Abstract
Associating changes in protein levels with the onset of cancer has been widely investigated to identify clinically relevant diagnostic biomarkers. In the present study, we analyzed sera from 205 patients recruited in the U.S. and Egypt for biomarker discovery using label-free proteomic analysis by liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). We performed untargeted proteomic analysis of sera to identify candidate proteins with statistically significant differences between hepatocellular carcinoma (HCC) and patients with liver cirrhosis. We further evaluated the significance of 101 proteins in sera from the same 205 patients through targeted quantitation by multiple reaction monitoring (MRM) on a triple quadrupole mass spectrometer. This led to the identification of 21 candidate protein biomarkers that were significantly altered in both the U.S. and Egyptian cohorts. Among the 21 candidates, 10 were previously reported as HCC-associated proteins (eight exhibiting consistent trends with our observation), whereas 11 are new candidates discovered by this study. Pathway analysis based on the significant proteins reveals up-regulation of the complement and coagulation cascades pathway and down-regulation of the antigen processing and presentation pathway in HCC cases versus patients with liver cirrhosis. The results of this study demonstrate the power of combining untargeted and targeted quantitation methods for a comprehensive serum proteomic analysis, to evaluate changes in protein levels and discover novel diagnostic biomarkers.
Keywords: Cancer biomarker discovery, Hepatocellular carcinoma, Liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS), Liver cirrhosis, Multiple reaction monitoring
1 Introduction
Hepatocellular carcinoma (HCC) is the third leading cause of cancer mortality worldwide with five-year relative survival rates less than 15% [1, 2]. While the combined cancer mortality rate has been declining for two decades, incidence and mortality rates of HCC are still increasing [3]. Survival rates of patients with HCC can significantly be improved if the diagnosis was made at earlier stages, when treatment is more effective [4]. Most of the risk factors for HCC including chronic infection with hepatitis B virus (HBV) or hepatitis C virus (HCV) lead to the development of liver cirrhosis, which is present in 80–90% of patients with HCC [5]. The malignant conversion of cirrhosis to HCC is often fatal in part because adequate biomarkers are not available for diagnosis during the progression stages of HCC. Alpha-fetoprotein (AFP), the serologic biomarker for HCC in current use, lacks the desired sensitivity for early diagnosis of HCC [6, 7]. Therefore, more potent biomarkers are needed for detection of HCC at its early stage when it can be intervened more effectively.
Proteomics is the comprehensive analysis of all proteins in a biological system. Emerging technologies are enabling delineation of changes in protein levels and associating these changes with various diseases has been widely investigated to identify clinically relevant biomarkers [8, 9]. In particular, there is a great potential in utilizing proteomics to identify diagnostic biomarkers circulating in biofluids [10–12]. With recent advances of mass spectrometry and separation methods, liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) has become an essential analytical tool in a variety of omic studies including proteomics [13, 14]. LC-MS/MS provides qualitative and quantitative analysis of proteins in a high-throughput fashion, and it has been widely used for biomarker discovery [15]. While untargeted analysis of biofluids by label-free LC-MS/MS methods gives a global characterization of proteomic signatures, the quantitative sensitivity and accuracy of these methods are not satisfactory to determine reliable biomarkers [16, 17]. Thus, subsequent confirmation of the findings from untargeted analysis is often desirable using more sensitive and reliable quantitation methods such as multiple reaction monitoring (MRM) [18, 19].
In the present study, we analyzed sera from HCC cases and patients with liver cirrhosis, recruited in the U.S. and Egypt. We performed untargeted proteomic analysis to identify proteins showing statistically significant differences in sera from HCC cases and patients with liver cirrhosis. These proteins were further analyzed through targeted quantitation by MRM, which yielded more sensitive and accurate quantitation results. A high-resolution mass spectrometer (LTQ Orbitrap Velos) was used for untargeted proteomic analysis, while targeted quantitation was performed by MRM on a triple quadrupole (QqQ) mass spectrometer. We confirmed 21 candidates that showed significant changes in protein expression between HCC cases and cirrhotic controls in both cohorts. The results of this study demonstrate the power of combining untargeted and targeted quantitation methods for a comprehensive serum proteomic analysis, to investigate changes in protein levels between HCC cases and patients with liver cirrhosis.
2 Materials and methods
2.1 Experimental design
Adult patients were recruited from the outpatient clinics and inpatient wards of the Tanta University Hospital (TU cohort) in Tanta, Egypt and from the hepatology clinics at MedStar Georgetown University Hospital (GU cohort) in Washington, DC, USA. The TU cohort consists of a total of 89 subjects (40 HCC cases and 49 patients with liver cirrhosis); both HCC cases and cirrhotic controls are from either outpatient or inpatient wards. The GU cohort comprises of 116 subjects (57 HCC cases and 59 patients with liver cirrhosis). The protocol of the study was approved by the respective Institutional Review Boards at Tanta University and Georgetown University. Through peripheral venipuncture, single blood sample was drawn into 10 mL BD Vacutainer sterile vacuum tubes without the presence of anticoagulant. The blood was immediately centrifuged at 1000g for 10 min at room temperature. The serum supernatant was carefully collected and centrifuged at 2500g for 10 min at room temperature. After aliquoting, serum was kept frozen at −80 °C until use. Primary tubes and serum aliquots were labeled using anonymous confidential code numbers with no personal identifiers. Identification codes were cross-referenced with clinical information in a pass code protected computer system. Characteristics of the patient populations in TU and GU cohorts are provided in Tables 1 and 2, respectively.
Table 1.
Characteristics of the TU study cohort.
| HCC (n=40) | Cirrhosis (n=49) | p-value | |
|---|---|---|---|
| Age | |||
| Mean (SD) | 53.2 (3.9) | 53.8 (7.6) | 0.3530 |
| BMI | |||
| Mean (SD) | 24.9 (3.1) | 24.5 (4.4) | 0.6513 |
| Gender | |||
| Male | 77.5% | 67.3% | 0.3474 |
| HCV serology | |||
| HCV Ab+ | 100.0% | 100.0% | 1.0000 |
| HBV serology | |||
| HBsAg+ | 0.0% | 6.1% | 0.2492 |
| MELD | |||
| Mean (SD) | 18.6 (7.7) | 18.9 (7.1) | 0.1328 |
| MELD ≤ 10 | 20.0% | 12.2% | 0.3863 |
| Child-Pugh grade | |||
| A | 15.0% | 0% | 0.0117 |
| B | 47.5% | 46.9% | |
| C | 37.5% | 53.1% | |
| AFP | |||
| Median (IQR) | 275.9 (1244.3) | ||
| HCC stage | |||
| Stage I | 72.5% | ||
| Stage II | 15.0% | ||
| Stage III | 5.0% | ||
| Unknown | 7.5% | ||
Table 2.
