Abstract
Background
Jalmagna is a popular deepwater rice variety with farmers of India because of its good yield under waterlogged condition. However, the variety is highly susceptible to bacterial blight (BB) disease. The development of resistant cultivars has been the most effective and economical strategy to control the disease under deepwater situation. Three resistance genes (xa5 + xa13 + Xa21) were transferred from Swarna BB pyramid line, using a marker-assisted backcrossing (MAB) breeding strategy, into the BB-susceptible elite deepwater cultivar, Jalmagna.
Results
Molecular marker integrated backcross breeding program has been employed to transfer three major BB resistance genes (Xa21, xa13 and xa5) into Jalmagna variety. During backcross generations, markers closely linked to the three genes were used to select plants possessing these resistance genes and markers polymorphic between donor and recurrent parent were used to select plants that have maximum contribution from the recurrent parent genome. A selected BC3F1 plant was selfed to generate homozygous BC3F2 plants with different combinations of BB resistance genes. The three-gene pyramid and two gene pyramid lines exhibited high levels of resistance against the BB pathogen. Under conditions of BB infection, the three-gene pyramided lines exhibited a significant yield advantage over Jalmagna. The selected pyramided lines showed all agro-morphologic traits of Jalmagna without compromising the yield.
Conclusion
The three major BB resistance genes pyramided lines exhibited high level of resistance and are expected to provide durable resistance under deep water situation where control through chemicals is less effective. High similarity in agro-morphologic traits and absence of antagonistic effects for yield and other characters were observed in the best pyramided lines.
Keywords: Deepwater rice, Marker-assisted selection, Bacterial blight gene pyramiding, Broad-spectrum resistance, Foreground selection, Background selection, xa5, xa13, Xa21
Background
Rice (Oryza sativa L.) is an important food crop that serves as a major carbohydrate source for nearly half of the world’s population. In India, it is grown in 43 million hectares accounting for 42% of food grain production and 55% of cereal production. To sustain self-sufficiency and to meet food grain requirement of future, India has to produce 135–140 million tones of rice by 2030. This has to necessarily meet from less land, less water, less labor and fewer chemicals, constant battle against new emerging pathogens and pests and possible adverse effects from climate change (Khush 2005). This ecosystem covers around 4 million hectares of which 3 million hectares are under deepwater ecology and 1 million hectare under very deepwater ecology (floating rice).Under deepwater ecology, the crop remains waterlogged for a period of more than a month with more than 50 cm water depth while in floating type the water depth remains more than one meter. The average productivity of deepwater ecosystem is around 1 t/ha while floating type is again very low yield. Bacterial leaf blight (BB) caused by Xanthomonas oryzae pv. oryzae (Xoo) is the most important disease of deepwater rice in India. In some areas of Asia, it can reduce crop yield by up to 50% (Khush et al. 1989) or even up to 80% (Singh et al. 1977). It also causes poor quality fodder. This affects photosynthetic areas and reduces the yield drastically and produce partial grain filling and low quality fodder yield.
Although a large number of rice varieties have been released for different agro-ecosystems in India, only a few are widely grown in deep water situations. Rice varieties, Jalmagna and Dinesh are very widely grown in deep water areas of India. Theses varieties are popular among rice farmers and consumers because of its high yield, medium slender grains and excellent cooking and eating qualities. Despite popularity, these varieties are highly susceptible to many pests and diseases including BB. BB is the most important disease of deepwater rice in India and has become a major production constraint. In absence of effective chemical or other control agents against the pathogen in deepwater situation, host plant resistance has gained enormous importance in controlling this disease (Devadath 1989). Therefore, host plant resistance offers the most effective, economical and environmentally safe option for management of BB pathogen in deepwater situation (Khush et al. 1989). In other ecology also, development of resistant cultivars carrying resistant genes have been the most effective and economical strategy to control BB disease and no environmental pollutions (Huang et al. 1997; Jena and MacKill 2008; Singh et al. 2001; Sundaram et al. 2008; Rajpurohit et al. 2011; Dokku et al. 2013; Suh et al. 2013). Globally, thirty eight BB resistance genes have been identified from diverse sources (Bhasin et al. 2012). A number of these resistance genes have been tagged by closely linked molecular markers (Yoshimura et al. 1995; Sonti 1998; Rao et al. 2002; Gu et al. 2008). A few of these genes like Xa4 have been incorporated widely in many high yielding varieties through conventional breeding (Khush et al. 1989). However, widespread cultivation of varieties with Xa4 has led to predominance of Xoo races that can overcome this gene (Mew et al. 1992). The deployment of rice cultivars that have multiple BB resistance genes is expected to lead to more durable resistance.
Pyramiding multiple R genes in a single line confers wide-spectrum and durable resistance. Tightly linked DNA markers have been developed for several BB resistance genes. The BB resistance genes, Xa1, xa5, xa13, Xa21, Xa26 and Xa27 have been cloned and used for breeding program. With the exception of xa5 and xa13, the BB resistance genes are dominant in nature and the markers developed from the sequencing information of these genes are widely used in MAS (Song et al. 1995; Yoshimura et al. 1998; Gu et al. 2005; Chu et al. 2006a). Using the gene pyramid approach, improved indica rice cultivars with broad spectrum durable BB resistance have been developed by combining different genes (Huang et al. 1997; Sanchez et al. 2000; Shanti et al. 2001; Singh et al. 2001; Joseph et al. 2004; Pha and Lang 2004; Bharatkumar et al. 2008; Hu et al. 2008; Perez et al. 2008; Sundaram et al. 2008; Rajpurohit et al. 2011; Dokku et al. 2013; Suh et al. 2013). A three-gene combination appeared to be the most effective; with Xa21 contributing the largest component of resistance. Therefore, incorporation of three BB resistant genes combination was taken up in the popular variety Jalmagna background by integrating marker-assisted backcrossing with phenotypic selection for development of pyramiding lines for the handicapped ecology.
Results
Pyramiding of bacterial blight resistance genes
The parent polymorphism was detected for the donor (CRMAS 2232–85) and recurrent parent (Jalmagna) with the markers pTA 248, RG 136 and xa5S, R (multiplex) for the genes Xa 21, xa13 and xa 5 respectively (Table 1). The parents were polymorphic with respect to these genes. In addition, the parents were screened with 236 rice microsatellite markers (Table 2) of which 120 were polymorphic and 60 were used for background selection.
Table 1.
Markers used for foreground selection of three bacterial blight resistance genes in marker-assisted backcross breeding
| Resistance gene | Chromosome number | Marker | Primer sequences used for gene detection | Expected size (bp) | Band type | reference | |
|---|---|---|---|---|---|---|---|
| Forward(5’-3’) | Reverse(5’-3’) | ||||||
| xa5 | 5 | xa5S (Multiplex) | GTCTGGAATTTGCTCGCGTTCG | TGGTAAAGTAGATACCTTATCAAACTGGA | 410 bp, 310 bp,180 bp | STS | Sundaram et al. 2011 |
| xa5SR/R (Multiplex) | AGCTCGCCATTCAAGTTCTTGAG | TGACTTGGTTCTCCAAGGCTT | |||||
| xa13 | 8 | RG136 | TCCCAGAAAGCTACTACAGC | GCAGACTCCAGTTTGACTTC | 530 bp, 490 bp | STS | Huang et al. 1997 |
| Xa21 | 11 | pTA248 | AGACGCGGAAGGGTGGTTCCCGGA | AGACGCGGTAATCGAAGATGAAA | 1000 bp | STS | Huang et al. 1997 |
Table 2.
