Summary
Nucleation promoting factors (NPFs) control the spatio-temporal activity of Arp2/3 complex in cells [1, 2]. Thus, WASP and the WAVE complex direct the formation of branched actin networks at the leading edge during cell motility and endo/exocytosis, whereas the WASH complex is involved in endosomal transport. Less understood are WHAMM and JMY, two NPFs with similar domain architecture. JMY is found in the nucleus and the cytosol, and is involved in transcriptional regulation [3], cell motility [4], and trans-Golgi transport [5]. WHAMM was reported to bind microtubules and to be involved in ER to cis-Golgi transport [6]. Here, we show that WHAMM directs the activity of Arp2/3 complex for autophagosome biogenesis through an actin-comet tail motility mechanism. Macroautophagy – the process by which cytosolic material is engulfed into autophagosomes for degradation and/or recycling – was recently shown to involve actin [7], but the mechanism is unknown. We found that WHAMM forms puncta that colocalize and comigrate with the autophagy markers LC3, DFCP1 and p62 through a WHAMM-dependent actin-comet tail mechanism. Under starvation, WHAMM and actin are observed at the interface between neighboring autophagosomes, whose number and size increases with WHAMM expression. Interfering with actin polymerization, inhibiting Arp2/3 complex, knocking down WHAMM, or blocking its interaction with Arp2/3 complex through mutagenesis, all inhibit comet tail formation and reduce the size and number of autophagosomes. Finally, JMY shows similar localization to WHAMM, and could be involved in similar processes. These results reveal a link between Arp2/3 complex-dependent actin assembly and autophagy.
Results and Discussion
WHAMM forms puncta and tubular structures that colocalize with the ER
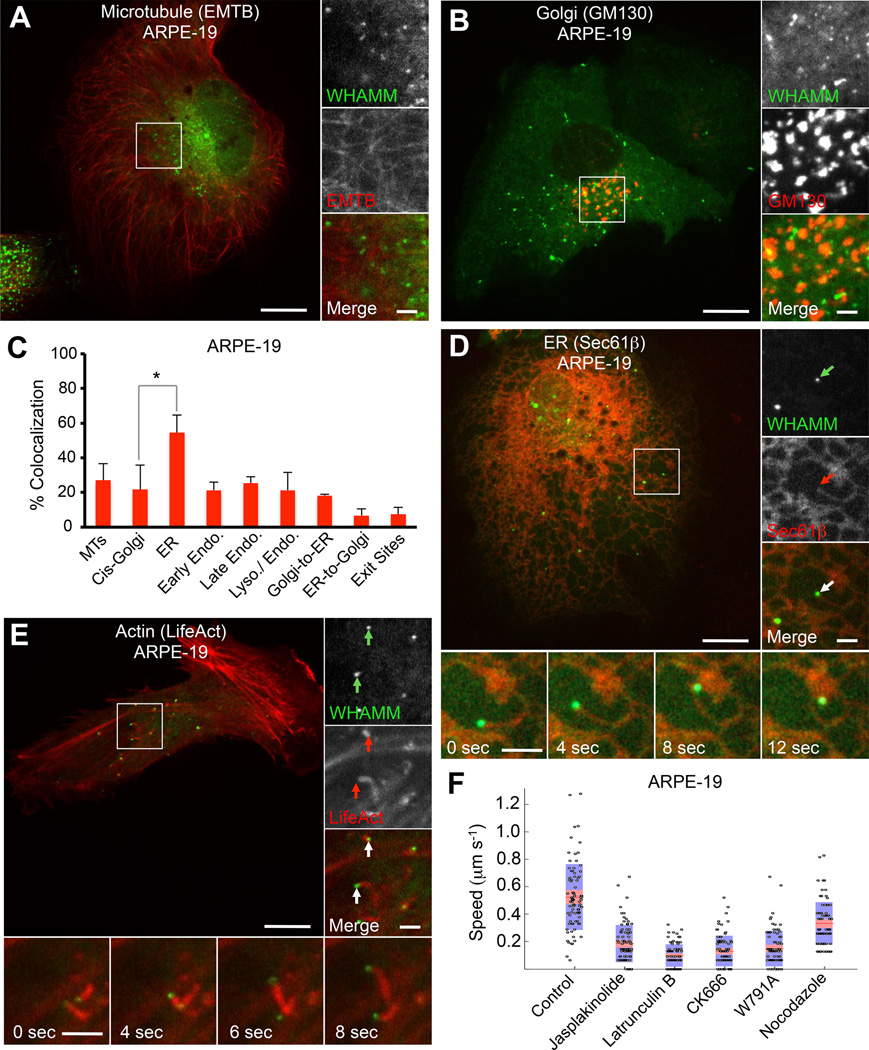
WHAMM was previously reported to bind microtubules (MTs) and to localize to the cis-Golgi apparatus and the ER-Golgi intermediate compartment (ERGIC), playing a role in ER to Golgi transport [6]. We found, however, that WHAMM localization depends strongly on protein expression levels post-transfection (Figure S1A). Thus, tubulation along MTs markedly increased 24 h after expression in ARPE-19 and HeLa cells (Figure S1B and S1C). (Note that most experiments here were performed in parallel in ARPE-19 and HeLa cells, and examples of both cell types are given throughout this work.) In contrast, 8–12 h post transfection WHAMM formed puncta that generally did not colocalize with and moved independently of the MT marker mCherry-EMTB and the cis-Golgi marker mCherry-GM130 (Figures 1A, 1B, S1D). Indeed, while WHAMM appeared static along MTs, the puncta were highly dynamic (Figure S1E), displaying two types of movement – directed motions reminiscent of MT-based transport (>0.8 µm s−1), or slower, spatially confined movement (0.2–0.7 µm s−1).