Characteristics of the GU study cohort.
| HCC (n=57) | Cirrhosis (n=59) | p-value | |
|---|---|---|---|
| Age | |||
| Mean (SD) | 59.7 (6.6) | 59.0 (7.1) | 0.602 |
| BMI | |||
| Mean (SD) | 30.0 (6.7) | 29.5 (6.5) | 0.6828 |
| Gender | |||
| Male | 71.9% | 74.6% | 1 |
| Caucasian | 57.9% | 66.1% | |
| Ethnicity | |||
| African American | 28.1% | 23.7% | 0.4469 |
| Others | 14.0% | 10.2% | |
| Current | 23.3% | 21.7% | |
| Smoker | |||
| Former | 49.1% | 49.2% | 1 |
| None | 26.3% | 27.1% | |
| Current | 24.6% | 22.0% | |
| Alcohol | |||
| Former | 51.7% | 51.7% | 1 |
| None | 33.3% | 30.0% | |
| HCV Serology | |||
| HCV Ab+ | 61.4% | 39.0% | 0.037 |
| HCV RNA+ | 54.3% | 37.3% | 0.8791 |
| HBV Serology | |||
| Anti HBC+ | 43.9% | 28.8% | 0.1574 |
| HBsAg+ | 12.3% | 5.1% | 0.2018 |
| MELD | |||
| Mean (SD) | 11.1 (3.7) | 16.4 (14.3) | 5.98E-03 |
| MELD < 10 | 47.4% | 15.3% | 8.31E-05 |
| Child-Pugh gradea) | |||
| A | 38.6% | 13.6% | |
| B | 36.8% | 54.2% | |
| C | 10.5% | 28.8% | |
| AFP | |||
| Median (IQR) | 28.8 (83.2) | 4.4 (9.7) | 3.22E-05 |
| HCC Stage | |||
| Stage I | 54.4% | ||
| Stage II | 31.6% | ||
| Stage III | 5.3% | ||
| Unknown | 8.7% | ||
Child-Pugh grade is unknown in eight HCC patients.
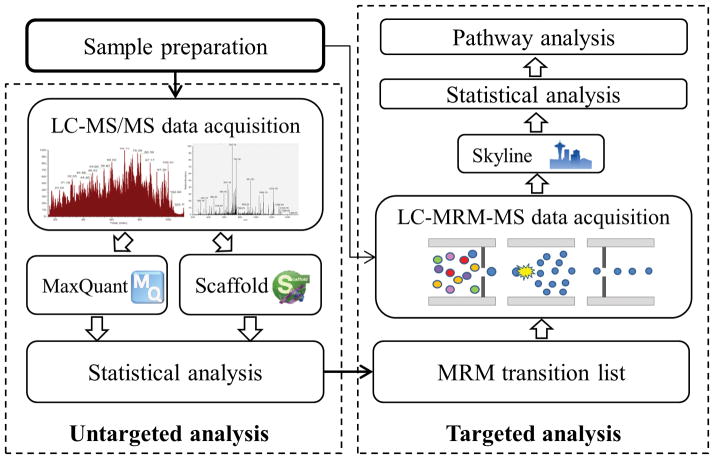
We analyzed sera from HCC cases and cirrhotic controls in TU cohort and GU cohort separately. We acquired LC-MS/MS data on a 3000 Ultimate nano-LC system interfaced to an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific, San Jose, CA). The analysis was performed following a randomized order to avoid systematic biases (Supporting Information Table 1). In addition to the patient samples, in the analysis of the TU cohort we included in-between quality control (QC) runs of technical replicates using pooled human blood serum purchased from Sigma Aldrich and full process replicates [20] using aliquots of a sample pooled from the patients’ sera. Technical replicates are generated by multiple injections of a single aliquot for which the protein assay/sample preparation procedure is performed once. Full process replicates, on the other hand, are generated from multiple aliquots of the same biological sample, where the protein assay/sample preparation of each aliquot is performed separately. Proteins with statistically significant differences between the two groups were identified as candidate biomarkers. On the same cohorts of patients, these findings were confirmed through targeted quantitation using MRM on a TSQ Vantage mass spectrometer (Thermo Scientific, San Jose, CA). All the patient samples were analyzed by both untargeted and targeted methods. Figure 1 summarizes the workflow of this biomarker discovery study.
Figure 1.
Workflow of the proposed biomarker discovery study involving untargeted and targeted analysis of sera.
2.2 Materials
HPLC-grade solvents, including methanol, isopropanol, and water, were procured from Macron Fine Chemicals™-Avantor Performance Materials (Center Valley, PA). HPLC grade acetonitrile (ACN) was purchased from Fisher Scientific (Pittsburgh, PA). Ammonium bicarbonate, DL-dithiothreitol (DTT), iodoacetamide (IAA), MS-grade formic acid (FA), and pooled human blood serum (H4522) were also purchased from Sigma-Aldrich. Mass spectrometry grade trypsin was obtained from Promega (Madison, WI).
2.3 Depletion of high-abundance proteins in serum
Sera were subjected to depletion using Agilent Plasma 7 Multiple Affinity Removal Spin Cartridge from Agilent Technologies (Santa Clara, CA). This cartridge depletes the seven most abundant human serum proteins, namely albumin, IgG, antitrypsin, IgA, transferrins, haptoglobin and fibrinogen. A 15-μl aliquot of serum was depleted as stated in the protocol provided by the manufacturer. The buffer of the depleted sample was exchanged into 50 mM ammonium bicarbonate (pH 8.0) using 3kDa MWCO Amicon Ultra 0.5mL centrifugal filters from Merck Millipore (Tullagreen, Carrigtwohill, Co. Cork). This buffer was used for tryptic digestion.
2.4 Trypsin digestion
Prior to trypsin digestion, the protein concentration of depleted serum was determined by micro BCA protein assay following the protocols recommended by the vendor (Thermo Scientific/Pierce, Rockford, IL). A 20-μg aliquot of depleted serum proteins that corresponds to 0.4 μl of original serum was transferred to an Eppendorf tube, to which 100-μl of 50 mM ammonium bicarbonate was then added. Thermal denaturation was performed at 65 °C for 10 min. DTT and IAA solutions were prepared in 50 mM ammonium bicarbonate. Sample was reduced by adding a 1.25-μl aliquot of 200 mM DTT solution and incubated at 60 °C for 45 min. The reduced proteins were then alkylated by adding of a 5-μl aliquot of 200 mM of IAA and incubated at 37.5 °C for 45 min. A second 1.25-μl aliquot of 200 mM DTT was added and followed by incubation at 37.5 °C for 30 min to consume excess IAA. A 0.8-μg aliquot of trypsin was added to the sample (enzyme/substrate ratio of 1:25 w/w), and then incubated at 37.5 °C overnight. This was followed by microwave-assisted digestion at 45 °C for 30 min at the power of 50 W. The enzymatic digestion was quenched by adding 0.5-μl neat FA to the samples. Then, the samples were speed-vacuum dried and re-suspended in 0.1% FA prior to LC-MS/MS and LC-MRM-MS analyses.
2.5 LC-MS/MS data acquisition by untargeted analysis
Analysis of sera was performed on a Dionex 3000 Ultimate nano-LC system (Dionex, Sunnyvale, CA) interfaced to an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific, San Jose, CA), which is equipped with a nano-ESI source. LC-MS/MS analysis was performed on a tryptic digest corresponding to 1 μg of proteins which was derived from 0.2 μl of original serum considering the whole depletion and digestion process. The samples were online-purified using Acclaim PepMap100 C18 cartridge (3 μm, 100Å, Dionex). The purified samples were then separated using Acclaim PepMap100 C18 capillary column (75 μm id × 150 mm, 2 μm, 100Å, Dionex). The separation of the digests was achieved at 350 nl/min flow rate, using the following gradient: 0–10 min sustaining 5% solvent B (98% ACN with 0.1%FA), 10–65 min ramping solvent B 5–20%, 65–90 min ramping solvent B 20–30%, 90–105 min ramping solvent B 30–50%, 105–106 min ramping solvent B 50–80%, 106–110 min maintaining solvent B at 80%, 110–111 min decreasing solvent B 80–5%, and 111–120 min maintaining solvent B at 5%. Solvent A was a 2% ACN aqueous solution containing 0.1% FA. The separation and scan time were set to 120 min.