Microsatellite markers those are polymorphic between Jalmagna and CRMAS 2232-85
| Chromosome | No. of markers analyzed | Total no. of polymorphic markers | Name of the polymorphic markers |
|---|---|---|---|
| 1 | 25 | 11 | RM23, RM48, RM212, RM272, RM575, RM428, RM488, SSR09, SSR 31, SSR 60 ,SSR 71 |
| 2 | 25 | 12 | RM154, RM211, RM233A, RM263, RM475, RM45, RM530,SSR11, SSR 14, SSR 44 ,SSR 71 ,SSR 85 |
| 3 | 21 | 10 | RM16, RM130, RM218, RM203,SSR 06, SSR 13, SSR 18, SSR 45, SSR 85, SSR 93 |
| 4 | 18 | 12 | RM241, RM307, RM401, RM55, RM471, RM518, RM470, SSR 04, SSR 10, SSR 19, SSR 32, SSR 40 |
| 5 | 19 | 12 | RM164, RM592, RM440, SSR 05, SSR 13, SSR 21, SSR 27, SSR 34,SSR 37, SSR 43, SSR 50, SSR 59 |
| 6 | 20 | 10 | RM225, RM276, RM340, RM402, RM586, RM589, RM588, SSR 21, SSR 31, SSR 54 |
| 7 | 21 | 9 | RM10, RM336, RM560, RM432, RM346, SSR 28, SSR 37, SSR 41, SSR 44 |
| 8 | 20 | 10 | RM223, RM241, RM407, RM3395, RM6208, RM22550, RM22506, RM8271, SSR 14 ,SSR 48 |
| 9 | 16 | 7 | RM219, RM242, RM257, RM410, RM3555,SSR 40, SSR 42 |
| 10 | 17 | 9 | RM171, RM216, RM333, RM330, SSR 03, SSR 06, SSR 11, SSR 25, SSR 30 |
| 11 | 18 | 11 | RM21, RM144, M202, RM206, RM209, RM260, RM287, SSR 3, SSR 4, SSR 11, SSR 27 |
| 12 | 17 | 7 | RM17, RM195, RM415, RM23, SSR 23, SSR 26, SSR 36 |
| Total | 236 | 120 |
Molecular markers were integrated in the backcross breeding programme upto BC3F2 generation. During the breeding procedure, foreground selection was practiced from F1 generation till BC3F3 generation at each stage to select the plants having resistance alleles of the three target genes and only progenies having the resistance alleles were advanced for the next generation (Figures 1, 2 and 3). Background selection was started from BC1F1 to BC3F1 generation and in each step genotype possessing highest genome content of the recipient parent was selected to hybridize for next backcross. A total of 650 F1 plants were produced and 150 F1 plants were tested for the hybridity and confirmed by their heterozygosity for the resistance gene linked markers of which 143 plants were observed to be true F1s. The true F1s were backcrossed using Jalmagna as a recurrent parent. These crossed seeds were raised (360 BC1F1 seeds) for further backcrossing with Jalmagna. Ninety three BC1F1 plants showed the presence of Xa21 resistance gene specific bands (1000 bp) while 91 plants showed the presence of xa13 resistance gene specific bands (490 bp and 530 bp). One hundred sixteen BC1F1 plants showed the presence of xa5 resistance gene specific bands (160 bp). Based on the amplification of resistance specific bands, 31 BC1F1 plants showed the presence of Xa21 and xa13 resistance genes while 42 BC1F1 plants showed the presence of Xa21 and xa5 resistance genes. Forty six BC1F1 plants showed the presence of xa13 and xa5 resistance genes. Only fourteen plants showed the presence of three BB resistance genes Xa21, xa13 and xa5. Out of these 14 BC1F1 progenies, plant showing 77.5% of recurrent genome (Plant No.53) was backcrossed with recurrent parent Jalmagna (Table 3).
Figure 1.
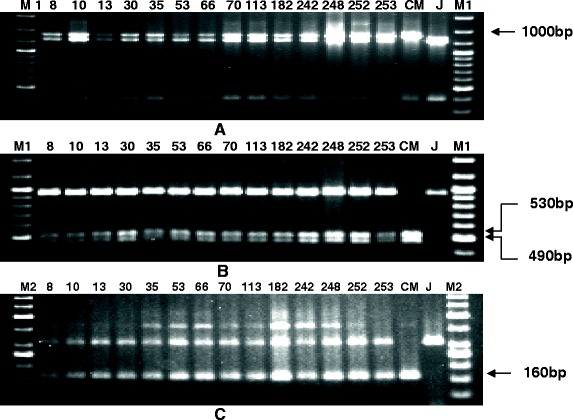
PCR amplification of markers linked to resistance genes Xa21, xa13 and xa5 using primers A) pAT248, B) RG136 and C) xa5S and xa5R of BC1F1. Lanes on the top of the gel shows the BC1F1 plant no., CM- CRMAS 2232-85, J-Jalmagna, M1-Molecular weight marker (100bp plus ladder), M2-Molecular weight marker (50bp ladder), Arrows indicate the resistance specific markers.
Figure 2.
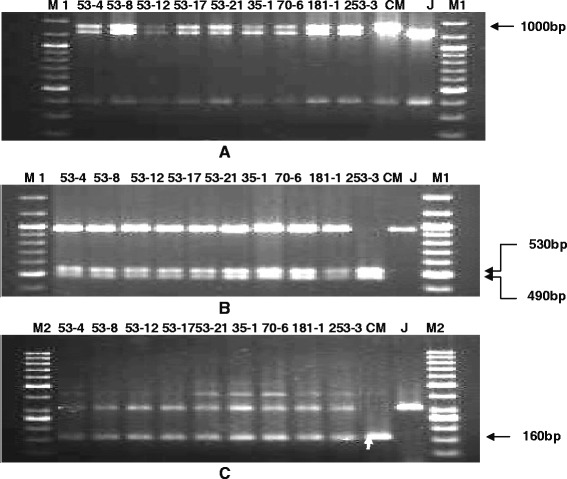
PCR amplification of markers linked to resistance genes Xa21, xa13 and xa5 using primers A) pAT248, B) RG136 and C) xa5S and xa5R of BC2F1. Lanes on the top of the gel shows the BC2F1 plant no., CM- CRMAS 2232-85, J-Jalmagna, M1-Molecular weight marker (100bp plus ladder), M2-Molecular weight marker (50bp ladder).
Figure 3.
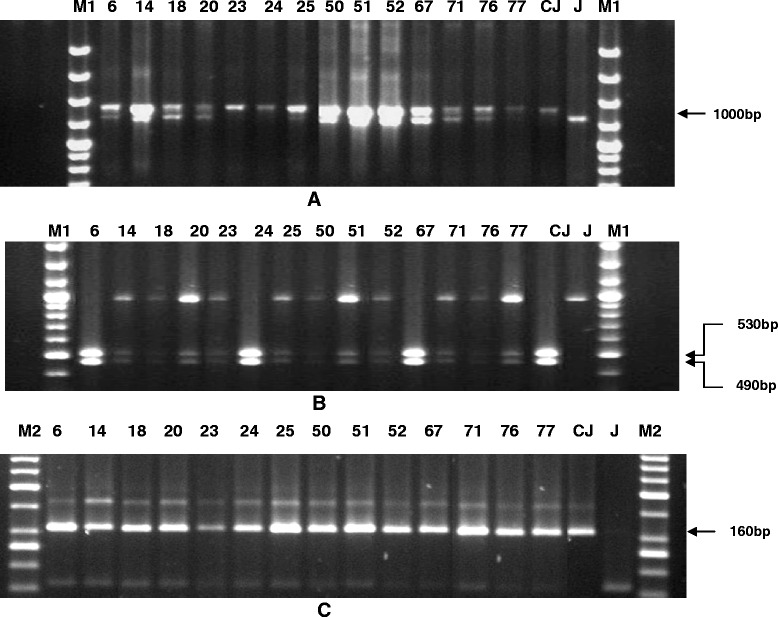
PCR amplification of markers linked to resistance genes, Xa21, xa13 and xa5 using primers A) pAT248, B) RG136 and C) xa5S and xa5R of BC3F1 plants. Lanes on the top of the gel shows the BC3F1 plant no. CM- CRMAS 2232-85, J-Jalmagna, M1-Molecular weight marker (100bp plus ladder), M2-Molecular weight marker (50bp ladder).
Table 3.
