Tuberous Sclerosis Consortium. Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell 1993;75:1305-1315. [DOI] [PubMed]
- 81.van Slegtenhorst M, de Hoogt R, Hermans C, et al. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 1997;277:805–808. doi: 10.1126/science.277.5327.805. [DOI] [PubMed] [Google Scholar]
- 82.Osborne JP, Fryer A, Webb D. Epidemiology of tuberous sclerosis. Ann N Y Acad Sci. 1991;615:125–127. doi: 10.1111/j.1749-6632.1991.tb37754.x. [DOI] [PubMed] [Google Scholar]
- 83.Smalley SL. Autism and tuberous sclerosis. J Autism Develop Disord. 1998;28:407–414. doi: 10.1023/a:1026052421693. [DOI] [PubMed] [Google Scholar]
- 84.Numis AL, Major P, Montenegro MA, et al. Identification of risk factors for autism spectrum disorders in tuberous sclerosis complex. Neurology. 2011;76:981–987. doi: 10.1212/WNL.0b013e3182104347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sepp T, Yates JR, Green AJ. Loss of heterozygosity in tuberous sclerosis hamartomas. J Med Genet. 1996;33:962–964. doi: 10.1136/jmg.33.11.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tee AR, Fingar DC, Manning BD, et al. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci U S A. 2002;99:13571–13576. doi: 10.1073/pnas.202476899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Takei N, Nawa H. mTOR signaling and its roles in normal and abnormal brain development. Front Mol Neurosci. 2014;7:28. doi: 10.3389/fnmol.2014.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Iossifov I, O'Roak BJ, Sanders SJ, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jiang YH, Yuen RK, Jin X, et al. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am J Hum Genet. 2013;93:249–263. doi: 10.1016/j.ajhg.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cukier HN, Lee JM, Ma D, et al. The expanding role of MBD genes in autism: identification of a MECP2 duplication and novel alterations in MBD5, MBD6, and SETDB1. Autism Res. 2012;5:385–397. doi: 10.1002/aur.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Morrow EM. Genomic copy number variation in disorders of cognitive development. J Am Acad Child Adolesc Psychiatry. 2010;49:1091–1104. doi: 10.1016/j.jaac.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 95.Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 96.Sebat J, Lakshmi B, Malhotra D, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Levy D, Ronemus M, Yamrom B, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 98.Sanders SJ, Hus V, Luo R, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11. 23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Talkowski ME, Rosenfeld JA, Blumenthal I, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mefford HC, Sharp AJ, Baker C, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Girirajan S, Dennis MY, Baker C, et al. Refinement and discovery of new hotspots of copy-number variation associated with autism spectrum disorder. Am J Hum Genet. 2013;92:221–237. doi: 10.1016/j.ajhg.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Weiss LA, Shen Y, Korn JM, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 103.Marshall CR, Noor A, Vincent JB, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pinto D, Pagnamenta AT, Klei L, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Moreno-De-Luca D, Mulle JG, Kaminsky EB, et al. Deletion 17q12 is a recurrent copy number variant that confers high risk of autism and schizophrenia. Am J Hum Genetics. 2010;87:618–630. doi: 10.1016/j.ajhg.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Glessner JT, Wang K, Cai G, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Turner DJ, Miretti M, Rajan D, et al. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat Genet. 2008;40:90–95. doi: 10.1038/ng.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pescosolido MF, Gamsiz ED, Nagpal S, Morrow EM. Distribution of disease-associated copy number variants across distinct disorders of cognitive development. J Am Acad Child Adolesc Psychiatry. 2013;52:414–430. doi: 10.1016/j.jaac.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Warnica W, Merico D, Costain G, et al. Copy number variable microRNAs in schizophrenia and their neurodevelopmental gene targets. Biol Psychiatry. 2015;77:158–166. doi: 10.1016/j.biopsych.