The LTQ Orbitrap Velos mass spectrometer was operated with two scan events. The first scan event was a full FTMS scan of 380–2000 m/z with a mass resolution of 15,000 at m/z of 400. The second scan event was collision induced dissociation (CID) MS/MS of parent ions selected from the first scan event with an isolation width of 3.0 m/z. Normalized collision energy was set to 35% with an activation Q value of 0.250 and an activation time of 10 ms. The CID MS/MS was performed on the five most intense ions observed from the first MS scan event.
2.6 LC-MS/MS data analysis
The LC-MS/MS data were analyzed using MaxQuant [21] (version 1.4.1.2) and Scaffold [22] (version Scaffold_3.6.3, Proteome Software Inc., Portland, OR), where the quantitation of proteins was based on ion intensity and spectral count, respectively.
In the MaxQuant analysis, MS/MS spectra were searched against the UniProt human protein database (20,259 protein entries, version of April 23, 2014) using Andromeda [23]. Decoy database of modified reversed protein sequences and 247 common contaminants were also considered in the searching. MaxQuant used an initial searching result for recalibration of masses and retention times, where the mass tolerance of precursor mass and fragment mass used in Andromeda were set as 6 ppm and 0.5 Da, respectively. Carbamidomethylation of cysteine was set as a fixed modification; methionine oxidation and protein N-terminal acetylation were set as variable modifications. Minimal peptide length was set to seven amino acids and at most two missed cleavages were allowed. Only proteins with more than two identified peptides were considered. The false discovery rate (FDR) was set at 0.01 for identification of peptides and proteins. Minimal numbers of razor and unique peptide were both set to one. Uniqueness means a peptide is unique to a single protein group. The “matching-between-runs” feature was enabled and the “label-free quantification” (LFQ) approach with a minimum of two ratio counts was used to compare and normalize protein intensities across runs [24]. This yielded LFQ intensities that were used in the subsequent statistical analysis.
The statistical analysis was performed using Perseus (version 1.4.1.3), an accompanying tool to MaxQuant. Identifications from reversed sequences and contaminants were first removed and only proteins detected in over half of the runs in either case or control group were retained. After the screening, the LFQ intensities were log-transformed and missing value imputation was applied in consideration of the estimated intensity distribution. The most relevant proteins with statistically significant differences between HCC cases and cirrhotic controls were selected using Welch’s test. We calculated p-values with the null hypothesis that means of the two groups (HCC and cirrhosis) are the same. In addition, we used a burden of illness test [25] to identify proteins differentially expressed between the two groups, by taking into account both the mean of the intensity values and the proportion of missing values (set to 0) in the two groups. The p-values associated with the tests were adjusted for multiple testing using the method of Benjamini and Hochberg [26] to control the FDR at a 0.05 level. Proteins with an adjusted p-value <0.05 by either test were selected as candidates for further analysis by MRM.
In the Scaffold analysis, Proteome Discoverer software (version 1.2, Thermo Scientific, San Jose, CA) was used to generate Mascot generic format files (*.mgf), which were subsequently employed for database searching using Mascot (version 2.4.0, Matrix Science, London, UK). Mascot was set up to search the UniProt human protein database. Peptides were searched with a parent ion tolerance of 10 ppm and a fragment ion mass tolerance of 0.8 Da. Trypsin was selected allowing two missed cleavages. Carbamidomethylation of cysteine was set as a fixed modification while oxidation of methionine was set as a variable modification. Scaffold was used to probabilistically assign peptide and protein identifications based on PeptideProphet [27] and ProteinProphet algorithms [28], respectively. Peptide identifications were accepted with a probability greater than 95%, while protein identifications were accepted with a probability greater than 99% and contained at least two identified peptides. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Identified proteins were then quantitated based on spectral counting. Missing values were set to 0 prior to subsequent statistical analysis. Proteins with statistically significant differences (p-value <0.05) between HCC cases and cirrhotic controls were selected using t-test.
2.7 Design of MRM transitions for targeted quantitation
In the untargeted LC-MS/MS analysis, candidate protein biomarkers were identified using MaxQuant and Scaffold. We merged the results by both approaches and evaluated these proteins through targeted quantitation by MRM. For each targeted protein, one or two associated peptides were selected using the following rules [29]: (1) identified in the untargeted analysis with a Scaffold probability greater than 95%, (2) completely digested by trypsin, (3) 7-25 amino acid residues, (4) excluding the first 25 amino acids at the N-terminus of proteins, (5) excluding peptides with M, RP, KP and glycosylation site (NXS/T), (6) excluding peptides with ragged ends (tryptic peptides cleaved between R/K, K/R, R/R and K/K), and (7) fixed carbamidomethylation of Cysteine. Next, five transitions of selected peptides were determined using the following rules: (1) precursor ions with charge states of two or three, (2) y series of fragment ions greater than y3 with a charge state of one, (3) the five most intense fragment ions in the MS/MS spectra from untargeted analysis, and (4) m/z values of precursor and transition ions between 300 and 1500.
Prior to MRM scheduling of individual samples, a 1-μl aliquot of each sample was pooled and subjected to MRM experiment to refine the transition list. A 3-μl aliquot of the pooled sample was analyzed by LC-MRM-MS. The expected retention time (RT) of each peptide and its transitions were manually checked using Pinpoint (Thermo Scientific, San Jose, CA) and compared with that in the untargeted analysis to confirm the targeted peptides. Their correlation plots in terms of RT are provided in Supporting Information Figure 1. An RT segment was set to 12 min for each targeted peptide with its expected RT in the center based on the pooled sample analysis. The three most intense transition ions of each peptide were selected as the final transitions. Peptides and transitions were removed from transition list if any of them was not detected in the pooled sample analysis. In total, 101 targeted proteins with 187 peptides and 561 transitions were scheduled and subjected to the LC-MRM-MS experiments. With the abovementioned 12-min RT segment, a minimum 30 ms dwell time was assigned to each transition. A complete list of the MRM transitions used in this study is provided in Supporting Information Table 2.
2.8 LC-MRM-MS data acquisition by targeted analysis
The LC conditions described previously in the untargeted analysis were used here for targeted quantitation by MRM on the TSQ Vantage mass spectrometer (Thermo Scientific, San Jose, CA), which was operated in positive mode with an ESI voltage of 1800V. 1.5 μg of serum peptides derived from 0.3 μl of original serum was injected to the LC system. Data independent acquisition mode was used for MRM experiment. Predefined precursor and transition ions were monitored to specifically select targeted peptides corresponding to each candidate protein with 10.0 sec chromatogram filter peak width. The MRM experiments were performed at a cycle time of 5.0 sec and a Q1 peak width (FWHM) of 0.70 Da. The collision energy (CE) value for each targeted peptide is predicted by Pinpoint (CE(+2) = 0.034 * m/z + 3.314(eV), CE(+3) = 0.044 * m/z + 3.314(eV)) with a collision gas pressure of 1.5 mTorr in Q2.