Number of triple resistant gene heterozygotes identified and estimation of recurrent parent genome contribution
| Generation |
# of plants
scored a |
# of plants that are
triple heterozygotes a, b |
Estimated maximum %
contribution of recurrent parent genome to selected backcross plant c |
Expected % contribution
of recurrent parent genome to selected backcross plant d |
|---|---|---|---|---|
| BC1F1 | 360 | 14 | 77.5 | 77.5 |
| BC2F1 | 122 | 9 | 91.8 | 91.8 |
| BC3F1 | 285 | 14 | 97 | 97 |
aAt each backcross generation, genomic DNA was isolated from derivative lines and genotyping was performed using primers that are tightly linked to BB resistant genes as described in Materials and methods.
bAt each backcross generation, fewer than expected triple heterozygotes were obtained. This is due to the fact that some of the
putative backcross progeny were obtained by inadvertent selfing of Jalmagna (the female parent in these backcrosses).
cAt each backcross generation, genomic DNA was isolated from derivative lines that are triple heterozygotes for BB resistant gene linked markers. Microsatellite markers that are polymorphic between the parental lines were then used, as described in Materials and Methods, to identify the plant with maximum recurrent parent genome contribution.
dAs per Mendelian ratios for independent gene action.
Total of 122 BC2F1 progenies were produced of which, twenty one, thirty three and thirty six BC2F1 plants showed presence of resistance genes, Xa21, xa13 and xa5, respectively. Based on the amplification pattern, 11 BC2F1 plants showed the presence of Xa21 and xa13 resistance genes while 13 BC2F1 plants showed the presence of Xa21 and xa5 resistance genes. Twenty three BC2F1 plants showed the presence of xa13 and xa5 resistance genes. Only nine plants exhibited the amplification of three resistance genes Xa21, xa13 and xa5. The background selection of these nine BC2F1 plants with sixty polymorphic SSR markers exhibited the presence of 88.13 % to 91.82 % with an average of 90.95% of recurrent genome content. The plant containing 91.82% genome content of Jalmagna (Plant No.53-21) was used for backcrossing (Table 3).
A total of 285 BC3F1 backcross derivative progenies were produced by backcrossing the plant showing 91.82% recurrent genome with the recipient parent, Jalmagna. Twenty eight BC3F1 plants were positive for Xa 21, 35 for xa 5 and 14 for xa 13. Eighteen BC3F1 plants showed the presence of Xa21 and xa13 resistance genes while 14 plants showed the presence of Xa21 and xa5 resistance genes and 14 plants showed the presence of xa13 and xa5 resistance genes. Only fourteen plants showed the presence of three resistance genes Xa21, xa13 and xa5. These BC3F1 plants showed recurrent genome content of Jalmagna ranging from 91 to 97% with an average of 92.38% (Table 3). BC3F1 derivative SPJ53-21-77 and and SPJ53-21- 25 showed more than 95% genome content of recipient parent were self pollinated to obtain the derivatives of BC3F2 generation. In BC3F2 generation, plants homozygous for three and two bacterial blight resistance gene combinations were identified. It is observed that 26 plants containing Xa21, xa13 and xa5 genes; 31 plants with Xa21 and xa5; 31 plants with Xa21, xa13 and 30 with xa13 and xa5 amongst the BC3F2 derivatives. The plants with three and two genes were grown as BC3F3 lines.
Bioassays
Bioassays conducted against eight isolates of Xoo confirmed the resistance and susceptible reaction of the donor (CRMAS 2232–85) and the recurrent (Jalmagna) respectively with the donor showing smaller range of average lesion lengths (2.1-2.8 cm) while on Jalmagna, the lesion lengths were longer (9.4-12.8 cm) (Table 4). The results indicated that the pyramided lines were better as compared to recurrent parent, Jalmagna with regard to bacterial leaf blight tolerance. Screening of the BC3F3 pyramided lines against Xoo isolates exhibited that all the pyramid lines were more effective in comparison to the recipient parent. The lesion lengths observed on the lines containing Xa21 + xa13 gene combination varied from 3.1 to 3.9 cm ; for Xa21 + xa5 combination, 3.5-4.8 cm ; for xa5 + xa13 combination 4.9 to 5.7 while 1.4 to 2.9 cm lesion length present in pyramided line containing xa5 + xa13 + Xa21 combination. The individual values for the donor parent and recurrent parent are in the range of 1.7–3.3 and 9.0–13.3 cm respectively. Though all the gene combinations tested did not show any susceptible reaction to any of the eight isolates employed, the gene pyramids with three genes displayed higher levels of disease resistance with shorter lesion lengths against all BB isolates. Results indicated that the degree of severity of the disease from the data, the order of gene combinations in conferring resistance was: xa5 + xa13 < xa5 + Xa21 < xa13 + Xa21 < xa5 + xa13 + Xa21.
Table 4.
Bacterial blight reaction of parental and BC 3 F 3 pyramided lines against different Xoo strains
|
Sl.
No. |