Figure 1. WHAMM Localizes to the ER and Induces the Formation of Actin Comet Tails.
(A) GFP-WHAMM puncta in ARPE-19 cells do not colocalize with the MT marker mCherry-EMTB.
(B) GFP-WHAMM forms dynamic puncta and small tubules in ARPE-19 cells 8 h post transfection. These puncta do not colocalize with the cis-Golgi marker mCherry-GM130.
(C) Quantification (±SEM; n=5–13 cells) of the colocalization of GFP-WHAMM in ARPE-19 cells with MTs (mCherry-EMTB), cis-Golgi (mCherry-GM130), ER (mCherry-Sec61β), early endosomes (mCherry-Rab5), late endosomes (mCherry-Rab7), enodo/lysosomes (RFP-Lamp1), Golgi-to-ER vesicles (mCherry-Arf1), ER-to-Golgi vesicles (mCherry-Sec-16L), and ER-exit sites (mCherry-Sec-24D).
(D) GFP-WHAMM puncta colocalize and comigrate with ER tubules (mCherry-Sec61β) in ARPE-19 cells cotransfected with GFP-WHAMM and the ER marker Sec61β. These tubules move with a mean speed of ~0.5 µm s−1. Insets show a WHAMM punctum (green arrow) comigrating with an ER tubule (red arrow).
(E) GFP-WHAMM puncta are propelled by actin comet tails in ARPE-19 cells, moving with an average speed of 0.5 µm s−1. Insets show WHAMM puncta (green arrows) leading actin comet tails (red arrows).
(F) Quantification of the speed of WHAMM puncta. Measured puncta speeds from five cells are presents as a ‘bee-swarm’ plot superimposed onto a box-and-whisker plot. The red area represents the middle half of the data, divided by the median value (red line). The blue area encompasses the data from the 5th to the 95th percentile.
Scale bars in whole cell and inset images are 10 and 2 µm, respectively.
*p < 0.05. See also Figure S1 and S2 and Movie S1.
To determine the identity of the organelle associated with the WHAMM puncta, we examined the colocalization of GFP-WHAMM with several organelle markers (Figure 1C). WHAMM did not colocalize with early endosomes (mCherry-Rab5), late endosomes (mCherry-Rab7), late endosome-lysosome intermediate vesicles (Lamp1-RFP), the cis-Golgi apparatus (GM130), or COPI vesicles budding from the Golgi (mCherry-Arf1) (Figures 1B, and S1F–S1I).
In contrast, the WHAMM puncta colocalized with two global ER markers, mCherry-Sec61β and mCherry-Calnexin (Figures 1C, 1D, S1J and S1K). However, WHAMM appeared to reside on specific sites of this organelle, distinct from COPII ER exit sites (mCherry-Sec16L or mCherry-Sec24D) and ER-mitochondria contact sites (DsRed2-Mito) (Figures S1L–S1N).
The WHAMM puncta appeared to lead the extension of tubular structures tethered to the ER (Figure 1D and Movie S1), moving with a mean speed of ~0.5 µm sec−1. Occasionally, WHAMM molecules coated these tubular structures along their length (Figure S1J and S1K). Such WHAMM-coated tubules elongated from the ER with a mean speed of ~0.03 µm sec−1, i.e. much slower than the motility of WHAMM puncta.
WHAMM promotes the movement of ER-tethered membranes through an actin comet tail mechanism
Because in vitro WHAMM functions as an activator of the Arp2/3 complex [6], we asked whether the motility of the WHAMM puncta and the deformation of the ER membrane proceeded through an actin polymerization-dependent mechanism. To test this idea, we coexpressed GFP-WHAMM with mCherry-LifeAct in ARPE-19 cells (Figure 1E and Movie S1). Long actin comet tails, which were also positive for the Arp2/3 complex-binding and branch-stabilizing protein cortactin (Figure S2A), were observed in association with WHAMM puncta that moved with a mean speed of ~0.5 µm sec−1 (Figure 1F). This form of motility is reminiscent of that of Listeria monocytogenes. Like commonly observed with Listeria [8], the movement of WHAMM puncta was locally confined, forming circles and spirals (Figure S2B). This was a surprising observation, because unlike Listeria, the WHAMM puncta remained tethered to the ER during motility (Figure 1D and Movie S1). It thus appears that the formation of such circles and spirals is a distinctive feature of actin comet tail motility. WHAMM-associated comet tails were also observed in Cos-7, C2C12 and HeLa cells (Figures S2C–S2E).
