Summary
The attenuation of protein synthesis via the phosphorylation of eIF2α is a major stress response of all eukaryotic cells. The Growth Arrest and DNA-damaged protein 34 (GADD34) bound to the serine/threonine protein phosphatase 1 (PP1) is the necessary eIF2α phosphatase complex that returns mammalian cells to normal protein synthesis following stress. The molecular basis by which GADD34 recruits PP1 and its substrate eIF2α are not fully understood, hindering our understanding of the remarkable selectivity of the GADD34:PP1 phosphatase for eIF2α. Here we report detailed structural and functional analyses of the GADD34:PP1 holoenzyme and its recruitment of eIF2α. The data highlight independent interactions of PP1 and eIF2α with GADD34, demonstrating that GADD34 functions as a scaffold both in vitro and in cells. This work greatly enhances our molecular understanding of a major cellular eIF2α phosphatase and establishes the foundation for future translational work.
Keywords: protein phosphatase 1, GADD34, mRNA translation, eukaryotic translation initiation factor-2, X-ray structure, NMR spectroscopy
Graphical abstract
Introduction
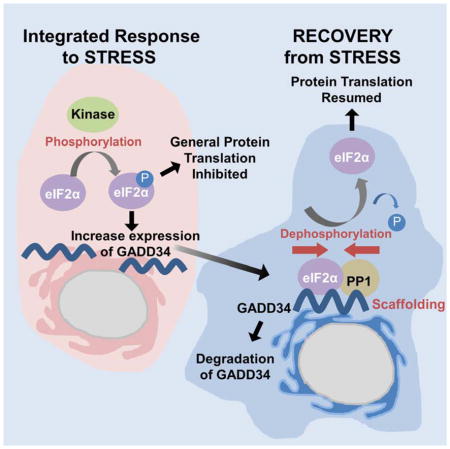
Protein synthesis in eukaryotes is actively controlled via the reversible phosphorylation of numerous translation initiation and elongation factors. In this context, the phosphorylation of the α-subunit of eIF2 (eukaryotic Initiation Factor 2), a trimeric complex composed α, β and γ subunits, is a pivotal down-regulator of translation initiation following changes in the cellular environment or stress (Walton and Gill, 1976). Under non-stressed conditions, eIF2α is largely unphosphorylated. In the presence of GTP, the eIF2 complex recruits Met-tRNA and ribosomal subunits to the translation start site to initiate mRNA translation. However, stresses, such as nutrient deprivation, heat shock, among other factors, activate one or more of four protein kinases, namely GCN2, PERK, PKR and HRI to phosphorylate eIF2α on a single serine, temporarily repressing general protein synthesis (Harding et al., 2000).
Some mRNAs are, however, preferentially translated following eIF2α phosphorylation. These include mRNAs encoding the transcription factors, ATF4 and CHOP, which together induce the stress response gene that encodes Growth Arrest and DNA-damaged protein 34 (GADD34; protein phosphatase 1 regulatory subunit 15A, PPP1R15A). As the stress declines, the GADD34 protein, by recruiting the ser/thr protein phosphatase 1 (PP1), facilitates the dephosphorylation of phospho-eIF2α, restoring normal protein synthesis (Connor et al., 2001; Novoa et al., 2001). The structural basis by which the GADD34:PP1 complex functions as an eIF2α phosphatase remains unclear.
GADD34 (674 residues, 73.5 kDa) possesses an endoplasmic reticulum (ER) targeting helix, four central PEST repeats and a C-terminal PP1-binding domain (Fig. 1A). Bioinformatics analyses suggest that GADD34 is largely unstructured (Fig. S1), making it impossible to assign function(s) simply by analysis of its primary structure. Phylogenetic analyses point to the existence of GADD34-like proteins in many multicellular organisms, from Caenorhabditis elegans to humans (Ceulemans et al., 2002). While some lower eukaryotes, like Drosophila, possess a single gene encoding dGADD34, mammals possess two genes encoding GADD34 and CReP (PPP1R15B) that share sequence homology solely in the C-terminal PP1-binding domain (Chen et al., 2015; Jousse et al., 2003). Finally, viral infection often results in PKR-mediated eIF2α phosphorylation, which inhibits protein synthesis and viral replication in the infected host cells. However, the genomes of selected poxviruses encode polypeptides, like ICP34.5 (also known as γ34.5), with sequence homology to the PP1–binding domain of GADD34 (He et al., 1997). By recruiting PP1, ICP34.5 promotes eIF2α dephosphorylation allowing these viruses to evade this host cell surveillance mechanism. Together, these data argued that the approximately 100 amino acids encompassing the conserved PP1-binding domain among distinct eIF2α phosphatases contain the critical determinants that direct eIF2α dephosphorylation.