Pyramided lines/Xoo strains | Gene combination | Mean lesion length in cm (Mean ± standard error) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Xa-17 | Xa-7 | xa-2 | xb-7 | xc-4 | xd-1 | xa-1 | xa-5 | Disease reaction | |||
| 1 | SPJ 53-21-06 | Xa21 + xa13 + xa5 | 2.8 ± 0.4 | 2.5 ± 0.8 | 2.7 ± 0.9 | 1.9 ± 1.1 | 2.7 ± 1.0 | 2.6 ± 0.75 | 2.4 ± 0.56 | 2.5 ± 0.47 | R |
| 2 | SPJ 53-21-14 | Xa21 + xa13 + xa5 | 2.6 ± 0.6 | 2.7 ± 0.4 | 2.5 ± 1.1 | 2.5 ± 0.8 | 2.9 ± 0.7 | 2.8 ± 0.64 | 2.3 ± 0.43 | 2.3 ± 0.64 | R |
| 3 | SPJ 53-21-18 | Xa21 + xa13 + xa5 | 2.7 ± 0.3 | 2.9 ± 0.5 | 2.8 ± 1.2 | 1.8 ± 0.9 | 2.1 ± 0.9 | 2.3 ± 0.37 | 1.9 ± 0.72 | 2.1 ± 0.75 | R |
| 4 | SPJ 53-21- 20 | Xa21 + xa13 + xa5 | 2.8 ± 0.3 | 2.5 ± 0.4 | 2.7 ± 0.7 | 1.6 ± 0.5 | 1.6 ± 1.2 | 1.9 ± 0.52 | 2.6 ± 0.54 | 1.7 ± 0.83 | R |
| 5 | SPJ 53-21- 23 | Xa21 + xa13 + xa5 | 2.7 ± 0.5 | 2.6 ± 0.6 | 2.6 ± 1.5 | 2.1 ± 0.7 | 1.8 ± 1.1 | 1.7 ± 0.93 | 2.8 ± 0.71 | 1.4 ± 0.85 | R |
| 6 | SPJ 53-21-24 | Xa21 + xa13 + xa5 | 2.9 ± 0.4 | 2.8 ± 0.7 | 2.8 ± 0.5 | 2.3 ± 0.4 | 1.9 ± 1.3 | 2.9 ± 0.33 | 2.5 ± 0.83 | 2.5 ± 0.52 | R |
| 7 | SPJ 53-21-25 | Xa21 + xa13 + xa5 | 2.9 ± 0.5 | 2.7 ± 0.9 | 2.9 ± 1.3 | 2.4 ± 0.6 | 2.8 ± 0.8 | 2.6 ± 0.84 | 2.4 ± 0.95 | 2.7 ± 0.44 | R |
| 8 | SPJ 53-21-50 | Xa21 + xa13 + xa5 | 2.8 ± 0.3 | 2.4 ± 0.5 | 2.7 ± 0.7 | 1.6 ± 1.1 | 2.6 ± 0.7 | 2.1 ± 0.72 | 1.7 ± 1.2 | 1.7 ± 0.92 | R |
| 9 | SPJ 53-21-51 | Xa21 + xa13 + xa5 | 2.9 ± 0.4 | 2.7 ± 0.4 | 2.5 ± 0.8 | 2.3 ± 0.7 | 2.1 ± 0.6 | 1.6 ± 0.94 | 2.9 ± 0.52 | 1.4 ± 1.1 | R |
| 1 | SPJ 53-21-52 | Xa21 + xa13 + xa5 | 2.7 ± 0.3 | 2.8 ± 0.7 | 2.8 ± 1.1 | 1.7 ± 0.8 | 2.9 ± 0.5 | 2.2 ± 0.88 | 2.9 ± 0.47 | 1.9 ± 1.2 | R |
| 11 | SPJ 53-21-67 | Xa21 + xa13 + xa5 | 2.5 ± 0.7 | 2.6 ± 0.9 | 2.6 ± 0.3 | 1.9 ± 0.9 | 2.8 ± 0.8 | 2.5 ± 1.12 | 1.8 ± 0.85 | 2.3 ± 0.53 | R |
| 12 | SPJ 53-21-71 | Xa21 + xa13 + xa5 | 2.6 ± 0.6 | 2.5 ± 0.5 | 2.5 ± 0.7 | 2.2 ± 1.0 | 2.8 ± 0.4 | 2.7 ± 0.85 | 2.3 ± 0.63 | 2.8 ± 0.44 | R |
| 13 | SPJ 53-21-76 | Xa21 + xa13 + xa5 | 2.8 ± 0.4 | 2.6 ± 0.7 | 2.7 ± 0.6 | 2.1 ± 0.6 | 2.9 ± 0.5 | 2.9 ± 0.67 | 2.6 ± 0.52 | 2.9 ± 0.52 | R |
| 14 | SPJ 53-21-77 | Xa21 + xa13 + xa5 | 2.9 ± 0.3 | 2.7 ± 0.9 | 2.8 ± 1.2 | 2.4 ± 0.5 | 2.8 ± 0.4 | 2.9 ± 0.69 | 2.8 ± 0.73 | 2.8 ± 0.55 | R |
| 15 | SPJ 53-21-27 | Xa21 + xa13 | 3.6 ± 0.4 | 3.4 ± 1.1 | 3.5 ± 1.1 | 3.1 ± 0.6 | 3.4 ± 0.7 | 3.7 ± 0.4 | 3.6 ± 1.2 | 3.7 ± 0.63 | MR |
| 16 | SPJ 53-21-15 | Xa21 + xa13 | 3.9 ± 0.5 | 3.8 ± 1.2 | 3.6 ± 1.0 | 3.7 ± 0.5 | 3.7 ± 0.9 | 3.8 ± 0.77 | 3.4 ± 1.1 | 3.85 ± 0.7 | MR |
| 17 | SPJ 53-21-38 | Xa21 + xa5 | 4.1 ± 0.5 | 3.7 ± 1.4 | 3.9 ± 0.7 | 3.5 ± 0.6 | 3.7 ± 1.2 | 4.3 ± 0.7 | 4.2 ± 0.4 | 3.9 ± 0.7 | MR |
| 18 | SPJ 53-21-66 | Xa21 + xa5 | 4.5 ± 0.8 | 3.9 ± 1.3 | 4.4 ± 0.6 | 4.8 ± 0.8 | 3.9 ± 0.9 | 4.3 ± 0.5 | 4.2 ± 0.63 | 4.1 ± 0.65 | MR |
| 19 | SPJ 53-21-13 | xa13 + xa5 | 5.6 ± 1.1 | 5.3 ± 0.8 | 5.1 ± 0.6 | 4.9 ± 0.7 | 5.5 ± 1.3 | 5.3 ± 0.49 | 5.7 ± 0.72 | 4.9 ± 0.53 | MR |
| 20 | SPJ 53-21-36 | xa13 + xa5 | 5.4 ± 0.7 | 5.1 ± 0.8 | 5.0 ± 0.7 | 5.2 ± 0.7 | 5.6 ± 0.9 | 5.1 ± 0.76 | 5.2 ± 0.84 | 5.3 ± 0.65 | MR |
| 21 | CRMAS 2232-85 |
Xa21 + xa13 + xa5 | 2.8 ± 0.4 | 2.5 ± 0.3 | 2.4 ± 0.4 | 2.1 ± 0.4 | 2.8 ± 0.5 | 2.4 ± 0.4 | 2.6 ± 0.8 | 2.2 ± 0.6 | R |
| 22 | Jalmagna | - | 12.6 ± 1.7 | 11.4 ± 1.4 | 12.8 ± 1.5 | 9.4 ± 1.2 | 11.6 ± 1.6 | 9.8 ± 1.8 | 10.2 ± 1.7 | 11.6 ± 1.8 | S |
Yield and agro-morphological traits of the pyramided lines
Fourteen three-gene pyramid and six two genes pyramid lines at BC3F3 generation along with the donor and recipient parents were evaluated during wet season, 2013 at CRRI, Cuttack. The recipient parent, Jalmagna recorded mean grain yield of 17.35 g/plant, while the donor parent (Swarna BB pyramided line) recorded 20.5 g/plant. The test entries viz., SPJ23, SPJ25, SPJ50, SPJ51, SPJ52 and SPJ77 showed grain yields higher than recurrent parent, Jalmagna (Table 5). Many test entries did not show any significant variation as compared to Jalmagna in terms of flowering duration, panicles/m2, plant stature as well as other characters that are considered under distinctness, uniformity and stability (DUS) tests. The genetic distance coefficient on 14 agro-morphologic traits of 20 pyramids and two parental lines revealed that two clusters were observed and it is interesting to note that all the pyramided lines are similar to the recipient parent, Jalmagna and are clubbed in cluster1 while in cluster 2, only solitary line the donor parent is accommodated. (Table 5; Figure 4A).
Table 5.
Agro-morphologic traits of pyramided and parental lines in BC 3 F 3 generation
| Pyramided lines | Plant height (cm) | Days to 50% flow | Panicles/plant | Panicle length(cm) | No of grains/ panicle | Fertility % | 1000- seed weight(g) | Single plant yield(g) | Grain length (mm) | Grain Breadth (mm) | Flag leaf length(cm) | Flag leaf breadth (cm) | 2 nd leaf length (cm) | 2 nd leaf breadth (cm) | Auricle colour | Collar colour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPJ 53-21-06 | 150 | 122 | 10 | 21.95 | 211.5 | 87 | 22.45 | 16.15 | 0.76 | 0.88 | 36.0 | 1.2 | 69.0 | 1.2 | Green | yellowish |