In cells treated with latrunculin B or jasplakinolide, two drugs that inhibit actin filament turnover by different mechanisms, actin comet tails were not observed, and the motility of WHAMM puncta was staled (Figures 1F, S2F and S2G). Treating cells with the Arp2/3 complex inhibitor CK666 [9] also inhibited the motility of WHAMM puncta (Figures 1F and S2H). Interestingly, cells treated with CK666 also showed a shift in WHAMM localization from puncta to tubular-coated structures that appeared to align with MTs (Figure S2H), somewhat analogous to the structures observed with prolonged WHAMM expression (Figures S1A–S1B). Together these results suggest that Arp2/3 complex-induced actin polymerization is required for the motility of ER-tethered WHAMM puncta. To determine whether WHAMM is directly responsible for Arp2/3 complex activation in this process, the conserved tryptophan residue within the A region, necessary for interaction with Arp2/3 complex, was mutated to alanine (WHAMMW791A). Compared to wild type WHAMM, expression of the mutant W791A dramatically reduced the number of comet tails in ARPE-19 and HeLa cells, and WHAMMW791A frequently localized to tubular structures (Figure S2I–S2K).
Treating cells with nocodazole, a drug that disrupts the MT network, had a limited effect on the mean speed of WHAMM puncta, and the comet tails appeared unaffected (Figures 1F and S2L). The slight reduction in the mean speed in this case is likely due to the loss of the small fraction of the puncta that undergo directed MT-based transport (Figure S1E). We thus conclude that the MT cytoskeleton is not involved in comet tail motility of the WHAMM puncta.
WHAMM colocalizes with autophagy markers
Having excluded the presence of WHAMM puncta at ER exit sites and ER-mitochondria contact sites (Figures S1L–S1N), we decided to explore a potential role for WHAMM in macroautophagy, a process often linked to the ER membrane [10] and recently shown to involve the actin cytoskeleton [7]. We thus investigated the colocalization of WHAMM with several markers along the autophagosome biogenesis pathway.
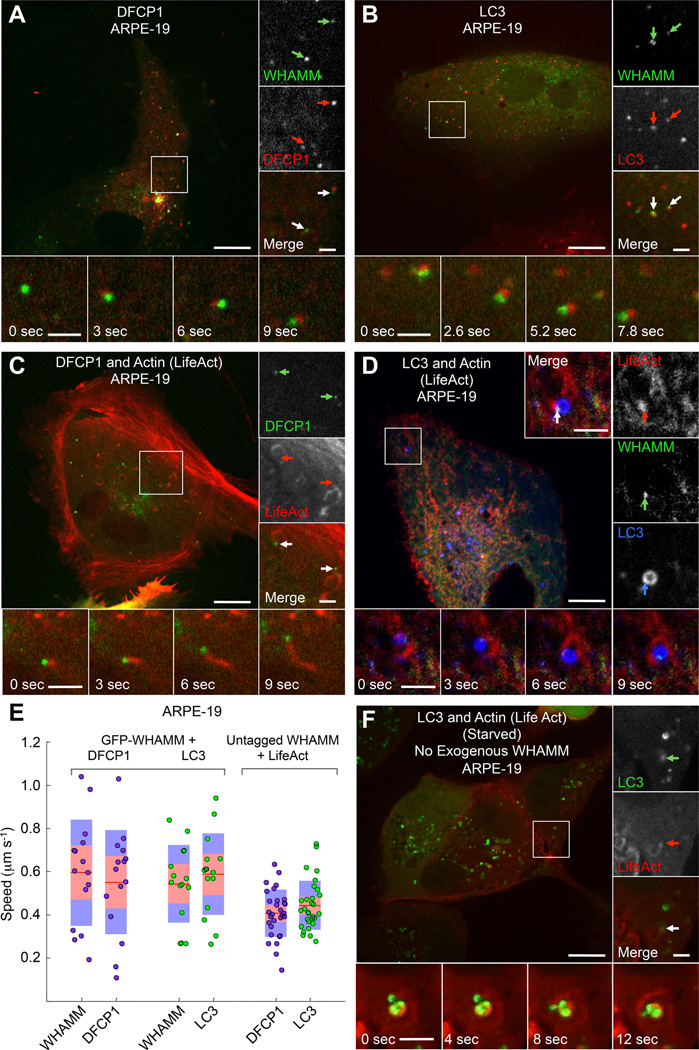
During starvation, a condition that upregulates autophagy [11], Ω-shaped membrane compartments (omegasomes) emerge from the ER at sites positive for ATG14, which is an early autophagy marker and an essential regulator of the PI3K complex [10, 12, 13]. The omegasomes then become coated with DFCP1 [10], followed by the formation of autophagosomes that accumulate the lipidated form of LC3 (LC3-II). While WHAMM puncta did not colocalize with the early marker ATG14 (Figure S3A), they did associate with vesicular compartments positive for both DFCP1 and LC3 (Figures 2A, 2B and Movie S2). Specifically, WHAMM puncta frequently appeared adjacent (possibly due to the relatively slow rate of image sampling compared to the fast speeds of the moving puncta) to and comigrated with puncta of these two autophagy markers. Moreover, in cells coexpressing untagged WHAMM and mCherry-LifeAct with either GFP-DFCP1 or GFP-LC3, DFCP1 and LC3 puncta associated with actin comet tails, which appeared to propel their movement (Figures 2C and S3B and Movie S2). In cells transfected with mCherry-LC3, GFP-WHAMM and BFP2-LifeAct, LC3-coated vesicles were typically associated with a single WHAMM punctum, from which an actin comet tail would invariably emerge (Figure 2D and Movie S2). The mean speed of DFCP1 and LC3 puncta associated with actin comet tails was similar to each other and to that of the WHAMM puncta (Figure 2E).