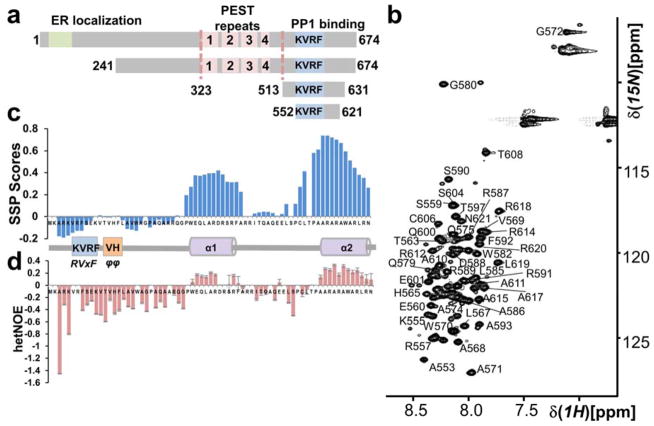
Figure 1. The GADD34 PP1-binding domain is intrinsically disordered.
(a) GADD34 domain structure; ER localization domain, PEST domain (4 PEST repeats; labeled 1 – 4; each ~40 aa) and a PP1-binding domain (includes canonical PP1-binding motif RVxF [KVRF]). GADD34241-674, GADD34513-631 and GADD34552-621 are constructs used. (b) Annotated 2D [1H,15N] HSQC of GADD34552-621. The narrow 1HN chemical shift dispersion is a hallmark of IDPs. (c) Secondary Structure Propensity (SSP) analysis of GADD34552-621 reveals two preferred α-helical secondary structures (helices α1 and α2). (d) Helices α1 and α2 have also reduced fast timescale motions.
Recent discoveries of small molecular inhibitors, namely Salubrinal (Boyce et al., 2005) and Guanabenz (Tsaytler et al., 2011), which target eIF2α phosphatases showed that they protected cells from cytotoxic cell death resulting from protein misfolding and aggregation (known as proteotoxicity). Indeed both compounds improved symptoms and prolonged life in animal models of human neurodegenerative diseases (Jiang et al., 2014; Saxena et al., 2009). Interestingly, Salubrinal inhibited the mammalian GADD34 and CReP-containing eIF2α phosphatases as well as the ICP34.5-containing viral phosphatase. By contrast, Guanabenz showed greater selectivity for the GADD34-containing eIF2α phosphatases. On the other hand, Guanabenz prevented prion protein aggregation and toxicity in both yeast and mammalian cells (Tribouillard-Tanvier et al., 2008). This is particularly remarkable as Saccharomyces cerevisiae does not express a GADD34 ortholog. Instead, the yeast eIF2γ subunit contains an N-terminal extension containing a PP1-binding RVxF motif that directly recruits PP1 (Glc7p) and catalyzes eIF2α dephosphorylation (Rojas et al., 2014). Thus, there is an urgent need for a better understanding of the structure of mammalian eIF2α phosphatases and the mode of action of inhibitors to facilitate the future development of therapies against human diseases associated with proteotoxicity.
Here, we utilized NMR spectroscopy, X-ray crystallography, biochemistry and cell biology to elucidate how GADD34 binds PP1 and recruits its substrate, eIF2α. The crystal structure of the GADD34:PP1 holoenzyme (the complex between PP1 and the PP1-binding domain of GADD34) shows that GADD34 uses both the RVxF and ΦΦ motifs to bind PP1. We also show that the selectivity for eIF2α as substrate is greatly enhanced by its binding to one or more of the central PEST domains in GADD34. The data establish that GADD34 functions as a scaffold, utilizing distinct domains to recruit PP1 and eIF2α in vitro and in living cells. Together, this work significantly advances our understanding of a protein phosphatase complex that is present at high levels in stressed or diseased cells but not detected in healthy cells, providing unique opportunities for future drug development.
Results
The GADD34 PP1-binding domain is intrinsically disordered
Previous studies identified the RVxF (555KVRF558) motif that is required for PP1 binding in the C-terminal domain of GADD34 (Brush et al., 2003) (Fig. 1A). To examine the interaction of GADD34 with PP1, we expressed GADD34513-631, a construct previously used in the analysis of the interaction of GADD34 with PP1 (Brush et al., 2003). Our studies showed that GADD34513-631 is heat stable (80°C, 15 min; Fig. S1) and that its 2D [1H,15N] HSQC spectrum lacked chemical shift dispersion in the 1HN dimension, confirming that the GADD34 PP1-binding domain is an intrinsically disordered protein (IDP).
As ~1/3 of the N-terminal 38 residues in GADD34513-631 are prolines and there was no experimental evidence that GADD34 residues 513-551 contribute to PP1-binding, we compared the 2D [1H,15N] HSQC spectrum of GADD34513-631 with that of GADD34552-621. The 2D [1H,15N] HSQC spectra of the two GADD34 polypeptides overlapped well but the quality of the data for GADD34552-621 was significantly better. Thus, we used GADD34552-621 for detailed NMR analysis. We achieved a ~95% sequence specific backbone assignment (4 prolines, Fig. 1B). Secondary structure propensity (SSP) analysis (Marsh et al., 2006) showed two regions with preferred helical secondary structure (helix α1, 582WEQLARDRS590, ~40% populated; helix α2, 610AARARAWARLRN621, ~70% populated; Fig. 1C). Consistently, helix α1 and α2 also showed reduced fast timescale motions in heteronuclear [15N]-NOE experiments (hetNOE), while the region surrounding the RVxF site was highly dynamic (Fig. 1D). Preferred secondary structure elements often contribute to protein:protein interactions. Thus, we used X-ray crystallography to determine the 3-dimensional structure of the GADD34:PP1 holoenzyme to evaluate the roles of helices α1 and α2 in PP1 binding and to reveal if and how this region of GADD34 dictates substrate selection by bound PP1.