| SPJ 53-21-14 | 178 | 124 | 12 | 22.1 | 204 | 86 | 22.05 | 16.85 | 0.73 | 0.875 | 39.0 | 1.2 | 49.0 | 1.2 | Green | yellowish |
| SPJ 53-21-18 | 163.5 | 121 | 11 | 26.25 | 205.5 | 84 | 21.6 | 17 | 0.67 | 0.89 | 41.0 | 0.9 | 59.0 | 0.9 | Green | yellowish |
| SPJ 53-21- 20 | 172.5 | 124 | 11 | 26.25 | 208 | 84 | 22.7 | 17.1 | 0.75 | 0.86 | 47.0 | 1.2 | 75.0 | 1.2 | Green | yellowish |
| SPJ 53-21- 23 | 158.5 | 123 | 12 | 24.2 | 207.5 | 83 | 21.5 | 16.5 | 0.74 | 0.88 | 45.0 | 1.1 | 58.0 | 1.1 | Green | yellowish |
| SPJ 53-21-24 | 153.5 | 122 | 9 | 21.25 | 200.5 | 84 | 23.25 | 16.65 | 0.75 | 0.88 | 35.0 | 1.1 | 41.0 | 1.1 | Green | Light purple |
| SPJ 53-21-25 | 151.5 | 123 | 8 | 22.8 | 199 | 82 | 21.35 | 16.65 | 0.75 | 0.87 | 47.5 | 1.2 | 56.0 | 1.2 | Green | Green |
| SPJ 53-21-50 | 177 | 125 | 10 | 24.95 | 203.5 | 86 | 23.05 | 16.85 | 0.75 | 0.87 | 35.0 | 1.2 | 62.0 | 1.2 | Green | Light purple |
| SPJ 53-21-51 | 152.5 | 123 | 10 | 24.95 | 208.5 | 82 | 21.5 | 17.95 | 0.745 | 0.87 | 43.0 | 1.1 | 69.0 | 1.1 | Green | yellowish |
| SPJ 53-21-52 | 177 | 121 | 11 | 21.9 | 200.5 | 83 | 23 | 16.65 | 0.75 | 0.85 | 34.0 | 1.0 | 43.0 | 1.0 | Green | yellowish |
| SPJ 53-21-67 | 178.5 | 122 | 9 | 24.95 | 203 | 85.5 | 21.05 | 17 | 0.745 | 0.86 | 36.0 | 1.2 | 54.0 | 1.2 | Green | yellowish |
| SPJ 53-21-71 | 176.5 | 120 | 10 | 25.6 | 208 | 85 | 20.1 | 16.85 | 0.74 | 0.875 | 46.0 | 1.1 | 63.0 | 1.1 | Green | yellowish |
| SPJ 53-21-76 | 182 | 123 | 11 | 25.45 | 205.5 | 84 | 21.65 | 16.55 | 0.74 | 0.88 | 38.0 | 1.0 | 70.0 | 1.0 | Green | yellowish |
| SPJ 53-21-77 | 183 | 124 | 12 | 23.4 | 203 | 86 | 19.5 | 18.2 | 0.74 | 0.89 | 39.0 | 1.2 | 54.0 | 1.2 | Green | yellowish |
| SPJ 53-21-27 | 161.5 | 123 | 11 | 24.75 | 200.5 | 85.5 | 21.8 | 17.15 | 0.725 | 0.88 | 32.5 | 1.5 | 55.0 | 1.5 | Green | yellowish |
| SPJ 53-21-15 | 158.5 | 122 | 9 | 26.05 | 196.5 | 83.5 | 20.1 | 18.3 | 0.74 | 0.86 | 33.6 | 1.4 | 47.0 | 1.4 | Green | yellowish |
| SPJ 53-21-38 | 170 | 119 | 10 | 26.05 | 196.5 | 87 | 22.05 | 17.15 | 0.72 | 0.89 | 34.2 | 1.3 | 56.2 | 1.3 | purple | Purple |
| SPJ 53-21-66 | 153.5 | 125 | 11 | 25.05 | 198.5 | 86 | 21.95 | 16.8 | 0.73 | 0.88 | 33.7 | 1.4 | 58.5 | 1.4 | Green | yellowish |
| SPJ 53-21-13 | 169.5 | 124 | 11 | 25.15 | 204.5 | 88 | 21.9 | 18.25 | 0.74 | 0.87 | 34.1 | 1.5 | 51.7 | 1.5 | Green | yellowish |
| SPJ 53-21-36 | 152 | 123 | 12 | 24 | 208.5 | 84 | 21.8 | 16.3 | 0.74 | 0.88 | 34.4 | 1.3 | 53.4 | 1.3 | Purple | Purple |
| CRMAS 2232-85 | 105.5 | 105 | 12 | 24.15 | 218 | 84.5 | 18.2 | 20.5 | 0.72 | 0.84 | 34.0 | 2.0 | 33.0 | 2.0 | Green | yellowish |
| Jalmagna | 174 | 130 | 9 | 24.95 | 186 | 85.5 | 23.2 | 17.35 | 0.75 | 0.88 | 28.0 | 1.5 | 52.0 | 1.5 | Purple | Purple |
| LSD5% | 15.85 | 3.075 | 1.68 | 46.67 | 7.01 | 1.42 | 4.47 | 0.86 | 0.108 | 6.048 | 0.424 | 7.76 | 0.47 | |||
| CV% | 4.7 | 14.1 | 3.3 | 11.0 | 4.0 | 3.2 | 12.5 | 5.6 | 6.0 | 7.7 | 16.2 | 6.7 | 17.8 |
Figure 4.
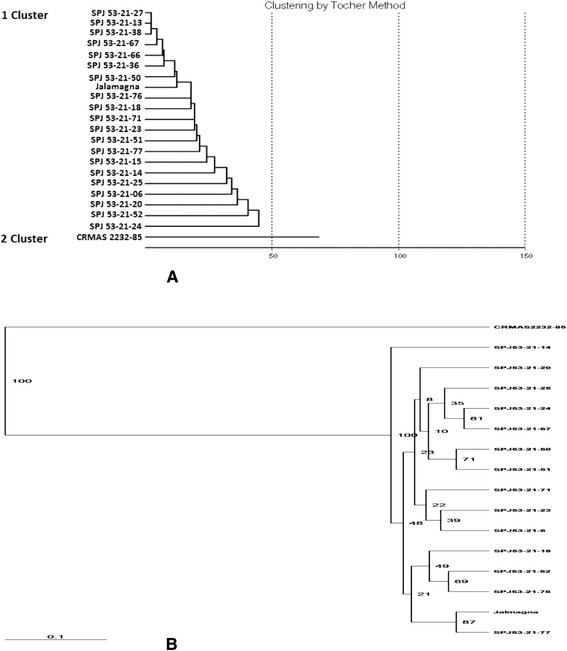
Dendrograms illustrating the genetic relationship between parents and pyramided lines (A) based on 14 agro-morphological traits; (B) based on microsatellite markers.
Background selection
The background selection was carried out for estimating the recurrent parent’s genome content in the pyramided lines. Background selection was performed by using 60 SSR markers among the lines possessing three resistance gene combinations in BC1F1, BC2F1 and BC3F1 generations. At BC3F1 generation, a total of 120 alleles from 60 markers were observed. The similarity co-efficiency among all lines ranged from 0.791 to 0.952 suggesting a high level of genetic similarity between the pyramids and Jalmagna. The dendrogram generated using the SSR data grouped the 14 three-gene pyramid lines into two major clusters (Figure 4B) with cluster I having CRMAS 2232–85 and rest 13 pyramided lines were clubbed in cluster II along with Jalmagna. Few members of cluster II viz., SPJ53-21-77, SPJ53-21-25, SPJ53-21-52 and SPJ53-21-18 were very close to Jalmagna with 97, 95.33, 92.48 and 92.48% respectively.
Analysis of genome introgression on the carrier and non-carrier chromosomes
In CRMAS2232-85/Jalmagna combination, 5–6 microsatellite markers on each of three carrier chromosomes in the genomic region flanking to xa5, xa13 and Xa21 were polymorphic. Based on six markers analysis, all the 14 lines showed heterozygosity for donor segment introgression of xa5 between marker HYV59 and HYV5-37 in BC3F1 generation while exhibited homozygocity for Jalmagna genome and no drag to xa5 gene was observed. In the flanking region of xa13, for five polymorphic markers, nine lines showed introgression of the donor segment of marker HYV14. In case of Xa21, six pyramid lines showed genetic drag of donor segment with the marker segment RM144 (Figure 5).
Figure 5.
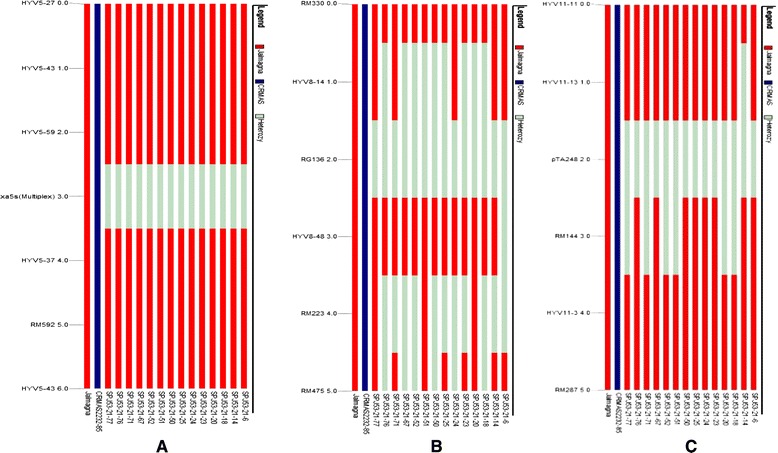
Analysis of genome introgression of 14 pyramided lines associated with resistance genes (A) xa5 on chromosome 5; (B) xa13 on chromosome 8 and (C) Xa21 on chromosome 11 in Jalmagna and CR MAS 2232-85 BC3F3 derivatives.