Figure 2. WHAMM-Dependent Comet Tails Propel the Movement of the Autophagosome Markers DFCP1 and LC3.
(A) GFP-WHAMM puncta comigrate with puncta of the omegasome marker mCherry-DFCP1 in fed ARPE-19 cells (color-coded arrows).
(B) GFP-WHAMM puncta comigrate with puncta of the autophagosome marker mCherry-LC3 in fed ARPE-19 cells.
(C) DFCP1 puncta are propelled by actin comet tails in ARPE-19 cells cotransfected with GFP-DFCP1, untagged WHAMM, and mCherry-LifeAct. Insets show two DFCP1 puncta (green arrows) with trailing actin comet tails (red arrows).
(D) WHAMM forms discrete puncta on LC3-coated vesicles (autophagosomes) that are propelled by actin comet tails in cells cotransfected with GFP-WHAMM, BFP2-LifeAct (red), and mCherry-LC3 (blue) in a fed ARPE-19 cell. Insets show a WHAMM punctum (green arrow) attached to an LC3 vesicle (blue arrow) and leading an actin comet tail (red arrow).
(E) Quantification of puncta speed for the indicated markers (n=30 puncta from 5 cells expressing GFP-WHAMM, and n=20 for 3 cells expressing untagged WHAMM).
(F) LC3 positive vesicles colocalize with actin-dense comet tails, which propel their movement in starved ARPE-19 cells expressing GFP-LC3 and mCherry-LifeAct but not exogenous WHAMM.
Scale bars in whole cell and inset images are 10 and 2 µm, respectively.
WHAMM positive puncta also colocalized and comigrated with p62 (Figure S3C), an autophagy marker recruited to emerging autophagosomes (phagophores) by interaction with LC3-II, and which serves as an adaptor for recruitment of ubiquitinated substrates destined for autolysosome degradation [14]. However, as pointed out above, WHAMM did not colocalize with Lamp1 (Figure S1H), suggesting that it dissociates from autophagosomes before they merge with lysosomes.
Comet tail formation on autophagosomes is independent of exogenous WHAMM expression
In starved ARPE-19 and HeLa cells treated with bafilomycin A1 (BafA), an inhibitor of autophagosome-lysosome fusion [15], LC3-positive vesicles form clusters, and their number and size increases. Under these conditions, mCherry-LifeAct accumulated at the interface between GFP-LC3-positive vesicles, and actin comet tails occasionally propelled the movement of autophagosomes in the absence of WHAMM expression (Figure 2F and Movie S3). Thus, exogenous WHAMM expression is not required for the formation of actin comet tails on autophagosomes. However, the number of DFCP1 and LC3 puncta associated with comet tails increased dramatically with WHAMM expression, whereas the mean speed of these puncta was mostly unaffected, both in ARPE-19 and HeLa cells (Figure S3D).