Crystal structure of GADD34:PP1 holoenzyme
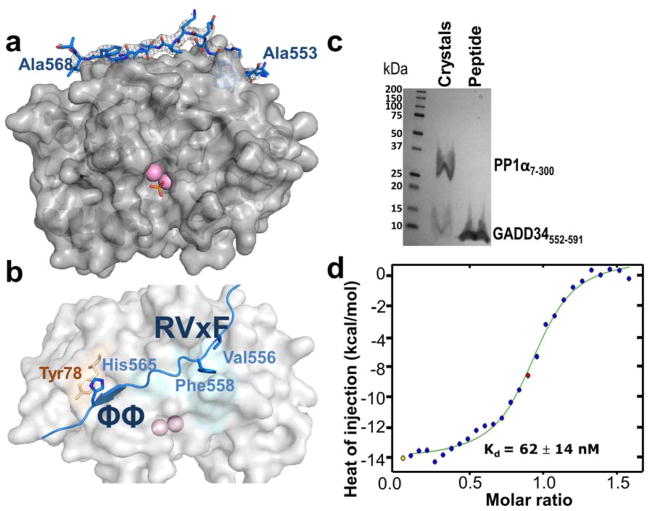
GADD34552-621:PP1 (GADD34552-621 was used in NMR studies) did not produce crystals. Additional GADD34 peptides (e.g. GADD34552-602; GADD34552-591; constructs based on the NMR analysis) were also screened for crystal formation, ultimately yielding crystals for GADD34552-602:PP1 and GADD34552-591:PP1, allowing for complete structure determination of the GADD34552-591:PP1 complex (Table S1; 2.29 Å). Clear electron density was observed for 16 GADD34 residues with the sequence 553ARKVRFSEKVTVHFLA568 (Fig. 2A, 2B). The complex buried ~1,500 Å2 of solvent accessible surface area. To ensure that GADD34552-591 was not proteolytically cleaved during crystallization, ~20 GADD34552-591:PP1 crystals were washed and subjected to SDS-PAGE. The molecular size, determined by the migration of the free GADD34552-591 (Fig. 2C), confirmed that GADD34552-591 was not degraded. Activity of the GADD34:PP1 holoenzyme was confirmed by the dephosphorylation of a model substrate, p-nitro-phenyl phosphate (pNPP) (Fig. S2).
Figure 2. The 3D structure of the GADD34:PP1 holoenzyme.
(a) GADD34 (residues 553-568, light blue, electron density 2Fo-Fc contoured at 1σ) and PP1α7-300 (gray, surface) form a complex. Two Mn2+ ions (pink spheres) are bound at the PP1 active site; bound phosphate is shown as sticks. No electron density was observed for GADD34 residues 552 or 569-591. (b) GADD34 PP1-binding domain with the two primary interaction sites (RVxF and ΦΦ residues are shown as sticks) highlighted. (c) SDS-PAGE of ~20 GADD34552-591:PP1α7-300 crystals; comparable migration of GADD34 from crystals and control (GADD34552-591 alone) shows that no proteolytic degradation of GADD34552-591 occurred during crystallization. (d) ITC of GADD34552-567 and PP1α7-330.
Isothermal titration calorimetry (ITC) of PP1α7-330 and GADD34552-567 confirmed a 1-to-1 binding ratio with a Kd of 62 ± 14 nM (Fig. 2D). The Kd of GADD34552-567 for PP1 was ~6.5-fold weaker than that determined for some PP1 regulators, namely PNUTS and spinophilin (Choy et al., 2014; Ragusa et al., 2010) (8.7 nM and 9.3 nM, respectively), but close to that for NIPP1 (Kd of 73 nM) (O’Connell et al., 2012). Competitive fluorescence anisotropy experiments with a longer GADD34 construct confirmed that GADD34552-567 constitutes the complete PP1 binding domain (Fig. S2).
The GADD34:PP1 holoenzyme
GADD34 bound PP1 via two recognized PP1-binding motifs (Bollen et al., 2010; Choy et al., 2014). First, the residues Val556 and Phe558, which constitute the RVxF-motif in GADD34, bind the hydrophobic RVxF-binding pocket on the surface of PP1 (Fig. 2B). Substituting Val556 and Phe558 with alanines results in complete loss of GADD34 binding by PP1 (Brush et al., 2003). Recently, we identified an RVxF motif ‘lid’ residue (Leu407 in PNUTS; Leu437 in spinophilin) that further stabilizes the RVxF interaction (Choy et al., 2014). However, as also observed for PP1 regulator NIPP1, no lid residue is present in the GADD34 (Fig. S2). Second, GADD34 binds PP1 via the ΦΦ-motif, a motif recently identified in several PP1 regulators, including spinophilin (Ragusa et al., 2010), NIPP1 (O’Connell et al., 2012) and PNUTS (Choy et al., 2014). The ΦΦ-motif of GADD34 is formed by Val564 and His565, where His565 in GADD34 forms a π-stacking interaction with Tyr78 in PP1 (Choy et al., 2014) (Fig. 2B). Interestingly, the partially populated GADD34 helices α1 and α2 do not contribute to the direct interaction with PP1; instead, they likely allow for the recruitment of additional proteins to GADD34 (Brush et al., 2003).