Discussion
Integration of molecular markers to the backcross breeding was highly effective for transfer of three bacterial blight resistance genes. Phenotypic selections in three backcrossing and two selfing generations coupled with SSR based background selection was sufficient for transfer of Xa21, xa13 and xa5 genes into popular deepwater variety Jalmagna background. Jalmagna is a very long duration and strongly photo-sensitive variety. Again under deepwater situation, control of the disease using chemicals was a very difficult task. Here, integrating molecular markers and using very less generations and advancement of some generations under RGA helped to obtain the broad spectrum BB resistant lines for deepwater ecology. The transferred genes in the recombinants did exhibit high level of resistance against the most virulent BB isolates that is comparable to the reaction level of CRMAS 2232–85, the donor parent and the results are similar to earlier reports (Huang et al. 1997; Sanchez et al. 2000; Singh et al. 2001; Shanti et al. 2001; Bharatkumar et al. 2008; Hu et al. 2008; Perez et al. 2008; Sundaram et al., 2008; Rajpurohit et al., 2011; Dokku et al., 2013; Suh et al., 2013). The three gene combination pyramided lines expressed higher levels of resistance in comparison to parental lines, two and single gene combination. The results suggest that two gene combinations with Xa21 + xa13 was most effective with shorter lesions lengths followed by Xa21 + xa5 while lines with xa13 + xa5 were relatively less effective. Lines with Xa21 in combination with either xa5, xa13, or both have shown promise advocating the utility of Xa21 in achieving higher levels of resistance in rice as reported earlier (Singh et al. 2001; Sanchez et al. 2000; Sridhar et al. 1999; Huang et al. 1997) suggesting that synergistic action and/or quantitative complementation between the resistant genes might result in enhanced levels of resistance (Sanchez et al. 2000).
All the three resistance genes that have been considered in the present work have been cloned and characterized. Xa21 is a dominant resistance gene that encodes a receptor kinase containing NBS-LRR domains (Song et al. 1995), while xa5 is a recessive resistance gene and encodes a variant form of transcription factor cIIa (Iyer and McCouch 2004). The xa13 resistance gene is also recessive in nature and has been shown to be a mutation in the promoter region of a gene that is a homolog of the nodulin MtN3 (Chu et al. 2006b). In rice lines containing the dominant (susceptibility) allele of the gene, the expression of the nodulin homolog is up regulated upon infection with Xoo. It appears that the increased expression of this gene is necessary for Xoo to grow on rice. This up regulation does not occur in rice lines containing the resistance (recessive) xa13 allele (Yang, 2006). The apparently different modes of action of the three resistance genes used in this work might contribute to make the resistance in the three-gene pyramid lines quite durable. There is a variation in the theoretically expected value of contribution from the recurrent parent genome to the BC1F1 plants and in other backcross generations. As per reports of Sundaram et al. 2008, there might be exercising a “pull” for introgression of the Xa21, xa13 and xa5 genes during selection, which favors inheritance of additional unlinked loci from the donor genome in BC1F1 plants and BC2F1 generation. But, we found no pull effect during the transfer of Xa21, xa13 and xa5 genes to different backcross generations.
Selection of plants similar to the recurrent parent from BC3F1 stage was the strategy followed in the study and the graphical genotyping data supports that view as genotype SPJ53-21-77 had 97% of the recurrent parent genome having donor segments of target resistance genes xa5, xa13 and Xa21 and further no linkage drag in regions flanking Xa21, xa13 and xa5 is observed. The high recurrent genome recovery observed in many pyramid lines may be due to the use of more number of polymorphic microsatellite markers. Similar results were obtained in case of Sundaram et al. 2008; Dokku et al. 2013; Suh et al. 2013 suggesting more number of background markers. No genetic linkage drag was observed for the transfer of genes Xa21, xa13 and xa5 (Figure 5) may be due to mega variety used as the donor source for BB resistance genes. The mega variety, Swarna is a highly adapted variety for the favorable ecology. Results indicated that a broad based highly adapted variety as source of donor may give better performance and less drag as compared to the wild and land races as donor. It is expected that all the favorable genes are accumulated in the mega variety and subsequently transfer of some of these genes is improving further the background of the pyramided lines. The approach used in the study ensured the realization of the major objective resulting in the release of a cultivar with enhanced resistance to BB and accelerated recovery of recurrent genome with better yield.
Yield and agro-morphologic data of 20 pyramided two parental lines revealed that the pyramided lines possessed excellent features of recurrent parent and also yielding ability with tolerance to bacterial blight resistance. This indicates that some pyramid lines are very close to the recurrent parent and some are even better than the recurrent parent with respect to yield. The higher yield of the pyramided lines may be due to inheritance of some yield traits or QTLs of mega variety (Swarna BB pyramid) used here as the donor parent, besides the recurrent parent Jalmagna to the pyramided line. The complete recovery of the yield and grain quality characters of Jalmagna along with transfer of three BB resistance genes of CRMAS 2232–85 is a very significant achievement. This is particularly so because yield and agro-morphologic traits is multigenic traits encoded by loci that are distributed across the rice genome. The traits recovery of Jalmagna was due to integration of many polymorphic markers in the backcross breeding program. As per our analysis, we find that there is a variation from the theoretically expected 75% contribution from the recurrent parent genome to the BC1F1 plants. All the selected BC1F1 plants had a recurrent parent genome contribution more than the expected 75%. Again in BC2F2, it had highest gemone content of 91.8% along with the target genes. During BC3F1, the genome content of Jalmagna in selected derivative (SPJ53-21-77) was as high as 97%. The background selection with many markers accelerated the recovery of recurrent genome suggests that selection for introgression of the Xa21, xa13 and xa5 genes has no antagonistic effects for yield and other traits.
The field evaluation of BC3F3 progenies showed that the best entry had better yielding than the Jalmagna parent. Besides, BB resistance, the pyramided line. SPJ53-21-77 was better yielder than its recurrent parent and equivalent to agro-morphological traits and grain quality features of the recurrent parent. The high levels of resistance to BB and the absence of any yield penalty due to accumulation of resistance genes in the pyramids provides us a successful example of the integrated approach of selection at both molecular and phenotypic levels for transfer of the desired trait(s) and recovery of the recurrent parental genome. Development of broad-spectrum resistance against BB in the Indian subcontinent is a major challenge due to the rich diversity of the agro-climatic zones where rice is cultivated, as well as the presence of a number of genetically distinct virulent Xoo strains in different geographical areas of India. Deployment of a three gene combination like xa5 + xa13 + Xa21 can achieve durable and broad-spectrum resistance in many BB prone rice growing areas in India including the deepwater ecosystem. The study clearly establishes the utility of MAS in pyramiding recessive genes like xa5 and xa13, and dominant gene Xa21 to present a multiple gene barrier against one of the most destructive diseases of rice in a long duration, photosensitive and deepwater rice.
Conclusion
Marker-assisted backcrossing using functional markers reduce the risk of false selection in recombination between the molecular marker and the gene of interest. We were successful in identifying superior recombinations for three BB resistance genes (Xa21, xa13 and xa5) in the homozygous condition in a long duration, photosensitive and deepwater rice variety. The pyramided genotypes can be further be used for multi-location testing to be released as variety in the country or be used as potential BB resistance donors. The BB pyramided deepwater breeding lines, which are developed through MAS and phenotypic selection, will be of practical value in providing durable bacterial blight resistance in the deepwater growing region where control through chemicals under deepwater situation was less effective. These BB pyramided lines are expected to have a high impact on the yield stability and sustainability of deep water rice production.
Methods
Plant materials and breeding method
The donor parent CRMAS 2232–85, a derivative of Swarna and IRBB 60 cross contains three BB resistance genes xa5, xa13 and Xa21 in the background of mega variety Swarna. The donor parent was developed at Central Rice Research Institute (CRRI), Cuttack, India (Sundaram et al. 2014). The recurrent parent was Jalmagna, a highly popular variety of deepwater ecosystem of India but highly susceptible to bacterial blight disease. Jalmagna was hybridized with CRMAS 2232–85 and F1 plants were backcrossed with recipient parent Jalmagna. Marker-assisted backcross method was followed up to BC3 generation and around 200 plants/lines were genotyped at each generation for the presence of the target genes and only positive plants having the resistance alleles were advanced to the next generation. Foreground selection continued till BC3F3 to identify pure homozygous lines for all three target genes while background selection was up to BC3F1 generation. Selection based on foreground, background and morphological traits was practiced from BC1F1 onwards for identification of lines that were similar to the recurrent parent. Rapid Generation Advancement (RGA) facility was used during dry season as Jalmagna was a strongly photo-sensitive and very long duration variety. The schematic diagram for development of BB pyramided lines is presented in Figure 6.