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cooper GM, Coe BP, Girirajan S, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McCarthy SE, Makarov V, Kirov G, et al. Microduplications of 16p11. 2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cook EH, Jr, Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 114.Brunetti-Pierri N, Berg JS, Scaglia F, et al. Recurrent reciprocal 1q21. 1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Davis JM, Searles VB, Anderson N, et al. DUF1220 dosage is linearly associated with increasing severity of the three primary symptoms of autism. PLoS Genet. 2014;10:e1004241. doi: 10.1371/journal.pgen.1004241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Berg JS, Brunetti-Pierri N, Peters SU, et al. Speech delay and autism spectrum behaviors are frequently associated with duplication of the 7q11.23 Williams–Beuren syndrome region. Genet Med. 2007;9:427–441. doi: 10.1097/gim.0b013e3180986192. [DOI] [PubMed] [Google Scholar]
- 117.Somerville MJ, Mervis CB, Young EJ, et al. Severe expressive-language delay related to duplication of the Williams-Beuren locus. N Engl J Med. 2005;353:1694–1701. doi: 10.1056/NEJMoa051962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pober BR. Williams–Beuren syndrome. N Engl J Med. 2010;362:239–252. doi: 10.1056/NEJMra0903074. [DOI] [PubMed] [Google Scholar]
- 119.Merla G, Brunetti-Pierri N, Micale L, Fusco C. Copy number variants at Williams–Beuren syndrome 7q11.23 region. Hum Genet. 2010;128:3–26. doi: 10.1007/s00439-010-0827-2. [DOI] [PubMed] [Google Scholar]
- 120.Adamo A, Atashpaz S, Germain P, et al. 7q11.23 dosage-dependent dysregulation in human pluripotent stem cells affects transcriptional programs in disease-relevant lineages. Nat Genet. 2015;47:132–141. doi: 10.1038/ng.3169. [DOI] [PubMed] [Google Scholar]
- 121.Luo R, Sanders SJ, Tian Y, et al. Genome-wide transcriptome profiling reveals the functional impact of rare de novo and recurrent CNVs in autism spectrum disorders. Am J Hum Genet. 2012;91:38–55. doi: 10.1016/j.ajhg.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cook EH, Jr, Lindgren V, Leventhal BL, et al. Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am J Hum Genet. 1997;60:928–934. [PMC free article] [PubMed] [Google Scholar]
- 123.Nicholls RD, Knepper JL. Genome organization, function, and imprinting in Prader–Willi and Angelman syndromes. Annu Rev Genomics Hum Genet. 2001;2:153–175. doi: 10.1146/annurev.genom.2.1.153. [DOI] [PubMed] [Google Scholar]
- 124.Hogart A, Wu D, LaSalle JM, Schanen NC. The comorbidity of autism with the genomic disorders of chromosome 15q11.2-q13. Neurobiol Dis. 2010;38:181–191. doi: 10.1016/j.nbd.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Veltman MW, Craig EE, Bolton PF. Autism spectrum disorders in Prader–Willi and Angelman syndromes: a systematic review. Psychiatric Genet. 2005;15:243–254. doi: 10.1097/00041444-200512000-00006. [DOI] [PubMed] [Google Scholar]
- 126.Veltman MW, Thompson RJ, Roberts SE, et al. Prader–Willi syndrome—a study comparing deletion and uniparental disomy cases with reference to autism spectrum disorders. Eur Child Adolesc Psychiatry. 2004;13:42–50. doi: 10.1007/s00787-004-0354-6. [DOI] [PubMed] [Google Scholar]
- 127.Schaaf CP, Gonzalez-Garay ML, Xia F, et al. Truncating mutations of MAGEL2 cause Prader–Willi phenotypes and autism. Nat Genet. 2013;45:1405–1408. doi: 10.1038/ng.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bijlsma EK, Gijsbers AC, Schuurs-Hoeijmakers JH, et al. Extending the phenotype of recurrent rearrangements of 16p11.2: deletions in mentally retarded patients without autism and in normal individuals. Eur J Med Genet. 2009;52:77–87. doi: 10.1016/j.ejmg.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 129.Shinawi M, Liu P, Kang SHL, et al. Recurrent reciprocal 16p11. 2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 2010;47:332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bachmann-Gagescu R, Mefford HC, Cowan C, et al. Recurrent 200-kb deletions of 16p11.2 that include the SH2B1 gene are associated with developmental delay and obesity. Genet Med. 2010;12:641–647. doi: 10.1097/GIM.0b013e3181ef4286. [DOI] [PubMed] [Google Scholar]