As previously shown, PP1 interaction motifs in regulatory proteins can contribute to PP1 binding and/or PP1 regulation (Choy et al., 2014; O’Connell et al., 2012; Ragusa et al., 2010). So far, both the RVxF- and ΦΦ-motifs have been ascribed roles primarily in PP1 binding (Choy et al., 2014). Indeed, phosphatase assays using the GADD34:PP1 specific substrate, eIF2α, showed that the complex readily dephosphorylated phosho-eIF2α (phosphorylated by PKR). Similar levels of dephosphorylation were observed regardless of the GADD34 peptide (GADD34513-631 or GADD34552-591) used for holoenzyme formation suggesting that residues adjacent to the core GADD34 PP1-interaction domain do not contribute to the activity of this heterodimeric complex (Fig. S2). However, this does not preclude the possibility that association of eIF2α with other regions of GADD34 restrict the substrate specificity and/or enhance the activity of GADD34-associated phosphatase.
GADD34 recruitment of the substrate eIF2α is mediated by the GADD34 PEST domain
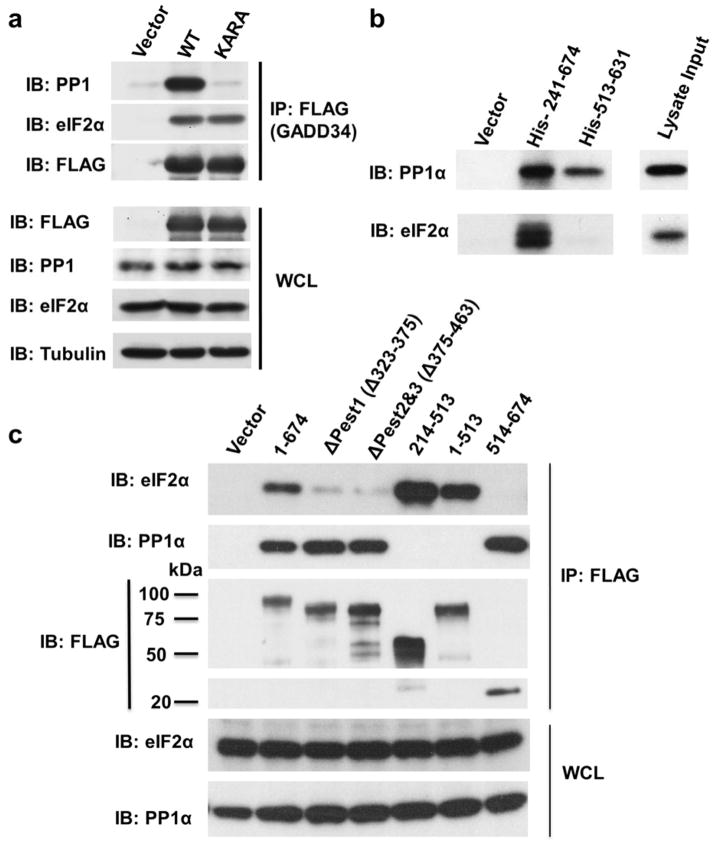
The GADD34:PP1 holoenzyme structure suggested that residues outside the minimal GADD34 PP1 binding domain recruit eIF2α. To test if PP1 is required for eIF2α recruitment by GADD34, we performed immunoprecipitation experiments (IP) using flag-tagged full-length GADD34 (WT and a PP1-binding deficient variant where the GADD34 555KVRF558 motif was mutated to KARA). eIF2α was sedimented to the same extent by both WT and KARA GADD34 (Fig. 3A), confirming that the eIF2α association with GADD34 is independent of PP1 binding.
Figure 3. Recruitment of eIF2α is mediated by the GADD34 PEST domain.
(a) Immuno-precipitation (IP) experiment using WT- and KARA-GADD34. GADD34 recruits eIF2α from the HEK293T cell lysate independent of PP1 binding. (b) IP experiment using His6-GADD34241-674 and His6-GADD34513-631. Only His6-GADD34241-674 binds eIF2α (HEK293T cells lysate), while both His6-GADD34241-674 and GADD34513-631 bind PP1. (c) FLAG vector or FLAG-GADD34 proteins were expressed in HEK293 cells and IP using anti-FLAG-conjugated to agarose beads. The IP and whole cell lysates (WCL) were subjected to immunoblotting (IB) with indicated antibodies. Molecular weight markers (kD) are shown.
We then used NMR spectroscopy to test for a direct interaction between the GADD34 PP1-binding domain and eIF2α. NMR-active 15N-labeled eIF2α4-184 (N-terminal domain) was produced and tested for chemical shift perturbations (CSPs) following the addition of unlabeled GADD34513-631. CSPs could result from direct interaction between the two proteins or from changes in the conformation of eIF2α resulting from eIF2α:GADD34 interactions. No CSPs were detected suggesting that the GADD34 PP1-binding domain does not bind eIF2α (Fig. S3).