WHAMM expression upregulates autophagosome biogenesis
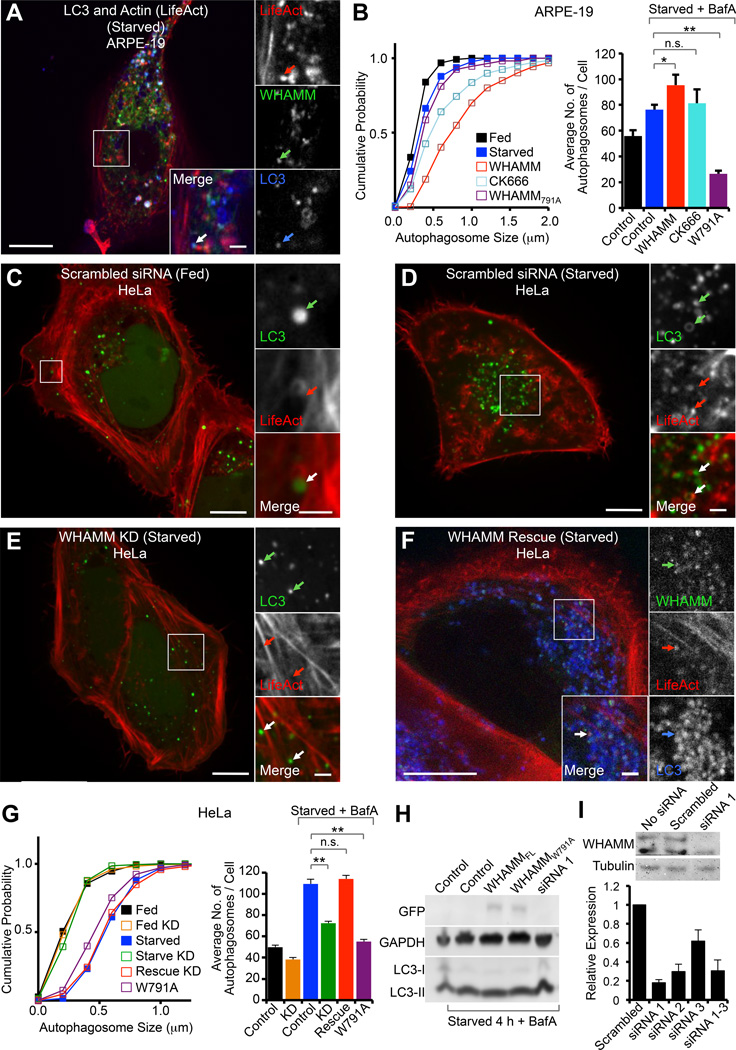
The smaller number of comet tails observed in WHAMM untransfected cells and cells expressing mutant W791A (Figure S2I) could be formed by either endogenous WHAMM or another NPF, such as JMY. Therefore, to further explore the role of WHAMM in comet tail formation and autophagy, GFP-WHAMM was coexpressed with BFP2-LifeAct and mCherry-LC3 in ARPE-19 cells starved for 4 h in the presence of BafA. Exogenous WHAMM expression markedly increased the number and size of autophagosomes (Figures 3A and 3B) compared to cells not expressing exogenous WHAMM (Figure 2F). We observed examples of bursts of actin assembly connected to WHAMM coated vesicles that immediately gave rise to LC3-positive tubular structures (Movie S4). We also often observed WHAMM tubules that appeared to tether autophagosomes together or to the ER (Figure S3E), suggesting that WHAMM could also play a role in autophagosome fusion, explaining the increase in their size.
Figure 3. Role of WHAMM in Autophagy.
(A) ARPE-19 cells starved for 4 h with 100 nM BafA, showing large autophagosomes positive for GFP-WHAMM, BFP2-LifeAct (red), and mCherry-LC3 (blue). Insets show a WHAMM tubule associated with LC3 and an actin comet tail.
(B) Cumulative autophagosome size distribution and average number of autophagosomes per cell (± SEM) for the indicated WHAMM expression experiments (n=7–10 cells from 2 independent experiments).
(C) Fed HeLa cells treated with scrambled siRNA and expressing GFP-LC3 and mCherry-LifeAct. Insets show an LC3 coated vesicle propelled by an actin comet tail.
(D) Starved HeLa cells (+BafA) treated with scrambled siRNA and expressing GFP-LC3 and mCherry-LifeAct show an increase in GFP-LC3-positive vesicles associated with actin (arrows in insets) near the center of the cell.
(E) WHAMM knockdown HeLa cells treated with siRNA 1 for 72 h prior to transfection with GFP-LC3 and mCherry-LifeAct and starved for 4 h (+BafA). These cells show fewer LC3-positive vesicles, which do not appear to associate with actin (arrows in insets).
(F) HeLa cells rescued for WHAMM expression with a siRNA resistant GFP-WHAMM (mouse) and transfected with BFP2-LifeAct (red) and mCherry-LC3 (blue). Cells were starved 4 h (+BafA) prior to imaging. These cells show numerous LC3-positive vesicles, which colocalize with WHAMM and actin.
(G) Cumulative autophagosome size distribution and average number of autophagosomes per cell (± SEM) for the indicated WHAMM expression experiments (n=15–20 cells from 2 independent experiments).
(H) Western blot showing the effect of exogenous expression on GFP-WHAMM, GFP-WHAMMW791A, and knockdown of WHAMM on LC3 maturation (conversion of LC3-I to LC3-II) during starvation.
(I) Validation of WHAMM knockdown. A Western blot shows that both the 100 kDa and 75 kDa WHAMM bands recognized by the anti-WHAMM antibody are reduced by siRNA treatment. qRT-PCR shows gene expression for cells treated with three different siRNAs or their mixture relative to untransfected HeLa cells (± SD; n=3 independent knockdowns). The estimated extent of the knockdown for siRNA 1 is >80%.
Scale bars in whole cell and inset images are 10 and 2 µm, respectively.
*p < 0.05, **p < 0.001, n.s. not significant. See also Figure S3 and Movie S4.
Since WHAMM is a NPF of Arp2/3 complex [6], we next asked whether autophagosome biogenesis depended on the activity of Arp2/3 complex. Treating WHAMM-expressing ARPE-19 cells with the Arp2/3 complex inhibitor CK666 marginally diminished the size and number of autophagosomes (Figures 3B and S3F), likely because autophagosomes were already formed prior to the treatment. As observed above, these cells also displayed a marked increase in WHAMM-positive tubular structures (Figures S2H and S3F).