- 131.Hanson E, Bernier R, Porche K, et al. The cognitive and behavioral phenotype of the 16p11.2 deletion in a clinically ascertained population. Biol Psychiatry. 2015;77:785–793. doi: 10.1016/j.biopsych.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kumar RA, KaraMohamed S, Sudi J, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 133.Qureshi AY, Mueller S, Snyder AZ, et al. Opposing brain differences in 16p11.2 deletion and duplication carriers. J Neurosci. 2014;34:11199–11211. doi: 10.1523/JNEUROSCI.1366-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jacquemont S, Reymond A, Zufferey F, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478:97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Maillard AM, Ruef A, Pizzagalli F, et al. The 16p11.2 locus modulates brain structures common to autism, schizophrenia and obesity. Mol Psychiatry. 2015;20:140–147. doi: 10.1038/mp.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Golzio C, Willer J, Talkowski ME, et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012;485:363–367. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Blumenthal I, Ragavendran A, Erdin S, et al. Transcriptional consequences of 16p11.2 deletion and duplication in mouse cortex and multiplex autism families. Am J Hum Genet. 2014;94:870–883. doi: 10.1016/j.ajhg.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Karayiorgou M, Morris MA, Morrow B, et al. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc Natl Acad Sci. 1995;92:7612–7616. doi: 10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jonas RK, Montojo CA, Bearden CE. The 22q11.2 deletion syndrome as a window into complex neuropsychiatric disorders over the lifespan. Biol Psychiatry. 2014;75:351–360. doi: 10.1016/j.biopsych.2013.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schneider M, Debbane M, Bassett AS, et al. Psychiatric disorders from childhood to adulthood in 22q11.2 deletion syndrome: results from the International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome. Am J Psychiatry. 2014;171:627–639. doi: 10.1176/appi.ajp.2013.13070864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Woodin M, Wang PP, Aleman D, et al. Neuropsychological profile of children and adolescents with the 22q11.2 microdeletion. Genet Med. 2001;3:34–39. doi: 10.1097/00125817-200101000-00008. [DOI] [PubMed] [Google Scholar]
- 142.Gothelf D, Eliez S, Thompson T, et al. COMT genotype predicts longitudinal cognitive decline and psychosis in 22q11.2 deletion syndrome. Nat Neurosci. 2005;8:1500–1502. doi: 10.1038/nn1572. [DOI] [PubMed] [Google Scholar]
- 143.Glaser B, Debbane M, Hinard C, et al. No evidence for an effect of COMT Val158Met genotype on executive function in patients with 22q11 deletion syndrome. Am J Psychiatry. 2006;163:537–539. doi: 10.1176/appi.ajp.163.3.537. [DOI] [PubMed] [Google Scholar]
- 144.Murphy KC, Jones LA, Owen MJ. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch Gen Psychiatry. 1999;56:940–945. doi: 10.1001/archpsyc.56.10.940. [DOI] [PubMed] [Google Scholar]
- 145.Hiramoto T, Kang G, Suzuki G, et al. Tbx1: identification of a 22q11.2 gene as a risk factor for autism spectrum disorder in a mouse model. Hum Mol Genet. 2011;20:4775–4785. doi: 10.1093/hmg/ddr404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Paylor R, Glaser B, Mupo A, et al. Tbx1 haploinsufficiency is linked to behavioral disorders in mice and humans: implications for 22q11 deletion syndrome. Proc Natl Acad Sci U S A. 2006;103:7729–7734. doi: 10.1073/pnas.0600206103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen YZ, Matsushita M, Girirajan S, et al. Evidence for involvement of GNB1L in autism. Am J Med Genet B. 2012;159b:61–71. doi: 10.1002/ajmg.b.32002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Paronett EM, Meechan DW, Karpinski BA, LaMantia AS, Maynard TM. Ranbp1, deleted in DiGeorge/22q11.2 deletion syndrome, is a microcephaly gene that selectively disrupts layer 2/3 cortical projection neuron generation. Cereb Cortex 2014 Dec 1 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 149.Meechan DW, Tucker ES, Maynard TM, LaMantia AS. Cxcr4 regulation of interneuron migration is disrupted in 22q11.2 deletion syndrome. Proc Natl Acad Sci U S A. 2012;109:18601–18606. doi: 10.1073/pnas.1211507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Phelan K, McDermid HE. The 22q13.3 deletion syndrome (Phelan–McDermid syndrome) Mol Syndromol. 2012;2:186–201. doi: 10.1159/000334260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cusmano-Ozog K, Manning MA, Hoyme HE. 22q13.3 deletion syndrome: a recognizable malformation syndrome associated with marked speech and language delay. Am J Med Genet C Semin Med Genet. 2007;145c:393–398. doi: 10.1002/ajmg.c.30155. [DOI] [PubMed] [Google Scholar]