To identify the GADD34 domain(s) that mediates eIF2α recruitment, we analyzed eIF2α binding to a GADD34 polypeptide that includes both the PEST repeats and the PP1-binding domain (His-GADD34241-674) and then compared it to a GADD34 polypeptide that includes only the PP1-binding domain (His-GADD34513-631). Only GADD34241-674 sedimented eIF2α from HEK293T cell lysates, establishing that GADD34 residues 241-513 are required for stable eIF2α recruitment (Fig. 3B). Further deletion analyses established that a peptide encompassing only the PEST repeats (GADD34241-513) bound eIF2α, with the PEST repeats 1, 2 and 3 (GADD34323-463) being prominent contributors to eIF2α recruitment (Fig. 3C).
GADD34 recruits both PP1 and eIF2α
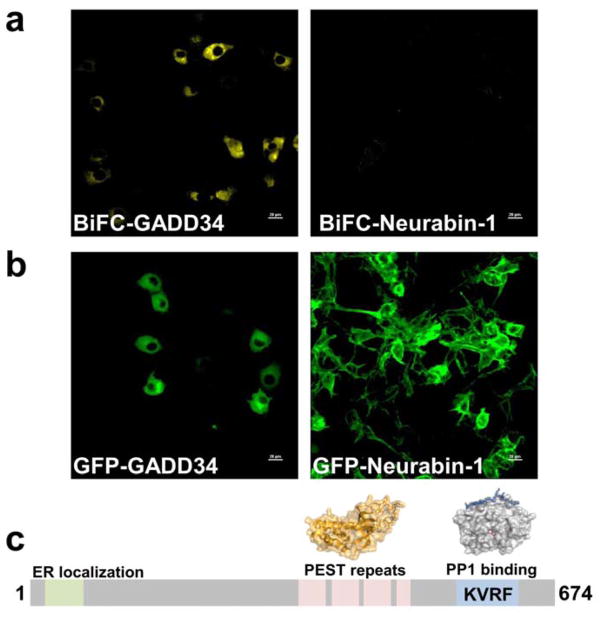
To study the association of GADD34 with both PP1 and eIF2α in cells, we used a bimolecular fluorescence complementation (BiFC) assay (Hu et al., 2002). Here, PP1α and eIF2α, each fused to distinct non-fluorescent halves of Yellow Fluorescent Protein (YFP), were transfected in COS-7 cells and the cells analyzed for the reconstitution of YFP fluorescence using confocal microscopy. When WT GADD34 was co-transfected in COS-7 cells containing the N-YFP-PP1 and C-YFP-eIF2α, YFP fluorescence was greatly enhanced, indicating the formation of the hetreotrimeric complex compared to cells not expressing GADD34 (vector control) or cells expressing another PP1 regulator, neurabin-1, which binds PP1 but not eIF2α (Fig. 4A). As anticipated, BiFC fluorescence was localized primarily at the ER, similar to that seen for GFP-GADD34 (Fig. 4B). A similar experiment, performed using the PP1-binding deficient variant of GADD34 (KARA), showed little or no YFP signal establishing that both eIF2α and PP1 must be recruited to GADD34 to reconstitute YFP fluorescence. These studies demonstrated that GADD34 functions as a protein scaffold to bring PP1 in proximity to its substrate, eIF2α (Fig. 4C, Fig. S4).
Figure 4. Bimolecular Fluorescence Complementation (BiFC) assays highlight the role of GADD34 as a scaffolding protein.
(a) Flag-tagged GADD34 enhances the BiFC signal that results from the complementation of YFP from N-YFP-PP1α and C-YFP-eIF2α. Over-expression of neurabin-1, a neuronal PP1 regulatory protein (negative control) does not assemble the BiFC complex. (b) Localization of GFP-tagged GADD34 and neurabin-1 (COS-7 cell overexpression). (c) Model: GADD34 domain structure and the proposed PP1α and eIF2α binding sites (PP1 and eIF2α structures are shown).
Discussion
Mammals possess two eIF2α phosphatases. The CReP:PP1 holoenzyme is constitutively expressed and likely controls basal eIF2α dephosphorylation to ensure continued protein synthesis in the absence of stress. However, under conditions of stress, increased phosphorylation of eIF2α results in significant repression of general protein synthesis allowing cells to conserve or redirect their energy sources to overcome the stress. The GADD34 mRNA is one of a handful of genes actively translated in the presence of phospho-eIF2α and functions in a feed-back loop to restore general protein synthesis and promote cell recovery from stress. Gene disruption studies suggested that inhibiting the CReP:PP1 complex may be associated with toxicity as CReP null mice are smaller and do not thrive after birth (Harding et al., 2009). By contrast, the GADD34:PP1 complex is only present in stressed or diseased cells and GADD34 null mice are largely normal (Harding et al., 2009; Kojima et al., 2003; Patterson et al., 2006). Thus, the major focus of drug discovery programs targeting eIF2α phosphatases to protect cells from protein misfolding disorders is to selectively target the GADD34:PP1 complex. However, structural information on the GADD34:PP1 complex that might guide the development of GADD34 specific drugs has been lacking.