2.9 LC-MRM-MS data analysis
The LC-MRM-MS data were analyzed using Skyline [30] (version 2.5.0.6079). Peptide search results from Andromeda, i.e., msms.txt and mqpar.xml, were used to recognize the monitored transitions from LC-MRM-MS data. The Skyline determined the RT location and integration boundaries for each peptide in each run independently. By comparing the same peptide across runs, we adjusted the RT location and integration boundaries to exclude interfering regions. We selected the peak closest to the RT center of segment if multiple peaks were detected. Each protein’s intensity was quantitated using the summation of intensities from its corresponding transitions. The difference between total area and background was assigned to quantify a transition [29]. Prior to the statistical analysis, the quantitated protein intensities were log-transformed and normalized by the summed intensity. The most relevant proteins with differential abundance between HCC cases and cirrhotic controls were selected using t-test, and the associated p-values were adjusted based on multiple testing correction (FDR <0.05).
3 Results and Discussion
3.1 Untargeted analysis
Data matrices of protein intensities were obtained by preprocessing LC-MS/MS runs from each cohort, based on ion intensity and spectral count. Only proteins with more than two identified peptides were considered. In the MaxQuant analysis, 269 and 252 proteins were identified in the TU and GU cohorts, respectively. The complete lists of the identified proteins are provided in Supporting Information Tables 3 (TU cohort) and 4 (GU cohort). In the Scaffold analysis, 231 and 227 proteins were identified in the TU and GU cohorts, respectively. The complete list of the identified proteins in each cohort is given in Supporting Information Table 5. Venn diagrams showing the number of proteins identified by MaxQuant and Scaffold are provided in Supporting Information Figure 2.
Prior to statistical analysis, we evaluated the experimental variability by comparing the quantitated protein intensities of QC runs obtained from technical replicates using a standard from Sigma Aldrich and full process replicates using aliquots of a sample pooled from the patients’ sera. The correlation coefficients between the technical replicates and the full process replicates ranged from 0.972–0.999 and 0.937–0.995, respectively. Supporting Information Figures 3 and 4 show the scatter plots between pairs of technical and full process replicates. The calculated correlation coefficients and the plots suggest that the untargeted proteomic analysis had a fairly good reproducibility.
Statistical analysis of the ion intensities obtained by MaxQuant revealed 80 proteins (38 in TU cohort and 54 in GU cohort) that are statistically significant (Supporting Information Table 6). Statistical analysis of the data obtained by spectral count using Scaffold identified 78 (42 in TU cohort and 48 in GU cohort) significant proteins (Supporting Information Table 7). The difference between the two cohorts may be attributed to their heterogeneous nature. For example, the participants in the TU cohort are all HCV positive and nearly all are HBV negative, whereas about half of the participants in the GU cohort are HCV positive and about a third are HBV positive. Also, while the TU cohort is homogeneous Middle Eastern, the GU cohort consists of approximately 56% Caucasian, 30% African American, 10% Asian, and 5% Hispanic. Moreover, about three quarter of the HCC cases in the TU cohort are stage I HCC, while stage I HCC accounts for about half of the HCC cases in the GU cohort. Through further evaluation of these candidate proteins on the TSQ Vantage mass spectrometer using pooled sera, we finalized a list of 101 proteins for further evaluation by MRM (Supporting Information Table 2).
3.2 Targeted quantitation
To evaluate the variability between untargeted analysis and targeted quantitation, we compared the coefficients of variation (CVs) of intensities for those proteins that were profiled by both quantitation methods and present in over 80% of the samples within the HCC or the cirrhotic groups. Since this evaluation was performed on the proteins that were found statistically significant in the untargeted analysis and further evaluated by targeted quantitation, we compared the CVs in HCC and cirrhotic groups separately. Therefore, the CVs we calculated reflect the combined biological and analytical variation for each protein within each disease group. In the TU cohort, the ranges of the CVs in the HCC and cirrhotic groups were 2.2–8.9% (with median at 3.8%) and 2.3–8.6% (with median at 4.4%) by untargeted analysis, and these numbers reduced to 0.6–6.5% (with median at 1.4%) and 0.5–7.2% (with median at 1.4%) by targeted quantitation. Similarly, in the GU cohort, the ranges of the CVs in the HCC and cirrhotic groups were 1.7–7.3% (with median at 3.2%) and 1.9–8.5% (with median at 3.6%) by untargeted analysis, and they reduced to 0.7–4.9% (with median at 1.7%) and 0.9–5.9% (with median at 1.8%) by targeted quantitation. The comparison is summarized by box plots in Supporting Information Figure 5. Furthermore, one-tailed paired Wilcoxon tests in every pair of group and cohort indicated that the CVs in the targeted quantitation are significantly smaller (p-value <0.05) than in the untargeted analysis.