- 152.Aldinger KA, Kogan J, Kimonis V, et al. Cerebellar and posterior fossa malformations in patients with autism-associated chromosome 22q13 terminal deletion. Am J Med Genet A. 2013;161a:131–136. doi: 10.1002/ajmg.a.35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Durand CM, Betancur C, Boeckers TM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Moessner R, Marshall CR, Sutcliffe JS, et al. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Betancur C, Buxbaum JD. SHANK3 haploinsufficiency: a "common" but underdiagnosed highly penetrant monogenic cause of autism spectrum disorders. Mol Autism. 2013;4:17. doi: 10.1186/2040-2392-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Baron MK, Boeckers TM, Vaida B, et al. An architectural framework that may lie at the core of the postsynaptic density. Science. 2006;311:531–535. doi: 10.1126/science.1118995. [DOI] [PubMed] [Google Scholar]
- 157.Roussignol G, Ango F, Romorini S, et al. Shank expression is sufficient to induce functional dendritic spine synapses in aspiny neurons. J Neurosci. 2005;25:3560–3570. doi: 10.1523/JNEUROSCI.4354-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yang M, Bozdagi O, Scattoni ML, et al. Reduced excitatory neurotransmission and mild autism-relevant phenotypes in adolescent Shank3 null mutant mice. J Neurosci. 2012;32:6525–6541. doi: 10.1523/JNEUROSCI.6107-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang X, McCoy PA, Rodriguiz RM, et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet. 2011;20:3093–3108. doi: 10.1093/hmg/ddr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Peca J, Feliciano C, Ting JT, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wilson HL, Crolla JA, Walker D, et al. Interstitial 22q13 deletions: genes other than SHANK3 have major effects on cognitive and language development. Eur J Hum Genetics. 2008;16:1301–1310. doi: 10.1038/ejhg.2008.107. [DOI] [PubMed] [Google Scholar]
- 162.Giza J, Urbanski MJ, Prestori F, et al. Behavioral and cerebellar transmission deficits in mice lacking the autism-linked gene islet brain-2. J Neurosci. 2010;30:14805–14816. doi: 10.1523/JNEUROSCI.1161-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Miller DT, Shen Y, Weiss LA, et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J Med Genet. 2009;46:242–248. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pagnamenta AT, Wing K, Akha ES, et al. A 15q13.3 microdeletion segregating with autism. Eur J Hum Genet. 2009;17:687–692. doi: 10.1038/ejhg.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ionita-Laza I, Xu B, Makarov V, et al. Scan statistic-based analysis of exome sequencing data identifies FAN1 at 15q13.3 as a susceptibility gene for schizophrenia and autism. Proc Natl Acad Sci U S A. 2014;111:343–348. doi: 10.1073/pnas.1309475110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sato D, Lionel AC, Leblond CS, et al. SHANK1 deletions in males with autism spectrum disorder. Am J Hum Genet. 2012;90:879–887. doi: 10.1016/j.ajhg.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kim HG, Kishikawa S, Higgins AW, et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Szatmari P, Paterson AD, Zwaigenbaum L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Jamain S, Quach H, Betancur C, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bourgeron T. A synaptic trek to autism. Curr Opin Neurobiol. 2009;19:231–234. doi: 10.1016/j.conb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 171.Talkowski ME, Mullegama SV, Rosenfeld JA, et al. Assessment of 2q23.1 microdeletion syndrome implicates MBD5 as a single causal locus of intellectual disability, epilepsy, and autism spectrum disorder. Am J Hum Genet. 2011;89:551–563. doi: 10.1016/j.ajhg.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Mullegama SV, Rosenfeld JA, Orellana C, et al. Reciprocal deletion and duplication at 2q23.1 indicates a role for MBD5 in autism spectrum disorder. Eur J Hum Genet. 2014;22:57–63. doi: 10.1038/ejhg.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Beunders G, Voorhoeve E, Golzio C, et al. Exonic deletions in AUTS2 cause a syndromic form of intellectual disability and suggest a critical role for the C terminus. Am J Hum Genet. 