Our work shows that GADD34 has little 3-dimensional structure and belongs to the family of IDPs (Fig. 1). This might, in part, explain the short half-life of the GADD34 protein in the cells, where it is degraded by the proteasome with a half-life of ≤1 hour. Common with many PP1 regulators, which also show disordered structure particularly within the PP1-binding domain, GADD34 readily and specifically interacts with PP1 to form a functional phosphatase holoenzyme (Fig. 2). To do this, GADD34 uses two well-established PP1-anchoring motifs: the canonical RVxF-motif found in ≥85% of PP1 regulators as well as the ΦΦ-motif, a newly identified motif predicted to be present in ~20% of PP1 regulators (Choy et al., 2014; Peti et al., 2013) (Fig. 2). While these motifs provide for tight binding of PP1 by GADD34 sufficient they do not account for the unique specificity of the GADD34:PP1 complex as an eIF2α phosphatase.
Here we showed that GADD34 interacts with its substrate eIF2α via one or more of the central PEST repeats (Fig. 3). Earlier studies established that the PEST sequences not only drive protein turnover but also mediate the association with regulatory proteins (Molinari et al., 1995). In that context, our earlier studies showed that PEST repeats in GADD34 did not determine the stability of the GADD34 protein in cells (Brush and Shenolikar, 2008), leading us to hypothesize that they represented sites of protein-protein interactions. Our cellular experiments in particular highlighted the necessity for GADD34-mediated scaffolding of both PP1 and eIF2α for efficient and selective dephosphorylation of phospho-eIF2α. The central PEST domain includes three repeats of ~15 hydrophobic and charged amino acids followed by long stretches of negatively charged poly-Glu and Asp residues. Notably, this domain is only present in GADD34; it is not present in CReP or the viral protein, ICP34.5, which share homology with GADD34 only in the PP1-binding domain. Yet, CReP and ICP34.5 can form active eIF2α phosphatases. Moreover, BiFC studies of ICP34.5 demonstrated the ability of the viral protein to recruit both PP1 and eIF2α and reconstitute YFP fluorescence (Brush and Shenolikar, 2008) albeit less efficiently than the fluorescence signal obtained with GADD34 in the current studies. Nevertheless, these studies hinted at a potential eIF2α-binding site within the PP1-binding domain of ICP34.5. However, our cellular and biochemical studies showed that the isolated PP1-binding domain, GADD34513-674, was unable to recruit eIF2α as effectively as WT GADD34 and identified the PEST domain as the primary, high affinity eIF2α-docking site in GADD34. We speculate that while the PP1-binding domain of GADD34 together with PP1 may bind eIF2α transiently to allow for eIF2α dephosphorylation, the evolution of the additional eIF2α association domain uniquely present in GADD34 may provide for more efficient reversal of the much higher levels of phospho-eIF2α seen in stressed cells. Indeed, earlier studies that compared a truncated GADD34 containing the central PEST repeats and the C-terminal PP1-binding domain suggested that this polypeptide generated a more active eIF2α phosphatase than the full-length GADD34 (Novoa et al., 2001), indicating the presence of both positive and negative structural elements that dictate the activity of GADD34-containing eIF2α phosphatase.
In recent years, there has been an increased interest in the development of small molecule inhibitors of the GADD34:PP1 holoenzyme, exploiting the cytoprotective effects of attenuating global protein synthesis in diseased cells and tissues experiencing chronic protein misfolding. These efforts have yielded two structurally unrelated compounds, Salubrinal (Boyce et al., 2005) and Guanabenz (Tsaytler et al., 2011). Unlike compounds that target the active site of PP1 and consequently modulate the covalent modifications on a large number of cellular phosphoproteins, the narrower specificity of Salubrinal and Guanabenz to selectively alter cellular eIF2α phosphorylation was interpreted as arising from their ability to dissociate the GADD34:PP1 complex (Boyce et al., 2005; Tsaytler et al., 2011). Indeed, the disruption of GADD34:PP1 was noted in the presence of millimolar concentrations of Guanabenz. Due to our ability to reconstitute a purified GADD34:PP1 holoenzyme in vitro, we examined the ability of both Guanabenz and Salubrinal to dissociate the GADD34:PP1 complex. No discernable dissociation of the complex was observed. Quite the contrary, high micromolar concentrations of Salubrinal may even enhance the interaction of GADD34 with PP1 (Fig. S4). Furthermore, we performed crystal trials of the GADD34:PP1 complex in the presence of 5 mM Guanabenz, but obtained no evidence for the disruption of the GADD34:PP1 holoenzyme by this drug, i.e. the GADD34:PP1 holoenzyme was intact and no electron density for Guanabenz was identified. Similarly, we have obtained no direct evidence for impaired PP1 or eIF2α binding by GADD34 in the presence of either Guanabenz or Salubrinal. Taken together, these data highlight the need for more work aimed at defining the mode of action of these drugs. The availability of structural information on the key contact points required to assemble GADD34:PP1 complex may also yield new strategies for the identification of small molecules that selectively disrupt this eIF2α phosphatase complex and provide a new avenue for drug development.
Methods
Briefly, proteins were expressed in E. coli BL21 (DE3) or BL21-CodonPlus (DE3)-RIPL competent cells (Agilent). Purification of PP1 was performed as previously described (Choy et al., 2014). GADD34 was purified using heat purification (80°C, 15 min).