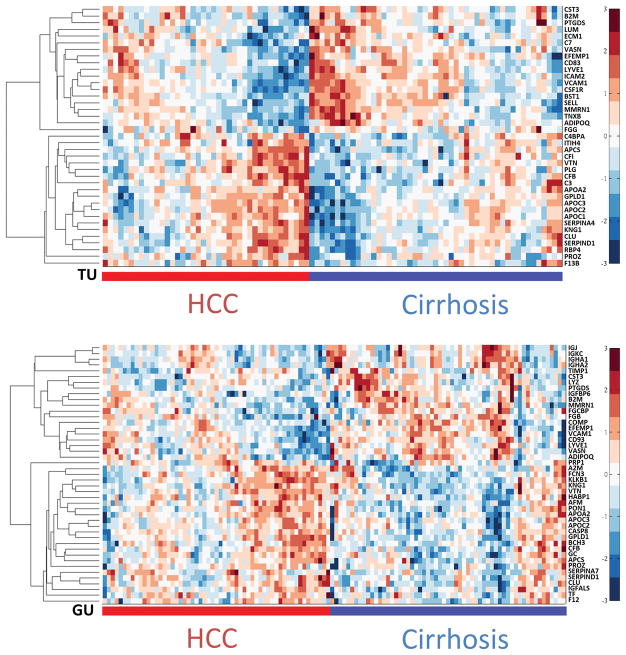
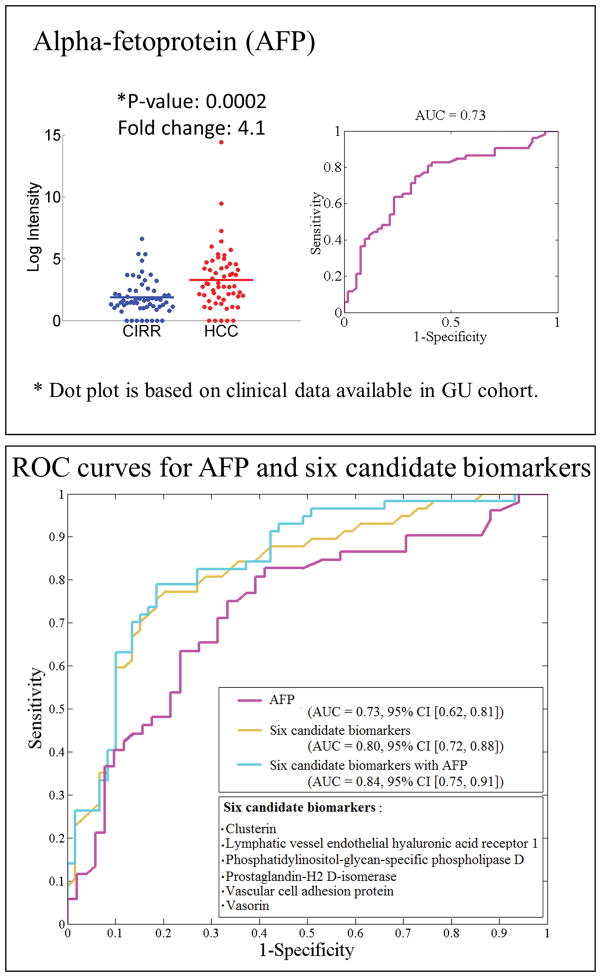
Through targeted quantitation of the 101 candidate proteins by MRM, we found 61 proteins that are statistically significant (adjusted p-value <0.05). These represent 39 and 43 significant proteins in the TU and GU cohorts, respectively, with 21 overlapping in both cohorts (Table 3). Heatmaps of hierarchical clustering results based on the significant proteins in each cohort are presented in Figure 2. The quantitation results from Skyline are provided in Supporting Information Table 8. Among the 21 proteins found significant in both cohorts, 11 are up-regulated in HCC versus cirrhosis, while 10 are down-regulated in HCC. All of their fold change directions are consistent with those in the untargeted analysis. The dot plots and ROC curves for these proteins in both cohorts are given in Supporting Information Figure 6, where the intensity values are based on MRM analysis and the fold changes are based on means. Other candidate biomarkers for the TU and GU cohorts are presented in Supporting Information Tables 9 and 10, respectively. While the reported AUC for each single biomarker is moderate, a panel selected by the SVM-RFE algorithm [31] from 21 proteins has led to a significant improvement over individual biomarkers including AFP. Specifically, the algorithm selected six proteins, namely, clusterin (CLU, P10909), vascular cell adhesion protein 1 (VCAM1, P19320), prostaglandin-H2 D-isomerase (PTGDS, P41222), phosphatidylinositol-glycan-specific phospholipase D (GPLD1, P80108), vasorin (VASN, Q6EMK4) and lymphatic vessel endothelial hyaluronic acid receptor 1 (LYVE1, Q9Y5Y7). Figure 3 depicts the ROC curves for AFP, a panel of six proteins, and the six proteins combined with AFP. We used a bootstrap method (1000 bootstrap replicates) to compute the 95% confidence interval (CI) of the area under each ROC curve. The six proteins in a panel achieved a higher AUC (95% CI [0.72, 0.88], with mean of 0.80) than AFP alone (95% CI [0.62, 0.81], with mean of 0.73). While the mean AUC was increased to 0.84 when the six proteins were combined with AFP in a panel, the addition of AFP does not improve the performance since the 95% CI [0.75, 0.91] overlaps substantially with the ROC based on the six proteins alone. An SVM classifier trained to minimize the misclassification rate by combining these markers and evaluated through cross-validation yielded higher sensitivity and specificity (0.75 and 0.77) compared with the performance of AFP alone (0.7 and 0.62). This comparison was performed using the GU study cohort, because the clinical measurement of AFP for the cirrhotic controls in the TU cohort was not available.
Table 3.
Protein candidate biomarkers identified by untargeted analysis and confirmed by targeted quantitation in both TU and GU cohorts. Fold change is the ratio of the mean intensity measured by MRM in the HCC group to the mean intensity in the cirrhotic group.
| Protein Name | UniProt ID | TU cohort | GU cohort | ||
|---|---|---|---|---|---|
|
| |||||
| Adjusted p-value | Fold Change | Adjusted p-value | Fold Change | ||
| Apolipoprotein A-II | P02652 | 0.025 | ↑1.4 | 0.006 | ↑1.3 |
| Apolipoprotein C-II | P02655 | 0.028 | ↑1.9 | 0.017 | ↑1.6 |
| Apolipoprotein C-III | P02656 | 0.032 | ↑1.8 | 0.045 | ↑1.6 |
| Clusterin | P10909 | 0.014 | ↑1.3 | 0.002 | ↑1.4 |
| Complement factor B | P00751 | 0.008 | ↑1.4 | 0.024 | ↑1.1 |
| Heparin cofactor 2 | P05546 | 0.025 | ↑1.5 | 0.011 | ↑1.4 |
| Kininogen-1 | P01042 | 0.014 | ↑1.3 | 0.023 | ↑1.2 |
| Phosphatidylinositol-glycan-specific phospholipase D | P80108 | 0.019 | ↑1.6 | 0.004 | ↑1.5 |
| Serum amyloid P-component | P02743 | 0.005 | ↑1.6 | 0.034 | ↑1.6 |
| Vitamin K-dependent protein Z | P22891 | 0.016 | ↑1.4 | 0.012 | ↑1.3 |
| Vitronectin | P04004 | 0.016 | ↑1.4 | 0.006 | ↑1.2 |
|
| |||||
| Adiponectin | Q15848 | 0.021 | ↓1.7 | 0.035 | ↓1.5 |
| Beta-2-microglobulin | P61769 | 0.025 | ↓1.3 | 0.042 | ↓1.7 |
| Complement component C1q receptor | Q9NPY3 | 0.040 | ↓1.4 | 0.011 | ↓1.3 |
| Cystatin-C | P01034 | 0.037 | ↓1.2 | 0.023 | ↓1.3 |
| EGF-containing fibulin-like extracellular matrix protein 1 | Q12805 | 0.019 | ↓1.3 | 0.008 | ↓1.4 |
| Lymphatic vessel endothelial hyaluronic acid receptor 1 | Q9Y5Y7 | 0.019 | ↓1.5 | 0.048 | ↓1.4 |
| Multimerin-1 | Q13201 | 0.019 | ↓1.5 | 0.017 | ↓1.5 |
| Prostaglandin-H2 D-isomerase | P41222 | 0.027 | ↓1.3 | 0.003 | ↓1.9 |
| Vascular cell adhesion protein 1 | P19320 | 0.006 | ↓1.5 | 0.048 | ↓1.3 |
| Vasorin | Q6EMK4 | 0.031 | ↓1.2 | 0.003 | ↓1.2 |
Figure 2.
Heatmaps for significant proteins measured by MRM in the TU (top panel) and GU (bottom panel) cohorts.
Figure 3.
Top panel: Dot plot and ROC curve for AFP. Bottom panel: ROC curves for AFP, a panel of six proteins, and a panel of six proteins combined with AFP (mean AUC and 95% confidence interval).