Figure 6.

Schematic diagram for Pyramiding bacterial blight resistance genes into variety, Jalmagna through MAS (Figures in parentheses indicate the number of hybrids/ lines raised in that generation).
Screening for bacterial blight resistance
For field evaluation against BB, the inoculums of eight predominant Xoo isolates of Orissa prepared by suspending the bacterial mass in sterile water to a concentration of aproximately109 cells/ml (Kauffman et al. 1973). Four leaves from four different plants of each entry were clip inoculated at the maximum tillering stage and lesion lengths (LL) were recorded after 15 days. The disease symptoms were scored as resistant (R, LL ≤ 3.0 cm), moderately resistant (MR, 3.0 cm < LL ≤ 6.0 cm), moderately susceptible (MS, 6.0 cm < LL ≤ 9.0 cm) or susceptible (S, LL > 9.0 cm) (Amante-Bordeos et al. 1992).
Characterization for agro-morphological traits
Thirty days’ old seedlings of the BC3F3 pyramid lines and the parents (Jalmagna and CRMAS 2232–85) were transplanted in three rows with twenty five plants per row per entry at 15 × 20 cm spacing under a randomized complete block design with two replications at the experimental farm of Central Rice Research Institute (CRRI), Cuttack. Data were recorded on ten plants from each entry and replication for agronomic traits like plant height, tillers/plant, panicle length, number of filled grains/panicle, 1000-grain weight, flag leaf, 2nd leaf length and breadth while days to 50% flowering was recorded on whole plot basis data analysis was performed using SAS statistical software (SAS Institute Inc. 2010).
DNA isolation and PCR amplification
Mini scale DNA isolation for PCR analysis was carried out as per Dellaporta et al. (1983). The PCR reaction mixture contained 50 ng templates DNA, 5 pico mole of each of the primers, 200 μM dNTPs, 1 X PCR buffer (10 mM Tris–HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, and 0.01 mg/ml gelatin) and 0.6 unit of Taq DNA polymerase in a volume of 20 μl and amplification of target sequences were as per earlier reports (Table 1). The PCR products of STS marker RG 136 were digested with restriction enzymes HinfI as per manufacturer’s instructions. The PCR products and the DNA fragments produced by restriction digestions were separated by gel electrophoresis and gel images were analyzed on gel documentation system (SynGene).
Marker analysis
The primers employed for the three target genes were all from published reports (Table 1). Of the 236 SSRs markers used for parental polymorphism survey, 120 were found to be polymorphic between the parents (range 4–6 per chromosome) and 60 were used for background selection. Data were analyzed and similarity matrix was constructed from binary data with Jaccard’s coefficients and dendrogram was generated with unweighted pair group method arithmatic average (UPGMA) algorithm, using FreeTree software (Hampl et al. 2001; Pavalíce et al. 1999) and the dendrograms were visualized by Treeview 32 software (Page 1996). Graphical Geno Types (GGT) Version 2.0 (Van Berloo 1999) software programme was used for the assessment of the genomic contribution of the parent in the selected recombinants based on SSR marker data.
Acknowledgments
The authors are highly grateful to the Director, Central Rice Research Institute, and Head, Crop Improvement Division, Central Rice Research Institute Cuttack for providing all the necessary facilities. The work was supported by a grant from Department of Biotechnology, Govt. of India.
Footnotes
Competing interests
We declare that the funding agencies and organizations are acknowledged and there is no financial or non financial competing interests.
Authors’ contributions
SKP conceived the study and participated in its design and coordination and helped to draft the manuscript. DKN and SM carried out the phenotyping and molecular experiments. LB was involved in designing and analysis of the molecular experiments. SRB and AA carried out the crossing work and statistical analysis. EP carried out the analysis and interpretation of data and contributed for preparation of manuscript. SL carried out the bioassays. All authors read and approved the final manuscript.
Authors’ information
Crop Improvement Division, Central Rice Research Institute, Cuttack, Odisha, 753006, India.
Contributor Information
Sharat Kumar Pradhan, Email: pradhancrri@gmail.com.
Deepak Kumar Nayak, Email: deepak_papu1984@yahoo.co.in.
Soumya Mohanty, Email: smmohanty.mohanty9@gmail.com.
Lambodar Behera, Email: l_behera@yahoo.com.
Saumya Ranjan Barik, Email: saumya_bt06@yahoo.co.in.
Elssa Pandit, Email: elsambio@gmail.com.
Srikanta Lenka, Email: srikantalenka@yahoo.in.
Annamalai Anandan, Email: anandanau@yahoo.com.
References
- Amante-Bordeos A, Sitch L, Nelson R, Dalmacio R, Oliva N, Aswidinnoor H, Leung H. Transfer of bacterial blight and blast resistance from the tetraploid wild rice Oryza minuta to cultivated rice, Oryza sativa. Theor Appl Genet. 1992;84:345–354. doi: 10.1007/BF00229493. [DOI] [PubMed] [Google Scholar]
- Bharatkumar S, Paulraj RSD, Brindha PV, Kavitha S, Gnanamanickam SS. Improvent of bacterial blight resistance in rice cultivars ajayothi and IR50 via marker-assisted backcross breeding. J Crop Improve. 2008;21:101–116. doi: 10.1300/J411v21n01_07. [DOI] [Google Scholar]
- Bhasin H, Bhatia D, Raghuvanshi S, Lore SJ, Gurpreet K, Sahi KG, Kaur B, Vikal Y, Singh K. New PCR-based sequence-tagged site marker for bacterial blight resistance gene Xa38 of rice. Mol Breeding. 2012;30:607–611. doi: 10.1007/s11032-011-9646-y. [DOI] [Google Scholar]
- Chu Z, Fu B, Yang H, Xu C, Li Z, Sanchez A, Park YJ, Bennetzen JL, Zhang Q, Wang S. Targeting xa13, a recessive gene for bacterial blight resistance in rice. Theor Appl Genet. 2006;112:455–461. doi: 10.1007/s00122-005-0145-6. [DOI] [PubMed] [Google Scholar]
- Chu Z, Yuan M, Yao J, Ge X, Yuan B, Xu C, Li X, Fu B, Li Z, Bennetzen JL. Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 2006;20:1250–1255. doi: 10.1101/gad.1416306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. doi: 10.1007/BF02712670. [DOI] [Google Scholar]
- Devadath S. Chemical control of bacterial blight of rice. In: IRRI, editor. Bacterial blight of rice. Manila, Philippines: IRRI; 1989. pp. 89–98. [Google Scholar]
- Dokku P, Das KM, Rao GJN. Pyramiding of four resistance genes of bacterial blight in Tapaswini, an elite rice cultivar, through marker-assisted selection. Euphytica. 2013;192:87–96. doi: 10.1007/s10681-013-0878-2. [DOI] [Google Scholar]
- Gu K, Yang B, Tian D, Wu L, Wang D, Sreekala C, et al. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature. 2005;435:1122–1125. doi: 10.1038/nature03630. [DOI] [PubMed] [Google Scholar]
- Gu K, Sangha JS, Li Y, Yin Z. High resolution genetic mapping of bacterial blight resistance gene Xa10. Theor Appl Genet. 2008;116:155–163. doi: 10.1007/s00122-007-0655-5. [DOI] [PubMed] [Google Scholar]
- Hampl V, Pavlicek A, Flegr J. Construction and bootstrap analysis of DNA fingerprinting based phylogenetic trees with the freeware program FreeTree: application to trichomonad parasites. Intl J Syst Evol Microbiol. 2001;51:731–735. doi: 10.1099/00207713-51-3-731. [DOI] [PubMed] [Google Scholar]