2013;92:210–220. doi: 10.1016/j.ajhg.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lionel AC, Tammimies K, Vaags AK, et al. Disruption of the ASTN2/TRIM32 locus at 9q33.1 is a risk factor in males for autism spectrum disorders, ADHD and other neurodevelopmental phenotypes. Hum Mol Genet. 2014;23:2752–2768. doi: 10.1093/hmg/ddt669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bucan M, Abrahams BS, Wang K, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS genetics. 2009;5(6) doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Neale BM, Kou Y, Liu L, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.O'Roak BJ, Vives L, Fu W, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.De Rubeis S, He X, Goldberg AP, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kong A, Frigge ML, Masson G, et al. Rate of de novo mutations and the importance of father's age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Rauch A, Wieczorek D, Graf E, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 181.Yu TW, Chahrour MH, Coulter ME, et al. Using whole-exome sequencing to identify inherited causes of autism. Neuron. 2013;77:259–273. doi: 10.1016/j.neuron.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Parrish JZ, Emoto K, Jan LY, Jan YN. Polycomb genes interact with the tumor suppressor genes hippo and warts in the maintenance of Drosophila sensory neuron dendrites. Genes Develop. 2007;21:956–972. doi: 10.1101/gad.1514507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Basel-Vanagaite L, Attia R, Yahav M, et al. The CC2D1A, a member of a new gene family with C2 domains, is involved in autosomal recessive non-syndromic mental retardation. J Med Genet. 2006;43:203–210. doi: 10.1136/jmg.2005.035709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Manzini MC, Xiong L, Shaheen R, et al. CC2D1A regulates human intellectual and social function as well as NF-kappaB signaling homeostasis. Cell Rep. 2014;8:647–655. doi: 10.1016/j.celrep.2014.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Noor A, Windpassinger C, Patel M, et al. CC2D2A, encoding a coiled-coil and C2 domain protein, causes autosomal-recessive mental retardation with retinitis pigmentosa. Am J Hum Genet. 2008;82:1011–1018. doi: 10.1016/j.ajhg.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Novarino G, El-Fishawy P, Kayserili H, et al. Mutations in BCKD-kinase lead to a potentially treatable form of autism with epilepsy. Science. 2012;338:394–397. doi: 10.1126/science.1224631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Gandin I, Faletra F, Faletra F, et al. Excess of runs of homozygosity is associated with severe cognitive impairment in intellectual disability. Genet Med. 2015;17:396–399. doi: 10.1038/gim.2014.118. [DOI] [PubMed] [Google Scholar]
- 188.Lin PI, Kuo PH, Chen CH, et al. Runs of homozygosity associated with speech delay in autism in a taiwanese han population: evidence for the recessive model. PLoS One. 2013;8:e72056. doi: 10.1371/journal.pone.0072056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pinto D, Delaby E, Merico D, et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet. 2014;94:677–694. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Poultney CS, Goldberg AP, Drapeau E, et al. Identification of small exonic CNV from whole-exome sequence data and application to autism spectrum disorder. Am J Hum Genet. 2013;93:607–619. doi: 10.1016/j.ajhg.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Voineagu I, Wang X, Johnston P, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Gupta S, Ellis SE, Ashar FN, et al. Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat Commun. 2014;5:5748. doi: 10.1038/ncomms6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Willsey AJ, Sanders SJ, Li M, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Parikshak NN, Luo R, Zhang A, et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Pasca SP, Portmann T, Voineagu I, et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med. 2011;17:1657–1662. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Griesi-Oliveira K, Acab A, Gupta AR, et al. Modeling non-syndromic autism and the impact of TRPC6 disruption in human neurons. Mol Psychiatry 2014 Nov 11 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1224 kb)