To more directly determine the role of Arp2/3 complex and WHAMM in autophagy, we expressed the mutant WHAMMW791A and starved cells for 4 h with BafA (Figure S3G). In this case, the cumulative autophagosome size distribution was not significantly different from that observed with LC3 expression alone, but the number of autophagosomes was markedly reduced (Figure 3B).
To more conclusively test WHAMM’s involvement in autophagosome formation we knocked down endogenous WHAMM expression in HeLa cells. In control experiments, fed HeLa cells treated with a scrambled siRNA showed few LC3-positive puncta, distributed uniformly throughout the cytosol (Figures 3C and 3G). Upon starvation for 4 h with addition of BafA these cells showed a marked increase in the number of LC3-positive puncta, which were now larger in size and clustered near the center of the cells (Figures 3D and 3G). In contrast, starved cells in which WHAMM was knocked down by siRNA treatment displayed an LC3 distribution similar to that of fed cells treated with scrambled siRNA (Figures 3E and 3G). Knocking down WHAMM also partially inhibited the conversion of LC3-I to LC3-II, characteristic of autophagosome formation (Figure 3H). In these experiments, the extent of WHAMM knockdown was estimated at more than 80% using both Western blotting and qRT-PCR (Figure 3I). Strikingly, rescuing WHAMM expression using siRNA-resistant GFP-mouse WHAMM restored the LC3 distribution observed in scrambled siRNA-treated starved cells (Figures 3F and 3G). What is more, WHAMM knockdown cells almost completely lacked actin comet tails, whereas rescuing WHAMM expression restored comet tail formation (Figure S3H). Mutant W791A had a similar phenotype in HeLa cell (Figure 3G) as observed in ARPE-19 cells (Figure 3B), displaying a reduced number of LC3 puncta with similar size to that of control starved cells. Together, these experiments strongly support a role for WHAMM in autophagosome biogenesis, through an Arp2/3 complex-dependent actin comet tail mechanism.
Domain architecture of WHAMM and relationship with JMY
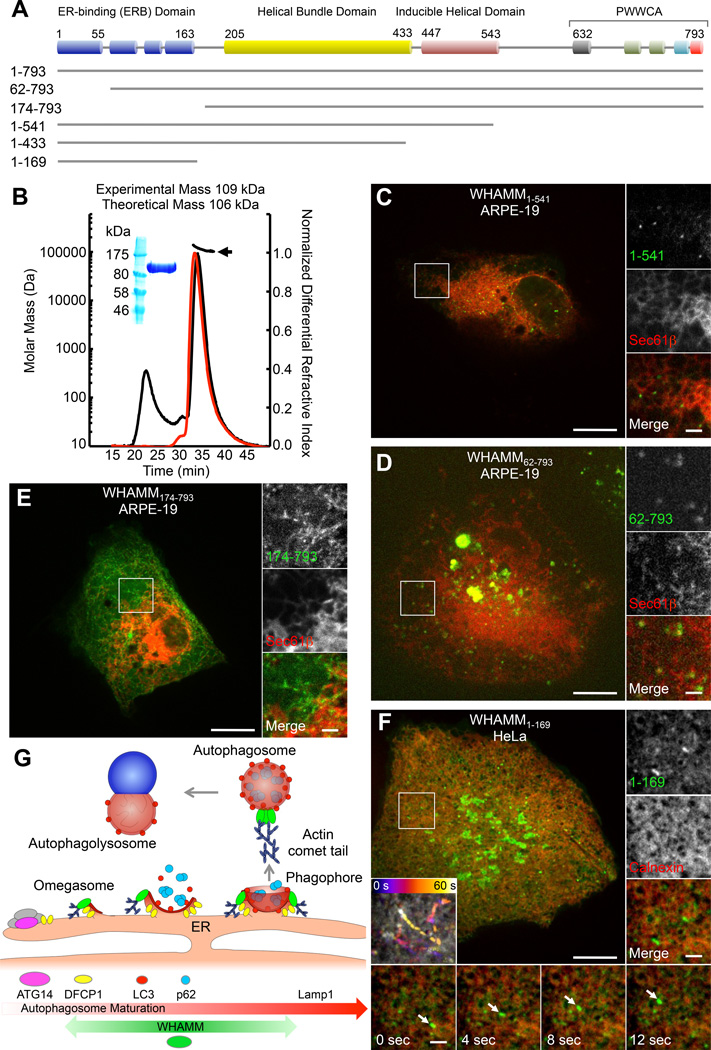
Through sequence analysis, four major domains can be identified in WHAMM and JMY (Figures 4A and S4A): a) an N-terminal domain consisting of a mixture of short β-strands and α-helices (WHAMM aa ~1–163), b) a helical bundle domain (aa ~205–433) predicted to comprise three or four large helices and reminiscent of the fold of Atg17, an essential autophagy factor in yeast [16], c) an inducible helical domain, whose structure may be stabilized by interaction with partners (aa ~447–543), and d) a C-terminal PWWCA domain involved in interactions with Arp2/3 complex, actin and possibly profilin (aa ~632–793). The N-terminal domain is less conserved than the rest of the sequence (Figure S4A), except for the first ~55-aa, which are highly conserved in WHAMM and JMY. This domain contains three highly variable loops, featuring several proline and glycine residues, and harboring some of the major differences between WHAMM and JMY. In particular, the first loop (WHAMM aa ~55–63) bears large insertions of up to 165-aa in some JMY sequences. JMY sequences also show shorter insertions before and after the Pro-rich region of the PWWCA domain, with the second of these insertions corresponding to an additional WH2 domain.