- Hu KM, Qiu DY, Shen XL, Li XH, Wang SP. Isolation and Manipulation of Quantitative trait Loci for Disease Resistance in Rice Using a Candidate Gene Approach. Mol Plant. 2008;1(5):786–793. doi: 10.1093/mp/ssn039. [DOI] [PubMed] [Google Scholar]
- Huang N, Angeles ER, Domingo J, Magpantay G, Singh S, Zhang G, Kumaravadevil N, Bennett J, Khush GS. Pyramiding of bacterial blight resistance genes in rice: marker assisted selection using RFLP and PCR. Theor Appl Genet. 1997;95:313–320. doi: 10.1007/s001220050565. [DOI] [Google Scholar]
- Iyer AS, McCouch SR. The rice bacterial blight resistance gene xa5 encodes a novel form of disease resistance. Mol. Plant Microbe Interact. 2004;17:1348–1354. doi: 10.1094/MPMI.2004.17.12.1348. [DOI] [PubMed] [Google Scholar]
- Jena KK, Mackill DJ. Molecular markers and their use in marker-assisted selection in rice. Crop Sci. 2008;48:1266–1276. doi: 10.2135/cropsci2008.02.0082. [DOI] [Google Scholar]
- Joseph M, Gopalakrishnan S, Sharma RK. Combining bacterial blight resistance and basmati quality characteristics by phenotypic and molecular marker assisted selection in rice. Mol Breed. 2004;13:377–387. doi: 10.1023/B:MOLB.0000034093.63593.4c. [DOI] [Google Scholar]
- Kauffman HE, Reddy APK, Hsien SPY, Merca SD. An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis Report. 1973;57:537–541. [Google Scholar]
- Khush GS. What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol Biol. 2005;59:1–6. doi: 10.1007/s11103-005-2159-5. [DOI] [PubMed] [Google Scholar]
- Khush GS, Mackill DJ, Sidhu GS. Breeding rice for resistance to bacterial leaf blight. In: IRRI, editor. Bacterial blight of rice. Manila, Philippines: IRRI; 1989. pp. 207–217. [Google Scholar]
- Mew TW, Vera Cruz CM, Medalla ES. Changes in the race frequency of Xanthomonas oryza pv. oryzae in response to the planting of rice cultivars in the Philippines. Plant Dis. 1992;76:1029–1032. doi: 10.1094/PD-76-1029. [DOI] [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Pavalíce A, Hrda S, Flegr J. Free Tree—freeware program for construction of phylogenetic trees on the basis of distance data and bootstrap/jackknife analysis of the tree robustness, Application in the RAPD analysis of genus Frenkelia. Folia Biol (Praha) 1999;45:97–99. [PubMed] [Google Scholar]
- Perez LM, Redona ED, Mendioro MS, Vera Cruz CM, Leung H. Introgression of Xa4, Xa7 and Xa21 for resistance to bacterial blight in thermosensitive genetic male sterile rice (Oryza sativa L.) for the development of two-line hybrids. Euphytica. 2008;164:627–636. doi: 10.1007/s10681-008-9653-1. [DOI] [Google Scholar]
- Pha PN, Lang NT. Marker assisted selection in rice breeding for bacterial leaf blight. Omonrice. 2004;12:19–26. [Google Scholar]
- Rajpurohit D, Kumar R, Kumar M, Paul P, Awasthi AA, Basha PO, Puri A, Jhang T, Singh K, Dhaliwal HS. Pyramiding of two bacterial blight resistance and a semi dwarfing gene in Type 3 Basmati using marker-assisted selection. Euphytica. 2011;178:111–126. doi: 10.1007/s10681-010-0279-8. [DOI] [Google Scholar]
- Rao KK, Lakshminarasu M, Jena KK. DNA markers and marker-assisted breeding for durable resistance to bacterial blight of rice. Biotechnol Adv. 2002;20:33–47. doi: 10.1016/S0734-9750(02)00002-2. [DOI] [PubMed] [Google Scholar]
- Sanchez AC, Brar DS, Huang N, Khush GS. Sequence tagged site markers-assisted selection for three bacterial blight resistance genes in rice. Crop Sci. 2000;40:792–797. doi: 10.2135/cropsci2000.403792x. [DOI] [Google Scholar]
- SAS Institute Inc. SAS ® 9.2 Language Reference: Concepts. Second. Cary, NC: SAS Institute Inc. USA; 2010. p. 626 978-1–60764–449–1. [Google Scholar]
- Shanti ML, George MLC, Vera Cruz CM, Bernardo MA, Nelson RJ, Leung H, Reddy JN, Sridhar R. Identification of resistance genes effective against bacterial leaf blight pathogen in eastern India. Plant Dis. 2001;85:506–512. doi: 10.1094/PDIS.2001.85.5.506. [DOI] [PubMed] [Google Scholar]
- Singh GP, Srivastava MK, Singh RM, Singh RV. Variation in quantitative and Xanthomonas oryzae. Plant Dis Rep. 1977;57:537–541. [Google Scholar]
- Singh S, Sidhu JS, Huang N, Vikal Y, Li Z, Brar DS, Dhaliwal HS, Khush GS. Pyramiding three bacterial blight resistance genes (xa-5, xa-13 and Xa-21) using marker-assisted selection into indica rice cultivar PR-106. Theor Appl Genet. 2001;102:1011–1015. doi: 10.1007/s001220000495. [DOI] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, Fauquet C, Ronald PC. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- Sonti RV. Bacterial leaf blight of rice: new insights from molecular genetics. Curr Sci. 1998;74:206–212. [Google Scholar]
- Sridhar R, Reddy JN, Singh UD, Agrawal PK. Usefulness of combinations of bacterial blight resistance genes at Cuttack, Orissa, India. IRRN. 1999;24(2):24–25. [Google Scholar]
- Suh JP, Jeung JU, Noh TH, Cho YC, Park SH, Park HS, Shin MS, Kim CK, Jena KK. Development of breeding lines with three pyramided resistance genes that confer broad-spectrum bacterial blight resistance and their molecular analysis in rice. Rice. 2013;6:5. doi: 10.1186/1939-8433-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram RM, Vishnupriya MR, Biradar SK, Laha GS, Reddy GA, Rani NS, Sarma NP, Sonti RV. Marker assisted introgression of bacterial blight resistance in Samba Mahsuri, an elite indica rice variety. Euphytica. 2008;160:411–422. doi: 10.1007/s10681-007-9564-6. [DOI] [Google Scholar]
- Sundaram RM, Laha GS, Viraktamath BC, Sujatha K, Natarajkumar P, Hari Y, Srinivasa Rao K, Reddy CS, Balachandran SM, Madhav MS, Hajira SK, Rani NS, Vishnupriya MR, Sonti RV. Marker Assisted Breeding For Development Of Bacterial Blight Resistant Rice. In: Muralidharan K, Siddiq EA, editors. Genomics and Crop Improvement: Relevance and Reservations, Institute of Biotechnology, Acharya NG Ranga Agricultural University, Hyderabad 500 030 India. 2011. pp. 154–182. [Google Scholar]
- Sundaram RM, Chatterjee S, Oliva R, Laha GS, Cruz LJE, Sonti RV. Update on Bacterial Blight of Rice: Fourth International Conference on Bacterial Blight. Rice. 2014;7:12. doi: 10.1186/s12284-014-0012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Berloo R. GGT: software for display of graphical genotypes. J Hered. 1999;90:328–330. doi: 10.1093/jhered/90.2.328. [DOI] [Google Scholar]
- Yang SH. Rice varieties registered at national level in 2005. Hybrid Rice. 2006;21(6):25–30. [Google Scholar]
- Yoshimura S, Yoshimura A, Iwata N, McCouch SR, Abenes SL, Baraoidan MR, Mew TW, Nelson RJ. Tagging and combining bacterial blight resistance genes in rice using RAPD and RFLP markers. Mol Breed. 1995;1(4):375–387. doi: 10.1007/BF01248415. [DOI] [Google Scholar]
- Yoshimura S, Yamanouchi U, Katayose Y, Toki S, Wang ZX, Kono I, Kurata N, Yano M, Iwata N, Sasaki T. Expression of Xa-1, a bacterial blight resistance gene in rice, is induced by bacterial inoculation. Proc Natl Acad Sci U S A. 1998;95:1633–1668. doi: 10.1073/pnas.95.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]