Crystallization of the GADD34:PP1 holoenzyme
Crystals of GADD34552-591:PP1α7-300 holoenzyme were grown using sitting drop (200 nl) vapor diffusion in 0.2 M ammonium phosphate dibasic, 20% w/v polyethylene glycol 3350. X-ray data to 2.29 Å were collected at the beamline 12.2 Stanford Synchrotron Radiation Lightsource (SSRL) at 100 K and a wavelength of 0.98 Å using a Pilatus 6M PAD detector. The GADD34552-591:PP1α7-300 structure was solved by molecular replacement using Phaser as implemented in Phenix, using PP17-300 (PDBID: 3E7A) as the search model (Kelker et al., 2009). A solution was obtained in space group P212121; clear electron density for the bound GADD34 was visible in the initial maps. The initial models of the GADD34552-591:PP1α7-300 were built using Phenix.AutoBuild, followed by iterative rounds of refinement in PHENIX and manual building using Coot.
NMR Spectroscopy
NMR measurements were performed at 298 K on a Bruker Avance 500 MHz spectrometer with a HCN TCI z-gradient cryoprobe. The NMR spectra were processed and analyzed using Topspin 3.1 (Bruker) and CARA software package (www.cara.nmr.ch). The hetNOE measurement and secondary structure propensity calculation were performed as previously described.
BiFC assay
For BiFC assays, plasmid DNAs encode the yellow fluorescence protein (YFP) pair (YN-PP1α & YC-eIF2α), together with the scaffolding proteins GADD34 and neurabin-1 were transfected into 60 – 70% confluence COS-7 cells grown in glass bottom Lab-Tek™ Chamber Slide™ (Nunc) using FuGENE 6. A vector control (Cyan Fluorescence Protein, CFP) was used to evaluate the transfection efficiency, and also to normalize and quantify the BiFC signal. To minimize non-specific BiFC signal, lower plasmid concentrations were used (250 ng for the BiFC pairs and scaffolding protein; 50 ng for the CFP vector control). Transfected cells were grown overnight in a 37°C incubator with 5% CO2. Two hours before imaging, cells were transferred to an incubator pre-set at 30°C for the maturation of the complemented YFP. The live cell images were captured using Nikon confocal microscope with motorized stage with environment chamber for temperature control and CO2 delivery. Image analysis was performed using MetaMorph (Molecular Devices).
Supplementary Material
Highlights.
Distinct domains in GADD34 are utilized to scaffold PP1 and eIF2α
GADD34 is an intrinsically disordered protein that binds PP1 via RVxF and ϕϕ motifs
PEST repeats in GADD34 represent an independent site for eIF2α binding
Acknowledgments
We thank Dr. Assen Marintchev (Boston University Medical School) for the generous gift of the eIF2α expression plasmids and Dr. Youjia Cao (Nankai University, Tanjin China) for the YN-PP1α and YC-eIF2α expression plasmids. GADD34241-674 was kindly provided by Dr. H. Imataka (Riken). We thank the Nikon Imaging Centre, Singapore for the use of the Nikon A1R confocal microscope for the BiFC experiments.
This work was supported by NIH grants R01NS091336 to WP, R01GM098482 to RP, Duke-NUS start-up funds provided by Singapore Ministry of Health, individual research grant (NMRC/GMS/1252/2010) from the National Medical Research Council, Singapore and a Translational Clinical Research Partnership grant from A*STAR (Agency for Science, Technology and Research, Singapore) to SS. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH. This research is based in part on data obtained at the Brown University Structural Biology Core Facility, which is supported by the Division of Biology and Medicine, Brown University.
Abbreviations
- IDP
intrinsically disordered protein
- NMR
nuclear magnetic resonance
- PP1
protein phosphatase 1
- GADD34
protein encoded by growth arrest and DNA damage-inducible transcript 34
Footnotes
Accession Numbers. NMR chemical shifts were deposited in the BioMagResBank (BMRBid: 25430). Atomic coordinates and structure factors were deposited in the Protein Data Bank (PDBID: 4XPN).
Supplemental Information. Supplemental Information includes detailed Experimental Procedures, 4 figures, and refinement table and can be found with this article online at XX.