Figure 4.
Domain architecture of WHAMM and ER localization and function
(A) Domain organization of WHAMM (see also Figure S4A).
(B) Multi-angle light scattering (MALS) analysis of MBP-WHAMM1–541. Purified protein shown in SDS-PAGE (inset). The mass distribution (indicated by the black arrow) of WHAMM1–541 was calculated from the peak where the light scattering profile (black curve) and normalized differential refractive index (red curve) overlap.
(C) GFP-WHAMM1–541 puncta localize to the periphery of ER tubules (mCherry-Sec61β) in ARPE-19 cells.
(D) GFP-WHAMM62–793 puncta localize to the periphery of ER tubules (mCherry-Sec61β), but also form large vesiclo-vacuolar aggregates in ARPE-19 cells.
(E) GFP-WHAMM171–793 forms elongated static tubules that partially colocalize with the ER (mCherry-Sec61β) in an ARPE-19 cell (see also Figures S4G–S4I).
(F) GFP-WHAMM1–169 extensively coats ER tubules (mCherry-Calnexin) in HeLa cells and occasionally forms dynamic puncta (insets).
(G) Model for role of WHAMM in autophagy. Autophagosome biogenesis is initiated by the ATG14-dependent PI3K complex on the ER, which then gives rise to a DFCP1 coated omegasome. LC3-I from the cytosol is converted to LC3-II through lipidation and anchored to the phagophore membrane forming on the omegasome. The cargo adaptor protein p62 is then recruited to the maturing phagophore by interaction with LC3, and serves to recruit ubiquitinated protein aggregates destined for lysosomal degradation. The phagophore closes off to form an autophagosome that then fuses with a lysosome (autophagolysosome), where its content is degraded. Autophagosome biogenesis and motility depend on an actin comet tail mechanism mediated by the Arp2/3 complex and WHAMM. WHAMM colocalizes with markers present during the maturation of autophagosomes (DFCP1, LC3, p62), but is not found in association with either ATG14 or Lamp1.
See also Figure S4.
While WHAMM [6] and JMY [4] have been described as coiled coil-containing proteins, we established here using multi-angle light scattering that WHAMM1–541, comprising the predicted coiled coil regions (Figure S4A), is monomeric (Figure 4B). Thus, these NPFs are unlikely to form coiled coil dimers, and their predicted large helical regions could instead be implicated in protein-protein interactions.
The similar domain architecture of these two NPFs prompted us to test whether JMY also colocalized with LC3 by cotransfecting ARPE-19 cells with GFP-JMY and mCherry-LC3. Strikingly, JMY formed puncta that occasionally colocalized and comigrated with LC3-positive vesicles (Figure S4B). However, contrary to WHAMM, the JMY-positive puncta did not appear to associate with actin comet tails within this context (Figure S4C). JMY has been shown to interact with the ER-resident protein VAP-A [5]. As a broad ER marker, VAP-A appeared to colocalize with WHAMM puncta in fed HeLa cells (Figure S4D). Yet, the colocalization was more striking in starved cells, where VAP-A appeared to accumulate to form puncta on the ER, which colocalized and comigrated with WHAMM-positive puncta (Figure S4E). Thus, the shared domain architecture and the colocalization and comigration of JMY and WHAMM with the autophagy marker LC3 and the ER-resident protein VAP-A all point to some degree of overlap in the function of these two NPFs.
Role of WHAMM domains in ER localization and function
To understand the role of each of the WHAMM domains in ER localization and function, we coexpressed the ER marker Sec61β along with N- and C-terminal WHAMM truncation mutants (Figures 4C–4F and S4F–S4I). WHAMM1–541, lacking the C-terminal PWWCA domain, showed the same localization as full-length WHAMM, consisting mainly of ER-associated puncta as well as some tubular structures (Figure 4C). As observed with WHAMMW791A, WHAMM1–541-positive puncta were either static or showed directed motility. Further deleting residues 434–540 from the C-terminus, WHAMM1–433, increased the tubulation phenotype (Figure S4F). Deleting the first 61 residues, WHAMM62–793, resulted in the formation of large vesiclo-vacuolar aggregates on the ER (Figure 4D). On the other hand, WHAMM174–793 mostly did not colocalize with the ER markers mCherry-Sec61β (Figure 4E) and mCherry-Calnexin (Figure S4G), and gave rise to static elongated tubular structures that colocalized with MTs (Figure S4H), similar to the phenotype observed with extended WHAMM expression (Figures S1B and S1C). Importantly, the different localizations of WHAMM174–793 and full-length WHAMM is not due to differences in expression levels, as confirmed by coexpression of these two constructs (Figure S4I). WHAMM174–793 and full-length WHAMM appeared mostly segregated in these cells, forming tubular structures or puncta, respectively, which is also consistent with the observation that WHAMM does not form coiled coil dimers (Figure 4B). Finally, WHAMM1–169 extensively coated the ER (Figure 4F), and occasionally produced highly dynamic puncta that shuttled along ER tubules. Because WHAMM1–169 localizes to the ER, whereas WHAMM174–793 is mostly found along MTs, we conclude that residues 1–169 are critical for ER localization, and we thus refer to this region as the ER-binding (ERB) domain (Figure 4A).