A number of these candidate biomarkers were reported as HCC-related biomarkers in previous studies. Among the candidate biomarkers found in both cohorts, five up-regulated proteins were reported in previous HCC studies, namely, apolipoprotein A-II (APOA2, P02652) [32, 33], clusterin (CLU, P10909) [34–36], complement factor B (CFB, P00751) [32, 35], serum amyloid P-component (APCS, P02743) [35], and vitronectin (VTN, P04004) [37, 38]. Alteration of proteins of biofluids in HCC compared with cirrhosis may potentially contribute to diagnostic signatures for HCC. It is noted that VTN was reported by two independent studies comparing serum/plasma proteins in HCC versus patients with cirrhosis [37, 38]. In addition, CLU, CFB and APCS were reported in a recent study by comparing the levels of plasma proteins in HCC patients, cirrhotic patients and healthy individuals from two African cohorts [35]. CLU was also found significant by comparing serum proteins in HCC versus HBV-related cirrhotic patients [36]. It is of interest to note that while this protein was found significant in both cohorts (Table 3), the analysis in GU cohort yielded greater statistical significance and fold change, where more patients recruited from this cohort were with HBV infection. However, the sample size in this study may not be sufficient enough to draw solid conclusion in this regard. The roles of CLU in HCC progression and metastasis have also been reported [39, 40]. Moreover, three down-regulated proteins, i.e., adiponectin (ADIPOQ, Q15848), EGF-containing fibulin-like extracellular matrix protein 1 (EFEMP1, Q12805), and lymphatic vessel endothelial hyaluronic acid receptor 1 (LYVE1, Q9Y5Y7), were supported by their previously reported prognostic implications. Decreased levels of ADIPOQ have been associated with poor prognostic in HCC patients based on in vitro and in vivo findings [41], and this protein was conjectured to prevent liver tumorigenesis. EFEMP1 gene was studied through expression profiling and karyotype analysis [42]. Decreased levels of EFEMP1 were found in HCC tumor tissues and closely associated with promoter hypermethylation and worse prognosis in HCC. In the context of postoperative survival analysis, LYVE1 was reported as a HCC prognostic biomarker [43].
In addition to the aforementioned eight candidate biomarkers, we found two previously reported protein biomarkers: cystatin-C (CST3, P01034) and beta-2-microglobulin (B2M, P61769). However, the reported up-regulation of these proteins contradicts with our observation (i.e., down-regulation in HCC cases versus patients with liver cirrhosis). Specifically, through analysis of SELDI-TOF MS, increased levels of CST3 were found in HCC versus cirrhosis [44]. It is noted, however, this protein has been frequently reported for its elevation in patients with hepatic diseases [45, 46]. Thus, additional investigation of this protein may further elaborate its role and functionality in the development of HCC. Another study that involves small sample size (six per group) showed that increased levels of B2M could be used for early diagnosis of HCC in cirrhotic patients [47]. Apart from these, 11 new candidates have been discovered in our study, including six up-regulated proteins: apolipoprotein C-II (APOC2, P02655), apolipoprotein C-III (APOC3, P02656), heparin cofactor 2 (SERPIND1, P05546), kininogen-1 (KNG1, P01042), phosphatidylinositol-glycan-specific phospholipase D (GPLD1, P80108), vitamin K-dependent protein Z (PROZ, P22891), and five down-regulated proteins: multimerin-1 (MMRN1, Q13201), prostaglandin-H2 D-isomerase (PTGDS, P41222), vascular cell adhesion protein 1 (VCAM1, P19320), complement and component C1q receptor (CD93, Q9NPY3), and vasorin (VASN, Q6EMK4). Among these new candidates, several are involved in blood coagulation (SERPIND1, KNG1, PROZ, MMRN1) and cell adhesion (MMRN1, VCAM1). KNG1 is also related to heparin binding. APOC2 and APOC3 have the functions of inhibiting lipoprotein lipase and hepatic lipase and decreasing the uptake of lymph chylomicrons by hepatic cells.
This study confirmed other previously reported HCC-associated biomarkers. These include four up-regulated proteins, complement C3 (C3, P01024) [32, 35, 48], C4b-binding protein alpha chain (C4BPA, P04003), complement factor I (CFI, P05156) [35], and plasminogen (PLG, P00747) [32] in the TU cohort (Supporting Information Table 9), and three up-regulated proteins, vitamin D-binding protein (GC, P02774) [32], alpha-2-macroglobulin (A2M, P01023) [35], and serotransferrin (TF, P02787) [32, 49] in the GU cohort (Supporting Information Table 10).
3.3 Gene ontology and pathway analysis
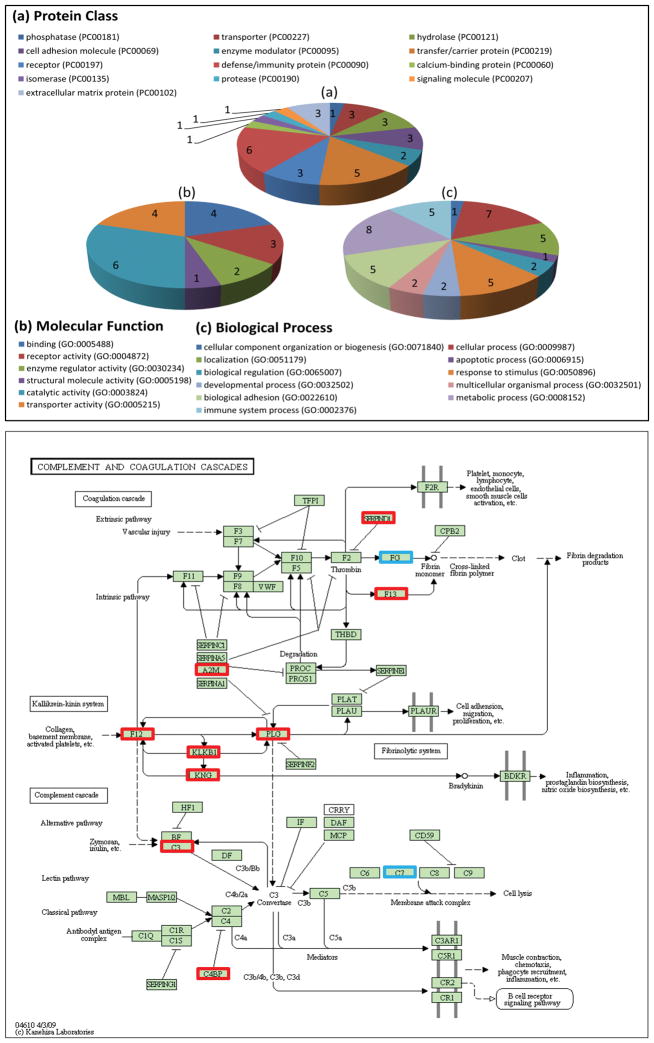
Gene ontology analysis was performed using PANTHER [50] based on the 21 biomarkers identified in both cohorts (Table 3). Figure 4 (a–c) illustrate the participation of these proteins in a diverse array of biological processes, including metabolic process, cellular process, biological adhesion, immune system process, localization, etc., emphasizing the systemic effects of HCC disease. The gene ontology analysis also reveals various molecular functions and pathways where the biomarkers are involved. For example, the new candidate biomarker heparin cofactor 2 (SERPIND1, P05546) is mapped to the pathway of blood coagulation and related to serine-type peptidase activity and peptidase inhibitor activity.
Figure 4.
Top panel: Gene ontology analysis by PANTHER (Protein ANalysis THrough Evolutionary Relationships). Bottom panel: Complement and coagulation cascades pathway involving both up-regulated (red) and down regulated biomarkers (blue) in KEGG database.