Author Contributions
M.C. designed and performed experiments reported in Figs. 1, 2, 4, S1–S4. P.Y., I.L. and C.G. designed and performed experiments reported in Fig. 3. J.N. designed and performed experiments reported in Fig. S2. W.P., S.S. and R.P. designed the study and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bollen M, Peti W, Ragusa MJ, Beullens M. The extended PP1 toolkit: designed to create specificity. Trends Biochem Sci. 2010;35:450–458. doi: 10.1016/j.tibs.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D, et al. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- Brush MH, Shenolikar S. Control of cellular GADD34 levels by the 26S proteasome. Mol Cell Biol. 2008;28:6989–7000. doi: 10.1128/MCB.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush MH, Weiser DC, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol Cell Biol. 2003;23:1292–1303. doi: 10.1128/MCB.23.4.1292-1303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceulemans H, Stalmans W, Bollen M. Regulator-driven functional diversification of protein phosphatase-1 in eukaryotic evolution. BioEssays: news and reviews in molecular, cellular and developmental biology. 2002;24:371–381. doi: 10.1002/bies.10069. [DOI] [PubMed] [Google Scholar]
- Chen R, Rato C, Yan Y, Crespillo-Casado A, Clarke HJ, Harding HP, Marciniak SJ, Read RJ, Ron D. G-actin provides substrate-specificity to eukaryotic initiation factor 2alpha holophosphatases. Elife. 2015;4 doi: 10.7554/eLife.04871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy MS, Hieke M, Kumar GS, Lewis GR, Gonzalez-DeWhitt KR, Kessler RP, Stein BJ, Hessenberger M, Nairn AC, Peti W, et al. Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code. Proc Natl Acad Sci U S A. 2014;111:4097–4102. doi: 10.1073/pnas.1317395111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor JH, Weiser DC, Li S, Hallenbeck JM, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 assembles a novel signaling complex containing protein phosphatase 1 and inhibitor 1. Mol Cell Biol. 2001;21:6841–6850. doi: 10.1128/MCB.21.20.6841-6850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Scheuner D, Chen JJ, Kaufman RJ, Ron D. Ppp1r15 gene knockout reveals an essential role for translation initiation factor 2 alpha (eIF2alpha) dephosphorylation in mammalian development. Proc Natl Acad Sci U S A. 2009;106:1832–1837. doi: 10.1073/pnas.0809632106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Gross M, Roizman B. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci U S A. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- Jiang HQ, Ren M, Jiang HZ, Wang J, Zhang J, Yin X, Wang SY, Qi Y, Wang XD, Feng HL. Guanabenz delays the onset of disease symptoms, extends lifespan, improves motor performance and attenuates motor neuron loss in the SOD1 G93A mouse model of amyotrophic lateral sclerosis. Neuroscience. 2014;277:132–138. doi: 10.1016/j.neuroscience.2014.03.047. [DOI] [PubMed] [Google Scholar]
- Jousse C, Oyadomari S, Novoa I, Lu P, Zhang Y, Harding HP, Ron D. Inhibition of a constitutive translation initiation factor 2alpha phosphatase, CReP, promotes survival of stressed cells. J Cell Biol. 2003;163:767–775. doi: 10.1083/jcb.200308075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelker MS, Page R, Peti W. Crystal structures of protein phosphatase-1 bound to nodularin-R and tautomycin: a novel scaffold for structure-based drug design of serine/threonine phosphatase inhibitors. J Mol Biol. 2009;385:11–21. doi: 10.1016/j.jmb.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima E, Takeuchi A, Haneda M, Yagi A, Hasegawa T, Yamaki K, Takeda K, Akira S, Shimokata K, Isobe K. The function of GADD34 is a recovery from a shutoff of protein synthesis induced by ER stress: elucidation by GADD34-deficient mice. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2003;17:1573–1575. doi: 10.1096/fj.02-1184fje. [DOI] [PubMed] [Google Scholar]
- Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari M, Anagli J, Carafoli E. PEST sequences do not influence substrate susceptibility to calpain proteolysis. J Biol Chem. 1995;270:2032–2035. doi: 10.1074/jbc.270.5.2032. [DOI] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell N, Nichols SR, Heroes E, Beullens M, Bollen M, Peti W, Page R. The Molecular Basis for Substrate Specificity of the Nuclear NIPP1:PP1 Holoenzyme. Structure. 2012;20:1746–1756. doi: 10.1016/j.str.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson AD, Hollander MC, Miller GF, Fornace AJ., Jr Gadd34 requirement for normal hemoglobin synthesis. Mol Cell Biol. 2006;26:1644–1653. doi: 10.1128/MCB.26.5.1644-1653.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peti W, Nairn AC, Page R. Structural basis for protein phosphatase 1 regulation and specificity. FEBS J. 2013;280:596–611. doi: 10.1111/j.1742-4658.2012.08509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragusa MJ, Dancheck B, Critton DA, Nairn AC, Page R, Peti W. Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites. Nat Struct Mol Biol. 2010;17:459–464. doi: 10.1038/nsmb.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas M, Gingras AC, Dever TE. Protein phosphatase PP1/GLC7 interaction domain in yeast eIF2gamma bypasses targeting subunit requirement for eIF2alpha dephosphorylation. Proc Natl Acad Sci U S A. 2014;111:E1344–1353. doi: 10.1073/pnas.1400129111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S, Cabuy E, Caroni P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat Neurosci. 2009;12:627–636. doi: 10.1038/nn.2297. [DOI] [PubMed] [Google Scholar]
- Tribouillard-Tanvier D, Beringue V, Desban N, Gug F, Bach S, Voisset C, Galons H, Laude H, Vilette D, Blondel M. Antihypertensive drug guanabenz is active in vivo against both yeast and mammalian prions. PloS one. 2008;3:e1981. doi: 10.1371/journal.pone.0001981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaytler P, Harding HP, Ron D, Bertolotti A. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science. 2011;332:91–94. doi: 10.1126/science.1201396. [DOI] [PubMed] [Google Scholar]
- Walton GM, Gill GN. Regulation of ternary (Met-tRNAf - GTP - eukaryotic initiation factor 2) protein synthesis initiation complex formation by the adenylate energy charge. Biochim Biophys Acta. 1976;418:195–203. doi: 10.1016/0005-2787(76)90069-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.