While we cannot rule out a connection between the tubular, MT-associated WHAMM localization observed previously [6] and the dynamic, ER-associated puncta emphasized here, we note that several factors appear to shift the equilibrium in favor of the tubular phenotype, including increased protein expression, inhibition of actin assembly through CK666 or the W791A mutation, and deletion of the N-terminal ERB or C-terminal PWWCA domains. Thus, the middle region of WHAMM (aa 174–541), implicated in MT binding [17], seems to be at least in part responsible for this phenotype.
In summary, we have shown here that WHAMM colocalizes with autophagy markers that emerge from the ER and develop into autophagosomes (Figure 4G). Specifically, WHAMM associates with markers present during the maturation of autophagosomes (DFCP1, LC3, p62), but is not found in association with ATG14 prior to the formation of omegasomes or with Lamp1 after autophagosomes fuse with lysosomes (Figure 4G). We have further shown that autophagosome biogenesis and motility depend on actin comet tail formation driven by the Arp2/3 complex and WHAMM. The various domains of WHAMM participate in this process by directing WHAMM to the ER though the N-terminal ERB domain, inducing membrane tubulation via the middle helical domains, and activating Arp2/3 complex nucleation and branching through the C-terminal PWWCA domain. The strong similarity between WHAMM and JMY and the fact that JMY also colocalizes with autophagy markers suggest that these two NPFs may play related or complementary roles in this process.
Supplementary Material
Highlights.
WHAMM, an NPF of Arp2/3 complex, forms puncta that localize to the ER
WHAMM puncta colocalize with autophagy markers (LC3, DFCP1, and p62)
WHAMM drives autophagy-marker motility trough an actin comet tail mechanism
WHAMM links Arp2/3 complex-dependent actin assembly to autophagosome biogenesis
Acknowledgments
This work was supported by NIH grants P01 GM087253 and R01 MH087950. D.J.K. was additionally supported by American Cancer Society grant PF-13-033-01-DMC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dominguez R. Structural insights into de novo actin polymerization. Current opinion in structural biology. 2010;20:217–225. doi: 10.1016/j.sbi.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rottner K, Hanisch J, Campellone KG. WASH, WHAMM and JMY: regulation of Arp2/3 complex and beyond. Trends Cell Biol. 2010;20:650–661. doi: 10.1016/j.tcb.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 3.Shikama N, Lee CW, France S, Delavaine L, Lyon J, Krstic-Demonacos M, La Thangue NB. A novel cofactor for p300 that regulates the p53 response. Mol Cell. 1999;4:365–376. doi: 10.1016/s1097-2765(00)80338-x. [DOI] [PubMed] [Google Scholar]
- 4.Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol. 2009;11:451–459. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schluter K, Waschbusch D, Anft M, Hugging D, Kind S, Hanisch J, Lakisic G, Gautreau A, Barnekow A, Stradal TE. JMY is involved in anterograde vesicle trafficking from the trans-Golgi network. Eur J Cell Biol. 2014;93:194–204. doi: 10.1016/j.ejcb.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Campellone KG, Webb NJ, Znameroski EA, Welch MD. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell. 2008;134:148–161. doi: 10.1016/j.cell.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aguilera MO, Beron W, Colombo MI. The actin cytoskeleton participates in the early events of autophagosome formation upon starvation induced autophagy. Autophagy. 2012;8:1590–1603. doi: 10.4161/auto.21459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shenoy VB, Tambe DT, Prasad A, Theriot JA. A kinematic description of the trajectories of Listeria monocytogenes propelled by actin comet tails. Proc Natl Acad Sci U S A. 2007;104:8229–8234. doi: 10.1073/pnas.0702454104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, Hartman J, Sakowicz R, Pollard TD. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 2009;460:1031–1034. doi: 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 11.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 13.Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 14.Bjorkoy G, Lamark T, Pankiv S, Overvatn A, Brech A, Johansen T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 2009;452:181–197. doi: 10.1016/S0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- 16.Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151:1501–1512. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen QT, Hsiue PP, Sindelar CV, Welch MD, Campellone KG, Wang HW. Structural insights into WHAMM-mediated cytoskeletal coordination during membrane remodeling. J Cell Biol. 2012;199:111–124. doi: 10.1083/jcb.201204010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.