We used PathwayLinker [51] and DAVID [52] (version 6.7) to further identify significant signaling pathways, where the UniProt IDs of the significant proteins confirmed by MRM and their interacting neighbors were considered. We used PathwayLinker to obtain the first neighbor interactors of the 286 proteins detected in this study, where three interaction databases, BioGrid [53], STRING [54], and HPRD [55], were considered. Using the detected proteins and their interacting neighbors as a reference, we analyzed the proteins found significant in this study to determine relevant signaling pathways in KEGG [56] through the DAVID functional annotation tool. By mapping 34 up-regulated proteins from both cohorts against the 286 detected proteins and their interacting neighbors, we found complement and coagulation cascades as the most significantly enriched signaling pathway, as shown in Figure 4. This pathway involves 11 proteins that were up-regulated in this study (A2M, F12, F13B, SERPIND1, CFI, KLKB1, KNG1, PLG, CFB, C3, and C4BPA). When we analyzed the 27 down-regulated proteins from the two cohorts, we found antigen processing and presentation, as the most significant pathway. Among the 27 down-regulated proteins, three (FGB, FGG, and C7) are involved in complement and coagulation cascades pathway. The proteins we used for pathway analysis, the interacting proteins obtained from PathwayLinker, and the signaling pathways found by DAVID are provided in the Supporting Information Table 11.
4 Concluding remarks
We analyzed serum proteins from HCC cases and cirrhotic controls in two study cohorts (TU and GU), where seven high-abundance proteins were depleted, allowing relative quantitation of hundreds of serum proteins. Candidate protein biomarkers were identified through LC-MS/MS based untargeted analysis and confirmed by targeted quantitation using MRM. The most relevant proteins in distinguishing HCC cases from cirrhotic controls were selected using three statistical tests. We identified 101 statistically significant proteins through untargeted analysis. Among these, 61 were confirmed through targeted analysis by MRM. In particular, there were 21 candidate protein biomarkers found significant in both the TU and GU cohorts. Our findings independently confirmed several previously reported HCC protein biomarkers and revealed a number of new candidate biomarkers. Furthermore, we performed multivariate analysis to identify a panel of six protein biomarkers, which yielded better performance in comparison with AFP.
Although less measurement variability has been observed by targeted quantitation compared with untargeted analysis, the reported fold changes of the candidate protein biomarkers are moderate and barely exceed two except for Ig alpha-1 chain C region, Ig alpha-2 chain C region, Ig kappa chain C region, and Immunoglobulin J chain in the GU cohort (Supporting Information Table 10). This may be attributed to the nature of the samples we collected. Instead of healthy controls, we used serum samples from cirrhotic patients as our controls. Also, the HCC cases in our study have liver cirrhosis and most are early stage HCC. Due to these subtle differences, substantial fold changes are not expected between HCC and cirrhosis. While the comparison has led to significantly altered proteins that can serve as candidate biomarkers, more potent computational and analytical strategies are required to identify biomarkers for clinical diagnosis. Further evaluation of the proteomic data at the peptide level might reveal more distinct signatures. Also, through more selective analysis using subsets of the serum proteome, it is expected to reduce the analytical complexity and enhance the power to identify clinically relevant biomarkers. In addition to aforementioned refinements, our future work will focus on evaluating if the observed proteomic signatures could be reliably used for diagnosis of HCC in high-risk population of cirrhotic patients. This will be accomplished by investigating candidate biomarkers discovered in this study on a larger population that includes healthy controls and that allows stratification of the patients on the basis of etiology and disease stage. Also, we will analyze the proteomic data to identify differences that may exist at the peptide level between HCC cases and cirrhotic patients. Finally, we plan to utilize additional omic measurements conducted on the same subjects [57–59] to integrate multi-omic signatures for a more comprehensive characterization of HCC.
Supplementary Material
Acknowledgments
This study was supported by NIH Grants R01CA143420 and R01GM086746 awarded to HWR.
Abbreviations
- AFP
alpha-fetoprotein
- AUC
area under curve
- CE
collision energy
- CV
coefficient of variation
- FWHM
full width at half maximum
- HCC
hepatocellular carcinoma
- LC-MS/MS
liquid chromatography coupled with tandem mass spectrometry
- MRM
multiple reaction monitoring
- QqQ
triple quadrupole
- ROC
receiver operating characteristic
- RT
retention time
Footnotes
The LC-MS/MS and LC-MRM-MS data in this paper have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (dataset identifier PXD001171) and PeptideAtlas repository (dataset identifier PASS00542), respectively.
The authors have declared no conflict of interest.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer. 2013;13:123–135. doi: 10.1038/nrc3449. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 4.Bialecki ES, Di Bisceglie AM. Diagnosis of hepatocellular carcinoma. HPB (Oxford) 2005;7:26–34. doi: 10.1080/13651820410024049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–27. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 6.Trevisani F, D’Intino PE, Morselli-Labate AM, Mazzella G, et al. Serum alpha-fetoprotein for diagnosis of hepatocellular carcinoma in patients with chronic liver disease: influence of HBsAg and anti-HCV status. J Hepatol. 2001;34:570–575. doi: 10.1016/s0168-8278(00)00053-2. [DOI] [PubMed] [Google Scholar]
- 7.Gupta S, Bent S, Kohlwes J. Test characteristics of alpha-fetoprotein for detecting hepatocellular carcinoma in patients with hepatitis C. A systematic review and critical analysis. Ann Intern Med. 2003;139:46–50. doi: 10.7326/0003-4819-139-1-200307010-00012. [DOI] [PubMed] [Google Scholar]
- 8.Srinivas PR, Verma M, Zhao Y, Srivastava S. Proteomics for cancer biomarker discovery. Clin Chem. 2002;48:1160–1169. [PubMed] [Google Scholar]
- 9.Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol. 2008;5:588–599. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 10.Hu S, Loo JA, Wong DT. Human body fluid proteome analysis. Proteomics. 2006;6:6326–6353. doi: 10.1002/pmic.200600284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diamandis EP. Point: Proteomic patterns in biological fluids: do they represent the future of cancer diagnostics? Clin. Chem. 2003;49:1272–1275. doi: 10.1373/49.8.1272. [DOI] [PubMed] [Google Scholar]
- 12.Poon TC, Yip TT, Chan AT, Yip C, et al. Comprehensive proteomic profiling identifies serum proteomic signatures for detection of hepatocellular carcinoma and its subtypes. Clin Chem. 2003;49:752–760. doi: 10.1373/49.5.752. [DOI] [PubMed] [Google Scholar]
- 13.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths WJ, Wang Y. Mass spectrometry: from proteomics to metabolomics and lipidomics. Chem Soc Rev. 2009;38:1882–1896. doi: 10.1039/b618553n. [DOI] [PubMed] [Google Scholar]
- 15.Diamandis EP. Mass spectrometry as a diagnostic and a cancer biomarker discovery tool: opportunities and potential limitations. Mol Cell Proteomics. 2004;3:367–378. doi: 10.1074/mcp.R400007-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 17.Bantscheff M, Lemeer S, Savitski MM, Kuster B. Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal Bioanal Chem. 2012;404:939–965. doi: 10.1007/s00216-012-6203-4. [DOI] [PubMed] [Google Scholar]
- 18.Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods. 2012;9:555–566. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 19.Boja ES, Rodriguez H. Mass spectrometry-based targeted quantitative proteomics: achieving sensitive and reproducible detection of proteins. Proteomics. 2012;12:1093–1110. doi: 10.1002/pmic.201100387. [DOI] [PubMed] [Google Scholar]
- 20.Carr SA, Abbatiello SE, Ackermann BL, Borchers C, et al. Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol Cell Proteomics. 2014;13:907–917. doi: 10.1074/mcp.M113.036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 22.Searle BC. Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics. 2010;10:1265–1269. doi: 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
- 23.Cox J, Neuhauser N, Michalski A, Scheltema RA, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 24.Cox J, Hein MY, Luber CA, Paron I, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang MN, Guess HA, Heyse JF. Reduction in burden of illness: a new efficacy measure for prevention trials. Stat Med. 1994;13:1807–1814. doi: 10.1002/sim.4780131803. [DOI] [PubMed] [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 27.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 28.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 29.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacLean B, Tomazela DM, Shulman N, Chambers M, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Mach Learning. 2002;46:389–422. [Google Scholar]
- 32.Chaerkady R, Harsha HC, Nalli A, Gucek M, et al. A quantitative proteomic approach for identification of potential biomarkers in hepatocellular carcinoma. J Proteome Res. 2008;7:4289–4298. doi: 10.1021/pr800197z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Sogawa K, Sunaga M, Umemura H, et al. Increased concentrations of apo A-I and apo A-II fragments in the serum of patients with hepatocellular carcinoma by magnetic beads-assisted MALDI-TOF mass spectrometry. Am J Clin Pathol. 2014;141:52–61. doi: 10.1309/AJCPBLFBNAP6N2UN. [DOI] [PubMed] [Google Scholar]
- 34.Feng JT, Liu YK, Song HY, Dai Z, et al. Heat-shock protein 27: a potential biomarker for hepatocellular carcinoma identified by serum proteome analysis. Proteomics. 2005;5:4581–4588. doi: 10.1002/pmic.200401309. [DOI] [PubMed] [Google Scholar]
- 35.Fye HK, Wright-Drakesmith C, Kramer HB, Camey S, et al. Protein profiling in hepatocellular carcinoma by label-free quantitative proteomics in two west African populations. PLoS One. 2013;8:e68381. doi: 10.1371/journal.pone.0068381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Liu YH, Mai SJ, He LJ, et al. Evaluation of serum clusterin as a surveillance tool for human hepatocellular carcinoma with hepatitis B virus related cirrhosis. J Gastroenterol Hepatol. 2010;25:1123–1128. doi: 10.1111/j.1440-1746.2009.06205.x. [DOI] [PubMed] [Google Scholar]
- 37.Paradis V, Degos F, Dargere D, Pham N, et al. Identification of a new marker of hepatocellular carcinoma by serum protein profiling of patients with chronic liver diseases. Hepatology. 2005;41:40–47. doi: 10.1002/hep.20505. [DOI] [PubMed] [Google Scholar]
- 38.Ferrin G, Ranchal I, Llamoza C, Rodriguez-Peralvarez ML, et al. Identification of candidate biomarkers for hepatocellular carcinoma in plasma of HCV-infected cirrhotic patients by 2-D DIGE. Liver Int. 2013 doi: 10.1111/liv.12277. [DOI] [PubMed] [Google Scholar]
- 39.Kang YK, Hong SW, Lee H, Kim WH. Overexpression of clusterin in human hepatocellular carcinoma. Hum Pathol. 2004;35:1340–1346. doi: 10.1016/j.humpath.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 40.Lau SH, Sham JS, Xie D, Tzang CH, et al. Clusterin plays an important role in hepatocellular carcinoma metastasis. Oncogene. 2006;25:1242–1250. doi: 10.1038/sj.onc.1209141. [DOI] [PubMed] [Google Scholar]
- 41.Saxena NK, Fu PP, Nagalingam A, Wang J, et al. Adiponectin modulates C-jun N-terminal kinase and mammalian target of rapamycin and inhibits hepatocellular carcinoma. Gastroenterology. 2010;139:1762–73. 1773.e1–5. doi: 10.1053/j.gastro.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nomoto S, Kanda M, Okamura Y, Nishikawa Y, et al. Epidermal growth factor-containing fibulin-like extracellular matrix protein 1, EFEMP1, a novel tumor-suppressor gene detected in hepatocellular carcinoma using double combination array analysis. Ann Surg Oncol. 2010;17:923–932. doi: 10.1245/s10434-009-0790-0. [DOI] [PubMed] [Google Scholar]
- 43.Kitagawa K, Nakajima G, Kuramochi H, Ariizumi SI, Yamamoto M. Lymphatic vessel endothelial hyaluronan receptor-1 is a novel prognostic indicator for human hepatocellular carcinoma. Mol Clin Oncol. 2013;1:1039–1048. doi: 10.3892/mco.2013.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zinkin NT, Grall F, Bhaskar K, Otu HH, et al. Serum proteomics and biomarkers in hepatocellular carcinoma and chronic liver disease. Clin Cancer Res. 2008;14:470–477. doi: 10.1158/1078-0432.CCR-07-0586. [DOI] [PubMed] [Google Scholar]
- 45.Chu SC, Wang CP, Chang YH, Hsieh YS, et al. Increased cystatin C serum concentrations in patients with hepatic diseases of various severities. Clin Chim Acta. 2004;341:133–138. doi: 10.1016/j.cccn.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 46.Takeuchi M, Fukuda Y, Nakano I, Katano Y, Hayakawa T. Elevation of serum cystatin C concentrations in patients with chronic liver disease. Eur J Gastroenterol Hepatol. 2001;13:951–955. doi: 10.1097/00042737-200108000-00013. [DOI] [PubMed] [Google Scholar]
- 47.Saito Y, Oba N, Nishinakagawa S, Mizuguchi Y, et al. Identification of beta2-microgloblin as a candidate for early diagnosis of imaging-invisible hepatocellular carcinoma in patient with liver cirrhosis. Oncol Rep. 2010;23:1325–1330. doi: 10.3892/or_00000767. [DOI] [PubMed] [Google Scholar]
- 48.Lee IN, Chen CH, Sheu JC, Lee HS, et al. Identification of complement C3a as a candidate biomarker in human chronic hepatitis C and HCV-related hepatocellular carcinoma using a proteomics approach. Proteomics. 2006;6:2865–2873. doi: 10.1002/pmic.200500488. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki Y, Aoyagi Y, Mori S, Suda T, et al. Microheterogeneity of serum transferrin in the diagnosis of hepatocellular carcinoma. J Gastroenterol Hepatol. 1996;11:358–365. doi: 10.1111/j.1440-1746.1996.tb01384.x. [DOI] [PubMed] [Google Scholar]
- 50.Thomas PD, Campbell MJ, Kejariwal A, Mi H, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farkas IJ, Szanto-Varnagy A, Korcsmaros T. Linking proteins to signaling pathways for experiment design and evaluation. PLoS One. 2012;7:e36202. doi: 10.1371/journal.pone.0036202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Da Huang Wei, Sherman Brad T, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 53.Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39:D698–704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jensen LJ, Kuhn M, Stark M, Chaffron S, et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–6. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–72. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–60. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ressom HW, Xiao JF, Tuli L, Varghese RS, et al. Utilization of metabolomics to identify serum biomarkers for hepatocellular carcinoma in patients with liver cirrhosis. Anal Chim Acta. 2012;743:90–100. doi: 10.1016/j.aca.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiao JF, Varghese RS, Zhou B, Nezami Ranjbar MR, et al. LC–MS based serum metabolomics for identification of hepatocellular carcinoma biomarkers in Egyptian cohort. Journal of proteome research. 2012;11:5914–5923. doi: 10.1021/pr300673x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsai T, Wang M, Di Poto C, Hu Y, et al. LC–MS Profiling of N-Glycans Derived from Human Serum Samples for Biomarker Discovery in Hepatocellular Carcinoma. Journal of proteome research. 2014;13:4859–4868. doi: 10.1021/pr500460k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.